Abstract
A protocol for quantification of human immunodeficiency virus type 1 (HIV-1) proviral DNA with the TaqMan technology was developed and validated. The assay was specific for HIV-1, with an analytic sensitivity of 10 copies and a linear dynamic range of >6 logs. Viral RNA levels, when at a stable state, were highly correlated with proviral DNA levels in 80 specimens of 18 HIV-infected children.
The use of highly active antiretroviral therapy (HAART) in human immunodeficiency virus (HIV)-infected patients can often significantly reduce the levels of viral RNA to undetectable levels in plasma in both adults and children (13-17). However, it is less clear how changes in proviral DNA levels respond to HAART in infected patients or what relative changes in proviral DNA levels precede changes in viral RNA levels after HAART induction. One of the current challenges is detection of low levels of proviral DNA in latently infected CD4+ lymphocytes and other reservoirs, which can replenish and revive viral infection upon activation (3-5). Thus, a highly reproducible and accurate assay for quantification of proviral DNA would enable more in-depth evaluations of the efficacies of antiviral therapies.
Several assays for the quantification of HIV type 1 (HIV-1) proviral DNA have previously been reported, and all of these were based on the principle of conventional PCR (1, 2, 6, 10, 11). The potential limitation associated with the traditional quantitative PCR is that the DNA copy numbers are calculated on the basis of the quantities of the final amplified gene products. Since DNA is amplified exponentially during PCR, a small variation in amplification efficiency early in the thermocycling process could potentially lead to large variations in the final quantities of amplified products.
The real-time PCR, which is also known as the TaqMan or the 5" exonuclease assay, quantifies PCR products cycle by cycle (in real time) as they accumulate (7-9). There are several advantages of using real-time PCR. (i) It does not rely on the final product of PCR amplification for quantification. DNA copy numbers are determined on the basis of the threshold cycle number (CT), which is directly proportional to the initial copy number. (ii) It is relatively specific. If the probe binds nonspecifically to some sequences other than the target sequence, it will not be cleaved or detected as part of the amplification. (iii) It allows a wide dynamic range of detection since the measured DNA copy number is directly proportional to the initial copy number. (iv) All real-time PCR tests are performed in 96-well and closed-tube formats, allowing high-throughput testing and a greatly reduced chance of cross-contamination.
In this report, a new protocol for quantification of HIV-1 proviral DNA with the TaqMan technology is described and validated. By using this method, HIV-1 proviral DNA levels were compared in parallel with the HIV-1 RNA levels at a stable state during HAART.
(This study was part of a thesis by Johann Miller in partial fulfillment of the requirements for the M.S. degree at Northwestern University.)
In order to obtain absolute quantification of HIV-1 proviral DNA, we constructed an internal quantification standard (QS) with known copy numbers. Four criteria were used to design the QS. (i) The nucleotide sequence of the QS probe [5"-(FAM)-TAACCCACTCGTGCACCCAACTGATCTT-(TAMRA)-3", where FAM is 6-carboxyfluoroscein and TAMRA is 6-carboxytetramethylrhodamine] is different from the gag probe sequence [5"-(FAM)-ACCATCAATGAGGAAGCTGCAGAATGGG-(TAMRA)-3"], so that cross-hybridization between the QS probe and the gag gene target does not occur, or vice versa, when they are amplified in the same tube. (ii) The same primers (primers SK452 and SK431) are used for both the QS and the gag sequences. This ensures identical amplification conditions when the PCR is initiated. (iii) The GCs contents (in percent) in both the gag and the QS DNA targets are identical. (iv) The QS amplicon is the same length (130 bp) as the gag amplicon. The resulting plasmid carrying QS was designated pZErO2-QS. The effective copy number of the QS was calculated on the basis of its molecular weight and was further calibrated with a PCR copy number panel provided by the AIDS Research and Reagent Reference Program of the National Institutes of Health (NIH) (12).
