Abstract
Porcine dermatitis and nephropathy syndrome (PDNS) is a sporadic, usually fatal disease of growing and finishing pigs that has been recognized in many pig-producing countries. Pasteurella multocida strains isolated from 15 pigs with PDNS and 51 pigs without PDNS were characterized by capsule and somatic antigen typing, random amplified polymorphic DNA (RAP-D) typing, and restriction analysis of genomic DNA using pulsed-field gel electrophoresis (PFGE). While capsular, somatic, and RAP-D typing did not discriminate PDNS isolates from non-PDNS isolates, all of the isolates from PDNS cases showed an identical ApaI PFGE restriction pattern. This pattern was also found in a high proportion (36%) of P. multocida strains isolated from non-PDNS cases. Isolation of a single variant of P. multocida from tissues of pigs with PDNS warrants further investigation into the possible role of these bacteria in the etiology of the disease.
Porcine dermatitis and nephropathy syndrome (PDNS) is a disease of unknown etiology that has been recognized in many pig-producing countries. The disease most commonly affects pigs between 2 and 7 months of age, and cases can occur sporadically or as an outbreak within herds. There is a high mortality rate (approximately 80%) in clinically affected pigs. The pathological changes are consistent with an immune complex disorder resulting in severe glomerulonephritis. Vasculitis occurs in the kidneys, skin, and other tissues with deposits of immunoglobulins and complement in the areas of lesion development (1, 8, 14, 15). A number of potential causes have been suggested, including viral or bacterial infections and dietary factors (8, 13). However, no definitive link has yet been established with any specific factors. In a study on bacterial isolations from PDNS-affected pigs, Pasteurella multocida was isolated from one or more tissue sites in 16 out of 20 cases, and P. multocida-specific antigen was demonstrated in affected kidney tissue from 17 of these cases (16). Other potential bacterial pathogens were isolated from some of the PDNS cases, but their isolation rates were much lower and there was little consistency between cases, compared with results for P. multocida (16). Since these studies suggested a strong association with P. multocida, a strain typing study was undertaken with representative P. multocida isolates arising from these cases. The aim was first to compare the strain types between PDNS cases and second to compare the PDNS strain types with those isolated from a cohort of non-PDNS cases. This paper reports on the comparative strain typing studies. An abstract summarizing the results of this study has been published previously (J. R. Thomson, F. A. Lainson, N. Thomson, and W. Donachie, Proc. 15th Int. Pig Vet. Soc. Congr., vol. 3, p. 396, 1998).
MATERIALS AND METHODS
PDNS cases.
Cases for this study were selected on the basis of their typical clinical histories and pathological lesions, the freshness of their carcasses, and because they each came from a different farm. The diagnostic criteria were consistent with previous descriptions of PDNS (1, 8, 14, 15). The pigs were between 10 and 20 weeks of age and had been ill for 2 to 14 days before they died. All of the pigs showed typical multifocal, irregular hemorrhagic skin lesions, sometimes coalescing to form widespread contiguous lesions over the sides of the body and lower limbs, in particular. One of the cases (case 3; Table 1) had healing skin lesions showing granulation, scab formation, and early scarring. Pigs had become anorexic and lethargic and showed a rapid loss of condition prior to death. None of the pigs that yielded P. multocida bacteria from internal organs had been treated with antimicrobial agents. On postmortem examination, in addition to the superficial skin lesions described above, some pigs had extensive dermal and subcutaneous hemorrhage and edema. In all cases the kidneys were very enlarged, with petechial stippling or ecchymotic hemorrhages of the cortex. Serosanguineous fluid was present in the abdominal cavity and hock joints in some cases. On histopathological examination, the skin lesions showed acute hemorrhagic dermatitis associated with fibrinoid vasculitis in the dermis and also the subcutis in some cases. Subcutaneous edema was present. The renal lesions were very dramatic, comprised of widespread acute glomerulonephritis with massive protein leakage into Bowman's capsules and renal tubules, which were distended by large hyaline casts. Abundant protein reabsorption droplet formation was present in proximal convoluted tubules. Interstitial mononuclear cell infiltration was present. Vasculitis affected the renal arcuate vessels and, variably, other medium- to small-sized arterioles in the spleen, liver, peripheral lymph nodes, synovial membranes, and stomach. In all cases, the lesions were consistent with PDNS.
