Abstract
Coagulase-negative staphylococci (CoNS) are a major cause of sepsis in the neonatal intensive care unit (NICU). We evaluated the hypothesis that the ica operon and biofilm production are associated with CoNS disease in this setting. CoNS associated with bacteremia or blood culture contamination and from the skin of infants with CoNS bacteremia or healthy controls were obtained during a prospective case-control study on a busy NICU. A total of 180 strains were identified, of which 122 (68%) were Staphylococcus epidermidis and the remainder were S. capitis (n = 29), S. haemolyticus (n = 11), S. hominis (n = 9), S. warneri (n = 8), and S. auricularis (n = 1). The presence of the genes icaA, icaB, icaC, and icaD was determined by PCR, and biofilm production was examined using qualitative (Congo red agar [CRA]) and quantitative (microtiter plate) techniques. There were no significant differences in the presence of the ica operon or CRA positivity among the four groups of strains. However, quantitative biofilm production was significantly greater in strains isolated from either the blood or the skin of neonates with S. epidermidis bacteremia. We conclude that the quantity of biofilm produced may be associated with the ability to cause CoNS infection. This conclusion suggests that the regulation of biofilm expression may play a central role in the disease process.
Coagulase-negative staphylococci (CoNS) are the major cause of late-onset sepsis in preterm infants (27, 39, 42, 44). Approximately one in six very-low-birth-weight (<1,500-g) neonates develops an episode of CoNS bacteremia (21, 44), an event that is associated with a significant increase in morbidity and mortality (44), duration of hospital stay (18, 21, 44), and overall cost of treatment (44). There is thus a need to reduce the risk of sepsis in this setting, a goal which depends in part on defining the pathogenesis of infection.
The role of biofilm formation as a determinant of CoNS infection has been the subject of ongoing study since the observation that some isolates of Staphylococcus epidermidis produced mucoid growth in vitro that adhered to the walls of culture tubes (2). A series of studies subsequently reported that this material, termed slime, was more commonly produced by isolates associated with sepsis, including intravenous-catheter-related bacteremia and other prosthetic device infections (5, 6, 10, 14, 28, 31). The relevance to the neonatal intensive care unit (NICU) setting of slime production by CoNS isolates has been studied in relation to both carriage flora and isolates causing infection. The proportion of slime-producing CoNS in carriage flora increased on repeated sampling of neonates during the first 4 weeks of age in one unit (9) but did not vary in two other units over either 2 weeks (35) or a mean of 8 weeks (24). Endemic clones of CoNS that were associated with disease and that were slime producers were demonstrated to persist over a period of 10 years in one unit (26); however, carriage flora was not examined, and an association between slime production and either biological fitness or virulence cannot be assumed. One study compared CoNS from carriage flora and blood culture isolates and found that 79% of blood culture isolates were slime producers, compared with 75 and 58% of skin flora isolates from infected and noninfected infants, respectively (22). Infants with bacteremia were more frequently colonized by S. epidermidis, and it is difficult to determine whether the higher rate of slime production by isolates colonizing sick neonates was a direct association or whether it reflected a relationship between slime production and species. Weekly mucocutaneous culturing for preterm infants over 3 months revealed that infants with slime-positive CoNS colonization were more likely to develop invasive CoNS disease than infants with slime-negative or no CoNS colonization (23).
The mechanism by which CoNS attach to prosthetic material and elaborate what is now termed biofilm is being increasingly understood. This is a complex and multistep process (33, 47), and the relevance of each major component will require study in relation to human disease. One important element in this process is the ica operon, a gene cluster encoding the production of polysaccharide intercellular adhesin (PIA), which mediates the intercellular adherence of bacteria and the accumulation of multilayer biofilm (25). The presence of this operon has been compared between unmatched strain collections of S. epidermidis associated with carriage and infection outside the NICU setting (1, 16, 19, 48). A study comparing a collection of 52 S. epidermidis isolates (comprising 51 isolates from blood cultures and 1 isolate from cerebrospinal fluid) with 36 isolates obtained from the skin and mucosa of healthy volunteers found ica genes to be present in 85 and 2%, respectively (48). A study of S. epidermidis associated with prosthetic-material-related joint infection demonstrated that 44 of 54 isolates were ica positive, compared with 2 of 23 isolates from eight healthy individuals (19). Similarly, 33 of 68 isolates associated with intravenous-catheter-related infection were ica positive, compared with none of 10 isolates from the skin or mucosa of healthy volunteers (1). Comparison of ica in S. epidermidis associated with either bacteremia, blood culture contamination, or colonized intravenous devices and S. epidermidis from normal flora of healthy volunteers who were not hospitalized showed that ica was more than twice as frequent in isolates associated with infection (16).