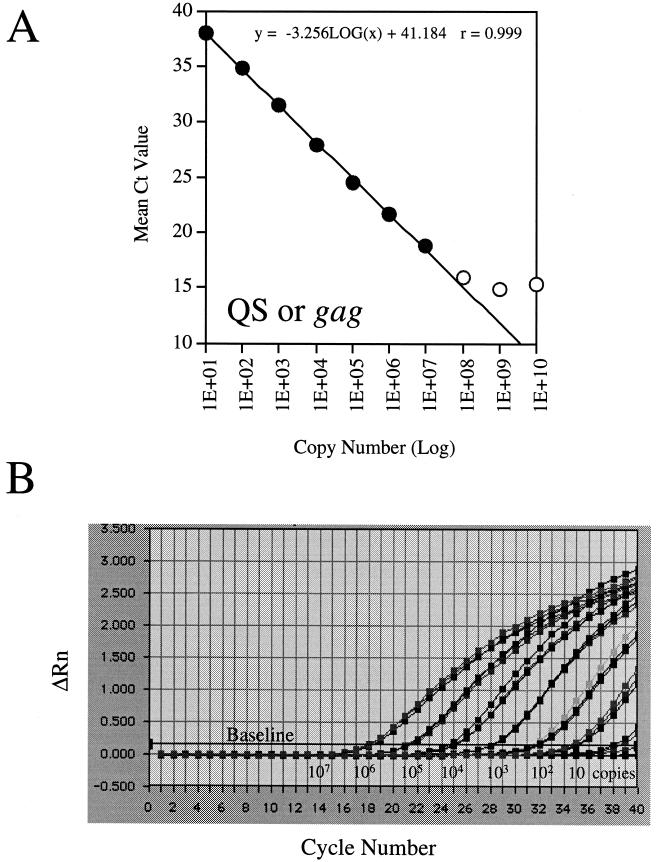
The linear dynamic range and the intra- and interassay variabilities of detection of the QS and the PCR-amplified HIV-1 gag gene were determined by preparing 10-fold serial dilutions with copy numbers ranging from 1.0 × 101 to 1.0 × 1010 copies/reaction mixture. The real-time PCR experiments were run in triplicate on an ABI 7700 Prism sequence detector and were performed three separate times. Each datum point on the concentration curve represents the average of nine CT values (Fig. 1A). Even though DNA was detected throughout the entire dilution range, the linear range was approximately 6 logs (1 × 101 to 1.0 × 107 copies; Fig. 1A). An initial copy number of 10 copies yielded a CT value of 37.9 ± 1.2 (n = 9), whereas an initial copy number of 1.0 × 107 copies yielded a CT value of 18.5 ± 0.5 (n = 9; Fig. 1A.) As expected, every 10-fold decrease in the copy number yielded an increase in the CT value of approximately 3, suggesting a linear relationship between the calculated and the initial DNA copy numbers (Perkin-Elmer, Foster City, Calif.). On the basis of the results of three experiments, the inter- and intra-assay variations were within 2.5-fold over the entire dynamic range (for a representative plot, see Fig. 1B).
FIG. 1.
Linear dynamic range and reproducibility as quantified by QS or the gag gene. (A) The mean CT plotted against the QS or the gag gene copy number yields a line which is linear over more than 6 logs (closed circles). Open circles represent the nonlinear portion of the curve. Each datum point represents the average of nine datum points collected from three experiments performed in triplicate (n = 9). The r value presents only the seven linear datum points (B). A representative amplification plot showing the reproducibility of a triplicate experiment. Only the seven linear datum points are shown. ΔRn, fluorescence intensity; cycle, PCR cycle numbers.
A PCR copy number panel, which was provided by the NIH AIDS Research and Reagent Reference Program, was used to test the detection limits of this assay. For 10 copies, HIV-1 gag DNA was detected 92% of the time (45 of 49 tests). For 5, 2.5, and 1.25 input copies of the gag gene, the detection frequencies were 83% (15 of 18 tests), 44% (8 of 18 tests), and 33% (6 of 18 tests), respectively, and the measured copy numbers were 5.2 ± 4.1, 1.9 ± 3.4, and 1.7 ± 3.5 copies, respectively. No gag DNA was detected in any gag-negative controls (n = 43), suggesting that this assay has a high degree of specificity.
A blindly coded HIV DNA quantification panel, which was provided by the Virological Quality Assurance program of the National AIDS Clinic Trials Group, was used to validate this assay. This panel consisted of 12 coded peripheral blood mononuclear cell (PBMC) samples that contained specified copy numbers including 3 HIV-negative specimens and 3 specimens each with 100, 320, and 1,000 copies per 106 PBMCs. Each sample was tested in triplicate three different times. The HIV-1 proviral DNA level was calculated as either the copy number per 106 cells or the number of micrograms of DNA. There was a close agreement (less than twofold) between the expected copy numbers and the actual copy numbers estimated on the basis of both the cell counts and the DNA concentration (Table 1). Even though estimates based on the cell counts were slightly higher than those based on the DNA concentration, there was no significant difference in the precision or the accuracy between these two estimates (P = 0.44) (Table 2). Therefore, both estimates can be used reliably to quantify HIV-1 proviral DNA.
TABLE 1.