TABLE 1.
Fifteen cases of PDNS showing the isolation sites of Pasteurella multocidaa
| Case no. | Tissues sites of isolates |
|---|---|
| 1 | Spleen,* ascitic fluid |
| 2 | Tonsil, hock synovial fluid* |
| 3 | Tonsil* |
| 4 | Lung,* spleen, bronchial lymph node |
| 5 | Retropharyngeal lymph node,* hock synovial fluid |
| 6 | Tonsil, bronchial lymph node, pleural fluid* |
| 7 | Tonsil, retropharyngeal lymph node, ascitic fluid, hock synovial fluid* |
| 8 | Tonsil, lung, bronchial lymph node,* pleural fluid |
| 9 | Tonsil,* hock synovial fluid |
| 10 | Spleen,* pleural fluid, ascitic fluid |
| 11 | Lung,* hock synovial fluid |
| 12 | Retropharyngeal lymph node, lung, kidney,* bronchial lymph node, pleural fluid |
| 13 | Tonsil, lung, bronchial lymph node* |
| 14 | Lung, spleen,* bronchial lymph node |
| 15 | Tonsil, lung, hock synovial fluid* |
Word(s) marked with an asterisk indicates the origin of the Pasteurella multocida isolate in each case that was used in the strain typing studies.
Bacterial isolates.
The P. multocida isolates recovered from tissue samples collected at necropsy were identified by cultural characteristics and biochemical testing, as detailed in an associated publication describing bacteriology and immunohistochemical studies with these cases (16). One representative strain per case was selected from different tissue sites for the typing studies (Table 1). For non-PDNS cases, lung samples from 52 pigs showing bacterial-type bronchopneumonia were collected from a local abattoir. The pigs were all 5 months of age and were derived from 13 different pig units (four samples were collected per herd). None of the pigs showed lesions of PDNS and none of the herds were known to have had cases of PDNS. Each sample was cultured on 5% sheep blood agar (Oxoid no. 2 base) and incubated under aerobic conditions at 37°C for up to 72 h. Isolates were identified using standard microbiological techniques and biochemical identification systems. Fifty-one P. multocida isolates were recovered from lung lesions. A total of 66 strains were analyzed by somatic antigen typing, pulsed-field gel electrophoresis (PFGE), and random amplified polymorphic DNA typing (RAP-D). Somatic antigen typing was carried out using the method of Heddleston et al. (11). Capsular typing was carried out using the method of Carter and Rundell (3) and Carter and Subronto (4).
PFGE.
PFGE conditions were modified from those used by Ramage et al. (12). Cultured P. multocida cells were washed six times in Pett IV buffer (1 M NaCl, 10 mM Tris-HCl [pH 7.5]) and suspended in a final concentration of 108 CFU/ml in 1% low-melting-point agarose. Subsequent treatments were carried out on cells suspended in the solidified gel matrix. The cell suspension was digested in EC lysis buffer (6 mM Tris-HCl [pH 7.6], 1 M NaCl, 0.05 M EDTA, 5 mM deoxycholate, 17 mM N-lauryl sarcosine, 0.1 mg of Brij58 ml−1, 1 mg of lysozyme ml−1, 20 μg of RNase ml−1) and incubated at 37°C for 16 h and then in ESP buffer (500 mM EDTA [pH 9.0], proteinase K at 1 mg ml−1, N-lauryl sarcosine at 10 mg ml−1) at 50°C for 48 h. They were then washed extensively in TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA). A gel slice was then equilibrated in ApaI digestion buffer (Boehringer) and digested with 20 units of ApaI in digestion buffer containing 10 mg of bovine serum albumin ml−1 and incubated at 30°C for 48 h. The gel slice was washed in TE buffer for 15 min and then loaded onto a 1% agarose gel.