The aim of this study was to evaluate the hypothesis that isolates of CoNS associated with disease in neonates are more likely to be positive for the ica operon and to produce biofilm than are isolates drawn randomly from NICU carriage flora. This aim was achieved by studying bacterial isolates collected from blood cultures and the skin of healthy and infected neonates during a prospective case-control study of CoNS sepsis.
MATERIALS AND METHODS
Study design.
The study was conducted on the NICU at John Radcliffe Hospital, Oxford, United Kingdom. This unit has the capacity to care for 27 neonates, with 20 high-dependency incubators or cots and 7 intensive care incubators. Ethical approval for the study was obtained from the Central Oxford Research Ethics Committee. A library of CoNS isolates was assembled during a prospective case-control study conducted between May 1999 and July 2000. This was performed to compare phenotypic characteristics between isolates associated with carriage and disease and to assess whether hypervirulent clones of CoNS exist in this setting (the results are reported in reference 13).
Blood samples for culturing were taken from peripheral sites rather than from previously placed lines. Neonates with blood cultures positive for CoNS were entered into the study, and the clinical relevance of all positive cultures was assigned as either invasive isolates (causing bacteremia) or contaminants. Bacteremia was defined as (i) two or more independent blood cultures positive for identical isolates of CoNS in association with a clinical picture consistent with sepsis or (ii) a single positive culture with no alternative explanation for the clinical picture of sepsis in an infant who responded to antistaphylococcal antibiotics. Skin specimens were taken by using a swab wash method from the ear lobe and axilla of (i) neonates with bacteremia (defined above) and (ii) two control neonates who had no features of sepsis and who were swabbed within 24 h of definition of a case of bacteremia. Control neonates were chosen at random from the ward list and, to avoid overmatching, were not matched for gestational age, location, or length of stay in the NICU.
Bacterial isolates and antibiotic susceptibility testing.
Blood culture isolates were obtained from the Department of Microbiology, John Radcliffe Hospital. Skin swab specimens were plated on blood agar and incubated at 37°C in air for 48 h. Presumptive CoNS, based on colonial morphology, were plated to purity, and identification was confirmed on the basis of a positive catalase test and negative coagulase and DNase tests. Species identification was performed by using the API ID32 system (bioMerieux Ltd.) according to the manufacturer's recommendations. Antibiotic susceptibility testing was performed by using a comparative disk diffusion method (20). Susceptibility to penicillin, oxacillin, gentamicin, netilmicin, amikacin, erythomycin, tetracycline, ciprofloxacin, vancomycin, fusidic acid, rifampin, and trimethoprim was tested. Isolates were stored in tryptic soy broth (TSB) with glycerol (15% [vol/vol]) at −80°C.
Detection of biofilm production.
Qualitative detection of biofilm formation was performed by using Congo red agar (CRA) as previously described (17). Strains were streaked onto the agar to obtain single colonies and incubated overnight at 37°C in air and a further 24 h at room temperature. PIA-positive strains appear as black colonies, and PIA-negative strains were red.
Quantitative determination of biofilm production was performed by using a microtiter assay as previously described (7), with the exception that the stain used was safranin rather than crystal violet. In brief, bacteria were inoculated into 10 ml of TSB with 0.25% glucose and incubated overnight with shaking at 37°C in air. This solution was diluted 1:100 in TSB with glucose, and 200 μl was inoculated into 96-well polystyrene microtiter plates. The plates were incubated overnight at 37°C in air, washed, and stained with 0.1% safranin. The optical density of the adherent biofilm was determined by using an enzyme-linked immunosorbent assay plate reader at 490 nm. Each plate contained S. epidermidis RP62A (ATCC 35984) and S. carnosus TM300 (kindly provided by Wilma Ziebuhr) as positive and negative biofilm-producing controls, respectively, and TSB but no bacteria (background control). Eight wells were inoculated per strain in a given experiment, and all strains were tested independently on three occasions. The absorbance was taken to be the optical density of the test isolate minus the mean of the background control values for the same plate.