Assay validation by use of a blindly coded HIV-1 DNA quantification panel
| Expected copy no. | Median estimates of actual copy no. based ona:
|
|
|---|---|---|
| Cell counts (per 106 PBMCs) | DNA concn (per μg of DNA) | |
| 0 | 0 | 0 |
| 100 | 137 | 127 |
| 320 | 604 | 540 |
| 1,000 | 2,029 | 1,933 |
Three samples with each expected copy number were tested.
TABLE 2.
Difference between expected and actual copy numbers (accuracy)a
| Method | Difference between expected and actual copy no.
|
|||
|---|---|---|---|---|
| Minimum | Median | Maximum | SD | |
| Cell counts | 0.06 | 0.29 | 0.61 | 0.20 |
| DNA concn | 0.08 | 0.26 | 0.72 | 0.24 |
The difference between the two methods (precision) was 0.02 ± 0.08 (mean ± standard deviation; P = 0.44; n = 9).
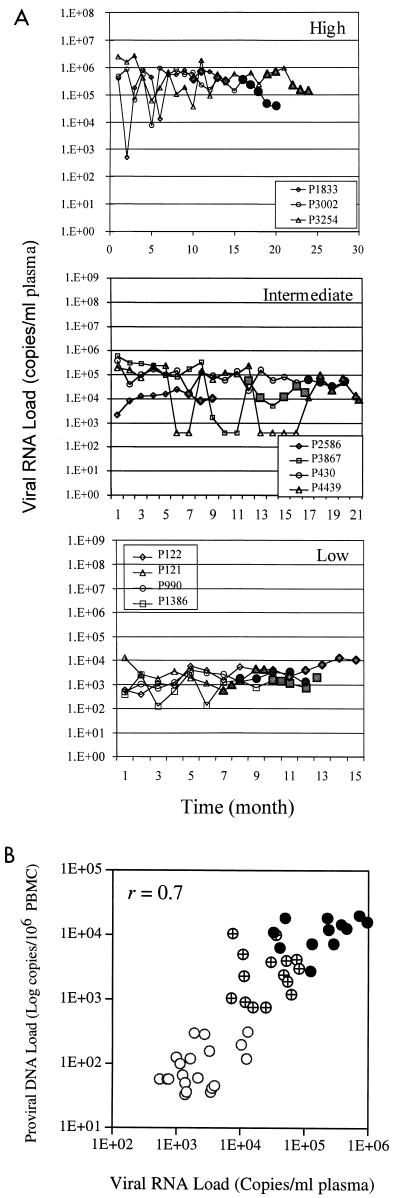
By using this method, HIV-1 proviral DNA levels were compared in parallel with viral RNA levels in a total of 80 specimens from 18 HIV-infected children who were stratified into four groups on the basis of their viral RNA levels (Fig. 2A), i.e., children with high (>5 log10 copies), intermediate (4 to 5 log10 copies), low (2.6 to 4 log10 copies), and nondetectable (<2.6 log10 copies) viral RNA levels. The inclusion criteria for each subject were that each had at least four consecutive samples with RNA levels within the defined range over a 6-month period. In addition, fluctuation of the viral RNA load during the 6-month period must have been less than 1 log. As shown in Fig. 2B, HIV-1 proviral DNA levels were positively correlated with the viral RNA load in the groups with high, intermediate, and low viral RNA levels (r = 0.7). Interestingly, of 18 samples that had nondetectable viral RNA for more than 6 months, HIV-1 proviral DNA was detected in all four patients, with only 3 of 18 samples (17%) having nondetectable viral DNA. The 15 DNA-positive samples had HIV-1 proviral DNA levels that varied over a range of approximately 2 logs, from 60 to 1,908 copies per 106 PBMCs. This observation suggests that proviral DNA could potentially be used as an additional surrogate marker to monitor the efficacy of antiretroviral therapy.
FIG.2.
Correlation of HIV-1 proviral DNA with viral RNA at stable stages. (A) Longitudinal measurement of viral RNA levels of HIV-infected children with viral RNA levels that are high (>5 log10 copies), intermediate (4 to 5) log10 copies), and low (2.6 to 4 log10 copies. Datum points with closed symbols represent results for samples for which the corresponding viral DNA level was measured by the TaqMan assay. (B) Positive correlation of the HIV-1 RNA level with the proviral DNA level when viral RNA levels were stabilized for at least 6 months.
In summary, we developed and validated a real-time PCR (TaqMan) assay for quantification of HIV-1 proviral DNA. This assay is highly specific, with an analytic sensitivity of 10 viral DNA copies and a linear dynamic range of more than 6 logs (Fig. 1A). This assay is also reproducible, with intra- and interassay variabilities of <2.5-fold over the entire linear range (Fig. 1B). Since this method has a much broader linear dynamic range of detection than the conventional PCR assays, it should provide a useful means for quantification of the HIV-1 proviral DNA load in HIV-infected patients receiving antiviral therapy.