Electrophoresis was carried out on a Bio-Rad CHEF II system using TBE buffer (44 mM Tris [pH 8.0], 44 mM boric acid, 1 mM EDTA) at 14°C and electrophoresis at 6 V/cm with an initial switch time of 1 s, increasing to 40 s at 23 h. Pulsed-field gels were stained with 1 mM ethidium bromide and recorded using an Image Master VDS (Pharmacia). Gel images were analyzed and banding profiles were compared using the Image Master Elite image analysis and database software (Pharmacia). A preliminary study was carried out to identify a suitable restriction endonuclease for analysis of P. multocida strains. ApaI was selected on the basis that it produced clearly distinguishable banding patterns with a capacity to discriminate between lung lesion strains.
RAP-D.
P. multocida strains were grown overnight in Luria-Bertani liquid medium at 37°C. Genomic DNA was prepared using the G-Nome reagent kit (Bio-101). Twenty-five nanograms of genomic DNA template was used in each RAP-D reaction. Analysis was carried out using Ready-To-Go RAPD Analysis Beads (Pharmacia) with a 25 pM concentration of each of the following primers: 1, GGTGCGGGAA; 2, GTTTCGCTCC; 3, GTAGACCCGT; 4, AAGAGCCCGT; 5, AACGCGCAAC; 6, CCCGTCAGCA (Pharmacia). PCR thermocycling was carried out with an initial denaturation step of 95°C for 5 min followed by 45 cycles of 95°C for 1 min, 36°C for 1 min, and 72°C for 2 min. Positive (Escherichia coli genomic DNA template) and negative (no template) controls were included to check the consistency of amplification. DNA fragment analysis was carried out on a 2% Tris acetate agarose gel containing 1 mM ethidium bromide.
RESULTS
Bacteriology.
Isolates from PDNS cases and non-PDNS cases could not be distinguished by morphological or cultural characteristics.
Capsule and somatic antigen typing.
Table 2 shows the analysis of P. multocida isolates from PDNS and non-PDNS cases. Four somatic antigen patterns were present among the 15 PDNS cases; the most prevalent type was 3, 4, 12 (11 of 15), whereas types 3, 3,4, and 3,12 occurred less frequently. The frequency of somatic antigen types among PDNS isolates was consistent with that found for non-PDNS isolates. With the exception of a single non-PDNS isolate, all of the P. multocida strains were capsule type A.
TABLE 2.
Electrophoretic profiles of Pasteurella multocida isolates from pigs with and without PDNSa
| Category and isolate no. | ApaI profile | Somatic type | Origin | RAP-D primer | Category and isolate no. | ApaI profile | Somatic type | Origin | RAP-D primer | |
|---|---|---|---|---|---|---|---|---|---|---|