Detection of the ica operon.
Chromosomal DNA was extracted by using a Puregene DNA extraction kit (Gentra Systems), with the modification that lysostaphin at 30 μg/ml (Sigma) was added at the cell lysis step. PCR was used to detect the presence of icaA, icaB, icaC, icaD, and the insertion sequence element IS256. The primer sequences and predicted product lengths for icaA, icaB, and icaC of S. epidermidis and IS256 of S. aureus were described by Ziebuhr et al. (49). The primer sequences for icaD were 5"-AGGCAATATCCAACGGTAA-3" (forward) and 5"-GTCACGACCTTTCTTATATT-3" (reverse), corresponding to bases 1891 to 2262 of the ica operon (GenBank accession number U43366). The PCR cycling conditions used were 30 cycles of 1 min of denaturation at 94°C and 2.5 min of elongation at 72°C for all reactions, with annealing for 1 min at 60°C (icaA), 59°C (icaB), 45°C (icaC), 59°C (icaD), or 59°C (IS256). PCR was performed by using a PTC-200 Peltier thermal cycler (MJ Research) with Taq polymerase (Bioline). The magnesium concentration was 3 mM for all genes. Reaction mixtures were analyzed by 1% agarose gel electrophoresis.
Analysis.
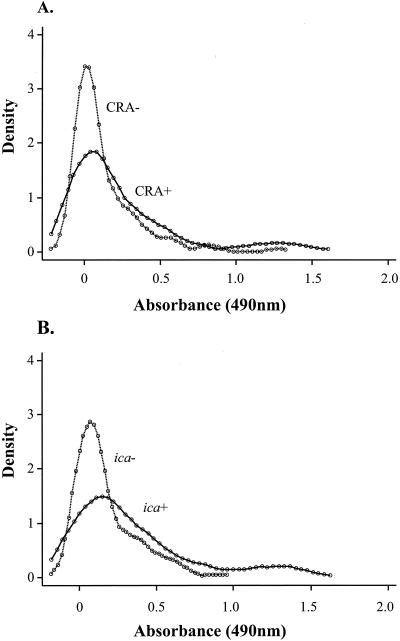
For neonates with multiple blood cultures positive for the same isolate, the results for testing of biofilm production and the presence of the ica operon were counted once, using the first isolate obtained. Isolate identity was based on pulsed-field gel electrophoresis (PFGE). Isolates with identical PFGE patterns were regarded as genotypically indistinguishable. Spectrophotometric absorbance measurements from the microtiter plate assay were found to be log transformable toward normality. Geometric mean absorbances were compared between groups by using analysis of variance or Student's t test. The distributions were displayed by using plots of the Epanechnikov kernel density, which are similar in interpretation to a histogram but give a continuous distribution curve. In addition, a histogram requires intervals or bins into which the data are divided, and the shape of the histogram can change considerably if the start point of these bins is altered. Kernel density plots do not have this disadvantage. Categorical variables were compared between groups by using Fisher's exact test. Spearman's rho test was used to assess nonparametrically the correlation between continuous variables. All analyses were performed using Stata 7 (StataCorp., College Station, Tex.).
RESULTS
Cases and isolates.
Sixteen neonates with CoNS bacteremia were recruited; 2 had a second episode with a new strain of CoNS, as determined with an antibiogram (and confirmed by PFGE), giving a total of 18 strains that were associated with infection. Skin swab samples obtained from these infected neonates resulted in the isolation of a further 35 strains of CoNS. A total of 71 strains were isolated from the skin of 32 well control babies. There were 39 neonates with blood cultures positive for CoNS strains that were considered to be contaminants; many of these cultures yielded multiple strains, so a total of 56 contaminating strains were identified. Thus, there was an overall total of 180 strains, of which 122 (68%) were S. epidermidis and the remainder were S. capitis (n = 29), S. haemolyticus (n = 11), S. hominis (n = 9), S. warneri (n = 8), and S. auricularis (n = 1). There was no statistical difference in the distribution of species among the four clinical groups (P = 0.97).
Presence of the ica operon.