Acknowledgments
We thank Donald Brambilla for statistical analysis. The PCR standard panel, which was contributed by Shirley Kwok and Cindy Christopherson, was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
This study was supported in part by Chicago Medical Research Institute Council, Junior Board, and John Lloyd Foundation (to Y.Z.).
REFERENCES
- 1.Bennett, J. M., S. Kaye, N. Berry, and R. S. Tedder. 1999. A quantitative PCR method for the assay of HIV-1 provirus load in peripheral blood mononuclear cells. J. Virol. Methods 83:11-20. [DOI] [PubMed] [Google Scholar]
- 2.Christopherson, C., Y. Kidane, B. Conway, J. Krowka, H. Sheppard, and S. Kwok. 2000. PCR-based assay to quantify human immunodeficiency virus type 1 DNA in peripheral blood mononuclear cells. J. Clin. Microbiol. 38:630-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 5.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 6.Guenthner, P. C., and C. E. Hart. 1998. Quantitative, competitive PCR assay for HIV-1 using a microplate-based detection system. BioTechniques 24:810-816. [DOI] [PubMed] [Google Scholar]
- 7.Higuchi, R., G. Dollinger, P. S. Walsh, and R. Griffith. 1992. Simultaneous amplification and detection of specific DNA sequences. Bio/Technology 10:413-417. [DOI] [PubMed] [Google Scholar]
- 8.Higuchi, R., C. Fockler, G. Dollinger, and R. Watson. 1993. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions Bio/Technology 11:1026-1030. [DOI] [PubMed] [Google Scholar]
- 9.Holland, P., R. Abramso, R. Watson, and D. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizating the 5"-3" exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Izopet, J., C. Tamalet, C. Pasquier, K. Sandres, B. Marchou, P. Massip, and J. Puel. 1998. Quantification of HIV-1 proviral DNA by a standardized colorimetric PCR-based assay. J. Med. Virol. 54:54-59. [DOI] [PubMed] [Google Scholar]
- 11.Jurriaans, S., J. T. Dekker, and A. de Ronde. 1992. HIV-1 viral DNA load in peripheral blood mononuclear cells from seroconverters and long-term infected individuals. AIDS 6:635-641. [DOI] [PubMed] [Google Scholar]
- 12.Kwok, S., and C. Christopherson. 2000. PCR standard, p. 93. In AIDS Research and Reference Reagent program catalog. U.S. Department of Health and Human Services, Rockville, Md.
- 13.Nachman, S. A., K. Stanley, R. Yogev, S. Pelton, A. Wiznia, S. Lee, L. Mofenson, S. Fiscus, M. Rathore, E. Jimenez, W. Borkowsky, J. Pitt, M. E. Smith, B. Wells, and K. McIntosh. 2000. Nucleoside analogs plus ritonavir in stable antiretroviral therapy-experienced HIV-infected children: a randomized controlled trial. Pediatric AIDS Clinical Trials Group 338 Study Team. JAMA 283:492-498. [DOI] [PubMed] [Google Scholar]
- 14.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582-1586. [DOI] [PubMed] [Google Scholar]
- 15.Sharkey, M. E., I. Teo, T. Greenough, N. Sharova, K. Luzuriaga, J. L. Sullivan, R. P. Bucy, L. G. Kostrikis, A. Haase, C. Veryard, R. E. Davaro, S. H. Cheeseman, J. S. Daly, C. Bova, R. T. Ellison, B. Mady, K. K. Lai, G. Moyle, M. Nelson, B. Gazzard, S. Shaunak, and M. Stevenson. 2000. Persistence of episomal HIV-1 infection intermediates in patients on highly active anti-retroviral therapy. Nat. Med. 6:76-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephenson, J. 1997. The art of “HAART': researchers probe the potential and limits of aggressive HIV treatments. JAMA 277:614-616. [PubMed] [Google Scholar]
- 17.Wiznia, A., K. Stanley, P. Krogstad, G. Johnson, S. Lee, J. McNamara, J. Moye, J. B. Jackson, H. Mendez, R. Aguayo, A. Dieudonne, A. Kovacs, M. Bamji, E. Abrams, S. Rana, J. Sever, and S. Nachman 2000. Combination nucleoside analog reverse transcriptase inhibitor(s) plus nevirapine, nelfinavir, or ritonavir in stable antiretroviral therapy-experienced HIV-infected children: week 24 results of a randomized controlled trial—PACTG 377. Pediatric AIDS Clinical Trials Group 377 Study Team. AIDS Res. Hum. Retrovir. 16:1113-1121. [DOI] [PubMed] [Google Scholar]