| Non-PDNS | ||||||||||
| 1 | 04 | 3,4 | 1 | 6 | ||||||
| 2 | 01 | 3,4,12 | 1 | |||||||
| 3 | 05 | 3,4,12 | 1 | 6 | ||||||
| 4 | 03 | 3,4 | 1 | |||||||
| 5 | 03 | 3 | 2 | 1-6 | ||||||
| 6 | 02 | 3 | 2 | |||||||
| 7 | 02 | 3,4 | 2 | 1-6 | ||||||
| 8 | 01 | 3,4,12 | 2 | 6 | ||||||
| 9 | 01 | 3,4,12 | 3 | |||||||
| 10 | 01 | 3,4,12 | 3 | 6 | ||||||
| 11 | 01 | 3,4,12 | 3 | |||||||
| 12 | 01 | 3,12 | 3 | 6 | ||||||
| 13 | 01 | 3,4,12 | 4 | |||||||
| 14 | 01 | 3,12 | 4 | 6 | ||||||
| 15 | 01 | 3,4,12 | 4 | |||||||
| 16 | 01 | 3,4,12 | 4 | |||||||
| 17 | 06 | 3,4 | 5 | 6 | ||||||
| 18 | 07 | 3,4 | 5 | 6 | ||||||
| 19 | 01 | 3,12 | 5 | |||||||
| 20 | 01 | 3,12 | 5 | |||||||
| 21 | 07 | 3,4 | 6 | |||||||
| 22 | 06 | 3,4 | 6 | |||||||
| 23 | 02 | 3,4 | 6 | |||||||
| 24 | 01 | 3,4,12 | 6 | |||||||
| 25 | 02 | 3,4 | 7 | |||||||
| 26 | 01 | 3,4,12 | 7 | |||||||
| 27 | 08 | 3,4 | 7 | 6 | ||||||
| 28 | 08 | 3,4 | 7 | |||||||
| 29 | 09 | 3,4,12 | 8 | |||||||
| 30 | 10 | 3,4 | 8 | |||||||
| 31 | 10 | 3,4 | 8 | 1-6 | ||||||
| 32 | 10 | 3,4 | 8 | |||||||
| 33 | 13 | 3,4 | 9 | |||||||
| 34 | 11 | 3,4 | 9 | 6 | ||||||
| 35 | 12 | 3,4 | 9 | |||||||
| 36 | 12 | 3,4 | 9 | |||||||
| 37 | 11 | 3,4 | 10 | |||||||
| 38 | 12 | 3,4 | 10 | |||||||
| 39 | 11 | 3,4 | 10 | |||||||
| 40 | 11 | 3,4 | 10 | 6 | ||||||
| 41 | 11 | 3,4 | 11 | |||||||
| 42 | 14 | 3,4 | 11 | 6 | ||||||
| 43 | 12 | 3,4,12 | 11 | 6 | ||||||
| 44 | 01 | 3,4,12 | 11 | 6 | ||||||
| 45 | 01 | 3,4,12 | 12 | 6 | ||||||
| 46 | 01 | 3,4,12 | 12 | |||||||
| 47 | 09 | 3,4,12 | 12 | 1-6 | ||||||
| 48 | 01 | 3,4,12 | 12 | 6 | ||||||
| 49 | 15 | 3,4 | 13 | 1-6 | ||||||
| 50 | 13 | 3,4 | 13 | 6 | ||||||
| 51 | 13 | 3,4 | 13 | 6 | ||||||
| PDNS | ||||||||||
| 52 | 01 | 3,4,12 | 14 | 6 | ||||||
| 53 | 01 | 3,4,12 | 15 | 6 | ||||||
| 54 | 01 | 3,4,12 | 16 | 6 | ||||||
| 55 | 01 | 3,4,12 | 17 | 6 | ||||||
| 56 | 01 | 3,4 | 18 | 6 | ||||||
| 57 | 01 | 3 | 19 | 6 | ||||||
| 58 | 01 | 3,4,12 | 20 | 6 | ||||||
| 59 | 01 | 3,4,12 | 21 | 6 | ||||||
| 60 | 01 | 3,4,12 | 22 | 6 | ||||||
| 61 | 01 | 3,4,12 | 23 | 6 | ||||||
| 62 | 01 | 3,12 | 24 | 1-6 | ||||||
| 63 | 01 | 3,4,12 | 25 | 6 | ||||||
| 64 | 01 | 3,12 | 26 | 6 | ||||||
| 65 | 01 | 3,4,12 | 27 | 6 | ||||||
| 66 | 01 | 3,4,12 | 28 | 6 |
Details of isolates derived from non-PDNS cases (bacterial pneumonia lesions in slaughtered pigs) (strains 1 to 51) or tissues or fluids from PDNS cases (strains 52 to 65). The geographic origin of the isolate is indicated by a farm number. ApaI restriction profiles, somatic antigen types, and primers used in RAP-D analysis are also indicated.
PFGE analysis.
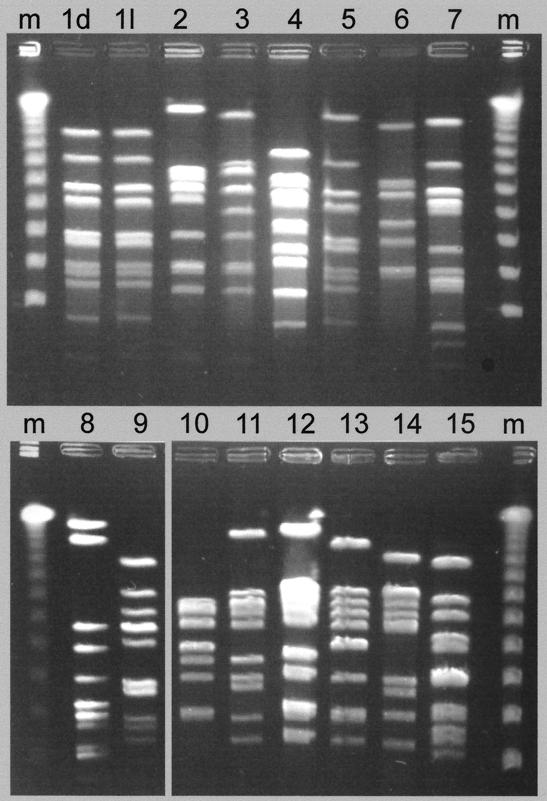
Restriction analysis with ApaI showed that the non-PDNS strains of P. multocida were genetically heterogeneous; 15 electrophoretic variants were identified among the 51 lung lesion isolates (Table 2 and Fig. 1). Type 01 was the most prevalent and comprised 36% of the total isolates (Table 3). In contrast, the PDNS isolates comprised only a single electrophoretic type, ApaI type 01, which was identical to 18 of 51 (36%) of the non-PDNS isolates (Table 2 and 3).
FIG. 1.
Representative PFGE types of P. multocida strains from pigs with and without PDNS. Lane m, lambda molecular weight markers. The strain types from non-PDNS cases are shown in lanes 1l and 2 to 15. The electrophoretic pattern shown in lane 1d is representative of all PDNS-derived strains.
TABLE 3.
Distribution of PFGE ApaI typesa
| ApaI type | No. (%) of occurrences (n = 51) | No. (%) of farm isolations (n = 13) |
|---|---|---|
| 01 | 18 (36) | 9 (69) |
| 02 | 4 (8) | 3 (23) |
| 03 | 2 (4) | 2 (15) |
| 04 | 1 (2) | 1 (8) |
| 05 | 1 (2) | 1 (8) |
| 06 | 2 (4) | 2 (15) |
| 07 | 2 (4) | 2 (15) |
| 08 | 2 (4) | 1 (8) |
| 09 | 2 (4) | 2 (15) |
| 10 | 3 (6) | 1 (8) |
| 11 | 5 (10) | 3 (23) |
| 12 | 4 (8) | 3 (23) |
| 13 | 2 (4) | 2 (15) |
| 14 | 1 (2) | 1 (8) |
| 15 | 1 (2) | 1 (8) |
This table lists the electrophoretic types (01 to 15) isolated from pigs without PDNS, the number of occurrences of each type (out of a total of 51), and the number of pig farms (out of a total of 13) where each electrophoretic type was isolated.
The relationship between non-PDNS strains.
The apparent relationship between the PDNS isolates and non-PDNS isolates is shown as a dendrogram (Fig. 2). This is based on the similarity of each of the restriction profiles shown in Fig. 1. A band-sharing index was calculated using the unweighted pair group method algorithm. Four major clusters were identified at a band-matching index of greater than 0.5 among the 15 distinct restriction profiles. Group 1 comprised restriction type (RT) 04; group 2, RT 08; group 3, RTs 06, 10, 11, 14, 02, 13, 03, and 12; and group 4, RTs 07, 01, 05, 15, and 09. On this criteria the 01 restriction profile was most closely related to RTs 05 and 07.
FIG. 2.
Dendrogram showing the relationship between isolates on the basis of common restriction fragments. Isolates from cases of PDNS are all electrophoretic type 01, while the isolates from non-PDNS cases comprise electrophoretic types 01 to 15. This uses the UPGMA algorithm and Dice coefficient as implemented in ImageMaster 1D gel analysis software (Non-Linear Dynamics, Newcastle, United Kingdom).
RAP-D analysis.
RAP-D provided an alternative measure of the genetic heterogeneity among isolates. Using six different primer sets to examine representative strains, no differences could be detected among PDNS strains and non-PDNS strains with any of the primers used. In each case a positive and negative control was carried out along with the test samples.
DISCUSSION
Genotyping techniques have been used extensively to differentiate epidemiologically significant strains of P. multocida. These include restriction endonuclease analysis (7, 10, 17, 18), ribotyping (5, 7, 9, 10, 18), and RAP-D analysis (5). In this study, restriction analysis with ApaI shows that considerable variation exists among P. multocida strains isolated from localized lesions of bronchopneumonia in pigs that were otherwise healthy (the non-PDNS cases). Fifteen electrophoretic types were identified among the 51 isolates examined from this cohort. The discriminating capacity of restriction analysis with ApaI makes it a potentially useful epidemiological tool in the analysis of P. multocida isolates derived from lung lesions. Buttenschon and Rosendal (2) similarly observed a high level of genetic diversity among P. multocida type A strains using the restriction endonuclease BamHI.
A genetic relationship between the non-PDNS strains can be inferred from comparison of their restriction profiles. Analysis of the 15 ApaI electrophoretic types identified here showed four clusters or individual strains with a band-sharing index of greater than 0.5. Electrophoretic type 01 has greatest similarity to types 05 and 07, with band-sharing indexes of greater than 0.9 and 0.7, respectively. This analysis suggests that genetic similarities exist among these strains, which presumably indicates a common lineage. Restriction patterns 04 and 08 are unique and do not appear to correspond with patterns in other strains.
In contrast to ApaI restriction analysis, RAP-D was found to be insensitive in detecting differences between P. multocida strains. Analysis of PDNS strains and non-PDNS strains with six different primers did not differentiate between these strains, indicating a genetic similarity between them. The differing results obtained with ApaI restriction analysis and RAP-D demonstrates that selection of methods for genotype analysis is largely empirical.
The 66 isolates of P. multocida from PDNS cases and non-PDNS cases could not be differentiated based on morphological or biochemical criteria. With the exception of a single non-PDNS strain, all of these isolates were capsular type A. Somatic antigen analysis identified three antigen types (3, 4, and 12) that were present in various combinations. No clear-cut correlation could be established between somatic antigen type and ApaI restriction type. The somatic antigen combination 3,4,12 occurred with high frequency in strains that were ApaI restriction type 01, but this correlation was not unique and its significance is unknown.
No clear association could be established between the non-PDNS P. multocida strains and the farm of origin. The ApaI type 01 was isolated most commonly, comprising 36% (18 of 51) of the strains examined, and had a wide geographical distribution, being identified in 9 of the 13 pig units sampled. The remaining ApaI types, 02 to 15, comprised 64% (33 of 51) of the isolates examined, and each type was represented by between 1 and 5 isolates. The range of types isolated differed between farms. On farms 3 and 4, a single ApaI type was present, whereas multiple ApaI types were isolated from farms 1, 6, and 11.
Conversely, all 15 of the PDNS isolates examined were ApaI type 01. This level of genetic similarity appears to be high considering the heterogeneity of ApaI types found in non-PDNS cases and that these strains originate with 15 geographically separate pig production units. The diverse origins of the PDNS strains makes it likely that they are truly representative of P. multocida isolated from PDNS cases and suggests that a specific association exists between PDNS strains and ApaI type 01.
Isolation of P. multocida ApaI type 01 strains from PDNS and non-PDNS cases suggests that they are closely related or are a common strain. The possibility that strains commonly involved in respiratory infections in pigs can give rise to PDNS must be considered, and a causal relationship must be established. Although the isolates studied from PDNS cases were derived from a wide range of tissue sites, the majority of affected pigs yielded isolates from the tonsil, respiratory tract, or tissues associated with the respiratory tract (retropharyngeal lymph node, bronchial lymph node, or pleural fluid). Six of the 15 PDNS cases had pneumonic lesions, and P. multocida bacteria were isolated from the lung tissue. In a larger study on the pathology of PDNS involving 43 cases from Scotland and 37 cases from England and Wales, pneumonia was found in 49 and 65% of cases, respectively (J. R. Thomson, R. J. Higgins, W. J. Smith, and S. H. D. Done, Proc. 15th Int. Pig Vet. Soc. Congr., vol. 3, p. 395). Thus, pneumonia is common but not always present in PDNS cases. In this present study, P. multocida isolates were selected from a wide range of tissue types in the PDNS cases in order to encompass invasive and potentially noninvasive strain types. It would have been desirable to determine if the isolates from all sites in PDNS cases were of a common electrophoretic type; however, this was beyond the scope of the study. Examination of the P. multocida isolates for dermonecrotic toxin was not carried out, as this test was not available to the investigators at the time of the study. However, dermonecrotic strains isolated from pigs are almost always of capsular type D and associated with clinical signs of progressive atrophic rhinitis (PAR) in herds (6). None of the herds with PDNS cases in this study had clinical histories of PAR, nor were the farmers using vaccines against toxigenic P. multocida to prevent PAR. These factors suggest that dermonecrotic strains of P. multocida were not present in the herds in question.
A previous analysis of paired isolates (2) suggests that dissemination of P. multocida may occur between the lungs and kidneys. Buttenschon and Rosendal used restriction analysis to examine the relationship between paired isolates of P. multocida from lung lesions and from focal nephritis in individual slaughtered pigs. In 8 of the 14 cases examined, P. multocida isolated from lung and kidney tissues showed identical BamHI restriction patterns. The authors concluded that kidney isolates represent blood-borne dissemination from primary bronchopneumonia lesions. The subjects of that study were apparently healthy pigs, and no overt disease or renal pathology was reported; however, that study supports the proposal in the present work that P. multocida infection in the lungs, nasopharynx, or tonsils may be disseminated to other organs.
While a specific single ApaI type of P. multocida appears to be associated with PDNS, this does not demonstrate that it is a primary pathogen in this disease. Systemic infection could equally arise from opportunistic colonization with P. multocida following infection with an unidentified infectious agent. However, the question remains as to why this pattern of invasiveness was specific to type 01 strains in this series of cases. Further studies are required to characterize the mechanisms of pathogenesis in these strains. In particular, the possible role of P. multocida toxins (especially dermonecrotic toxin) in the pathogenesis of the disease should be investigated. Comparative studies on toxins produced by PDNS and non-PDNS strains of P. multocida would provide preliminary information of their potential involvement. In addition, epidemiological studies are needed to determine the relationship between the P. multocida type 01 PDNS and non-PDNS strains and whether the presence of type 01 strains in an individual pig or in a herd is an indicator of susceptibility to PDNS.
Acknowledgments
Moredun Research Institute and Scottish Agricultural College, Veterinary Science Division, receive financial support from the Scottish Executive Rural Affairs Department.
We thank L. Henderson and C. Meikie of the SAC Veterinary Science Division, Edinburgh, for bacterial isolations, Kirsten Begg of Moredun Research Institute for technical support, and Mark Wilson, Ames, Iowa, for somatic antigen typing.
REFERENCES
- 1.Bourgault, A., and R. Drolet. 1995. Spontaneous glomerulonephritis in swine. J. Vet. Diagn. Investig. 7:122-126. [DOI] [PubMed] [Google Scholar]
- 2.Buttenschon, J., and S. Rosendal. 1990. Phenotypical and genotypical characteristics of paired isolates of Pasteurella multocida from the lungs and kidneys of slaughtered pigs. Vet. Microbiol. 25:67-75. [DOI] [PubMed] [Google Scholar]
- 3.Carter, G. R., and S. Rundell. 1975. Identification of type A strains of Pasteurella multocida using staphylococcal hyaluronidase. Vet. Rec. 96:343-345. [DOI] [PubMed] [Google Scholar]
- 4.Carter, G. R., and P. Subronto. 1973. Identification of type D strains of Pasteurella multocida with acriflavine. Am. J. Vet. Res. 34:293-295. [PubMed] [Google Scholar]
- 5.Chaslus-Dancla, E., M. C. Lesage-Decauses, S. Leroy-Setrin, J. L. Martel, P. Coudert, and J. P. Lafont. 1996. Validation of random amplified polymorphic DNA assays by ribotyping as tools for epidemiological surveys of Pasteurella from animals. Vet. Microbiol. 52:91-102. [DOI] [PubMed] [Google Scholar]
- 6.de Jong, M. F. 1999. Progressive and non-progressive atrophic rhinitis, p. 355-385. In B. E. Straw et al. (ed.), Diseases of swine, 8th ed. Iowa State University Press, Ames, Iowa.
- 7.Donnio, P. Y., A. Allardet-Servent, M. Perrin, F. Escande, and J. L. Avril. 1999. Characterisation of dermonecrotic toxin-producing strains of Pasteurella multocida subsp. multocida isolated from man and swine. J. Med. Microbiol. 48:125-131. [DOI] [PubMed] [Google Scholar]
- 8.Drolet, R., S. Thibault, S. D'Allaire, J. R. Thomson, and S. H. Done. 1999. Porcine dermatitis and nephropathy syndrome (PDNS): an overview of the disease. Swine Health Prod. 7:283-285. [Google Scholar]
- 9.Fussing, V., J. P. Nielsen, M. Bisgaard, and A. Meyling. 1999. Development of a typing system for epidemiological studies of porcine toxin-producing Pasteurella multocida subsp. multocida in Denmark. Vet. Microbiol. 65:61-74. [DOI] [PubMed] [Google Scholar]
- 10.Gardner, I. A., R. Kasten, G. J. Eamens, K. P. Snipes, and R. J. Anderson. 1994. Molecular fingerprinting of Pasteurella multocida associated with progressive atrophic rhinitis in swine herds. J. Vet. Diagn. Investig. 6:442-447. [DOI] [PubMed] [Google Scholar]
- 11.Heddleston, K. L., J. E. Gallagher, and P. E. Rebers. 1972. Fowl cholera: gel diffusion precipitin test for serotyping Pasteurella multocida from avian species. Avian Dis. 16:925-936. [PubMed] [Google Scholar]
- 12.Ramage, C. P., J. C. Low, J. McLauchlan, and W. Donachie. 1999. Characterization of Listeria ivanovii isolates from the UK using pulsed-field gel electrophoresis. FEMS Microbiol. Lett. 170:349-353. [DOI] [PubMed] [Google Scholar]
- 13.Rosell, C., J. Segales, J. A. Ramos-Vara, J. M. Folch, G. M. Rodriguez-Arrioja, C. O. Duran, M. Balasch, J. Plana-Duran, and M. Domingo. 2000. Identification of porcine circovirus in tissues of pigs with porcine dermatitis and nephropathy syndrome. Vet. Rec. 146:40-43. [DOI] [PubMed] [Google Scholar]
- 14.Sierra, M. A., J. M. Delas Mulas, R. F. Molenbeek, C. Van Maanen, J. H. Vos, M. Quezada, and E. Gruys. 1997. Porcine immune complex glomerulonephritis dermatitis (PIGD) syndrome. Eur. J. Vet. Pathol. 3:63-70.
- 15.Smith, W. J., J. R. Thomson, and S. Done. 1993. Dermatitis/nephropathy syndrome of pigs. Vet. Rec. 132:47.. [DOI] [PubMed] [Google Scholar]
- 16.Thomson, J. R., N. MacIntyre, L. E. A. Henderson, and C. M. Meikle. 2001. Detection of Pasteurella multocida in pigs with porcine dermatitis and nephropathy syndrome. Vet. Rec. 149:412-417. [DOI] [PubMed] [Google Scholar]
- 17.Wilson, M. A., M. J. Morgan, and G. E. Barger. 1993. Comparison of DNA fingerprinting and serotyping for identification of avian Pasteurella multocida isolates. J. Clin. Microbiol. 31:255-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao, G., C. Pijoan, M. P. Murtaugh, and T. W. Molitor. 1992. Use of restriction endonuclease analysis and ribotyping to study epidemiology of Pasteurella multocida in closed swine herds. Infect. Immun. 60:1401-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]