The primers used to determine the presence of genes in the ica operon were based on the sequence of the S. epidermidis genome. Of the 122 strains of S. epidermidis, 40% were positive for the ica operon, with PCR products being obtained for the icaA, icaB, icaC, and icaD genes in all of these strains (Table 1). The same primers were used to evaluate whether amplification would occur from DNA isolated from the five other CoNS species. No products were obtained for strains of S. haemolyticus, S. hominis, S. warneri, and S. auricularis. However, 12 of 29 S. capitis strains yielded a positive PCR result for the icaC and icaD genes (although all were negative for icaA and icaB), suggesting that a homolog of the S. epidermidis operon exists in some members of this species.
TABLE 1.
Presence of genes in the ica operon and results of qualititative (CRA) and quantitative (microtiter plate) assays for biofilm production
| Species(no. of strains) | No. (%) of strains positive
|
Geometric mean OD490 (95% CI) in the microtiter plate assay for the following strainsa:
|
|||||
|---|---|---|---|---|---|---|---|
| For icaA, icaB, icaC, and icaD | For icaC and icaD | CRA test | icaCD negative | icaCD positive | CRA test negative | CRA test positive | |
| S. epidermidis (122) | 49 (40) | 49 (40) | 29 (24) | 0.07 (0.04-0.10) | 0.22b (0.16-0.30) | 0.09 (0.06-0.13) | 0.22c (0.15-0.36) |
| S. capitis (29) | 0 | 12 (41) | 12 (41) | 0.03 (−0.01-0.08) | 0.01 (−0.04-0.06) | 0.02 (−0.01-0.06) | 0.02 (−0.04-0.9) |
| S. haemolyticus (11) | 0 | 0 | 0 | 0.01 (−0.04-0.09) | 0.01 (−0.04-0.09) | ||
| S. hominis (9) | 0 | 0 | 4 (44) | 0.02 (−0.03-0.08) | 0.05 (−0.2-0.13) | −0.01 (−0.10-0.17) | |
| S. warneri (8) | 0 | 0 | 0 | 0.07 (−0.01-0.16) | 0.07 (−0.01-0.16) | ||
| S. auricularis (1) | 0 | 0 | 0 | 0.01 | 0.01 | ||
OD490, optical density at 490 nm; CI, confidence interval.
The P value was <0.0001 in a comparison with the icaCD-negative group.
The P value was 0.0005 in a comparison with the CRA test-negative group.
Biofilm production.
Biofilm production assessed by using CRA was expressed by 45 strains (25%), with positive CRA reactions being found among S. epidermidis, S. capitis, and S. hominis strains but not among strains of the other species tested (Table 1). Quantitative biofilm production determined by a microtiter plate assay was significantly higher in strains which were found to be biofilm producers by the CRA method (geometric mean [95% confidence interval] optical density, 0.14 [0.08 to 0.21] versus 0.08 [0.06 to 0.11]; P = 0.04), although there was considerable overlap in the distributions (Fig. 1). Part of this overlap can be explained by the finding that although CRA-positive strains were detected for three species of CoNS, the difference in quantitative biofilm production between CRA-positive and CRA-negative strains was significant only for S. epidermidis (P = 0.0005) (Table 1). Thus, an association between CRA positivity and biofilm production assessed by a microtiter plate assay does not hold true for CoNS species, other than S. epidermidis, isolated during this study.
FIG. 1.
Density plots showing results of assessment of biofilm production. (A) Distribution of biofilm production in CRA-positive CoNS strains and CRA-negative strains. (B) Distributions of biofilm production in S. epidermidis strains with and without the ica operon. Absorbance in the microtiter plate assay is plotted against the Epanechnikov kernel density (see the text for details).
Relationships between presence of the ica operon and phenotype.
Of the 49 S. epidermidis strains found positive for the ica operon, 29 (59%) were found to be biofilm producers by the CRA method. All of the 73 S. epidermidis strains which lacked the ica operon were found to be CRA negative. As might be predicted, quantitative biofilm production was significantly higher in the S. epidermidis strains found positive for the ica operon (P < 0.0001) (Fig. 1). There was a significant association between a positive PCR result for icaC and icaD and a positive CRA test result for S. capitis (P = 0.001) but not between PCR positivity and quantitative biofilm production. This result suggests that the CRA test can detect the gene products of icaC and icaD in this species but that the microtiter plate assay cannot. For the four species consistently found negative for the ica operon by PCR, the absorbance results for the biofilm microtiter plate assay were uniformly low and the CRA test was negative for all but four strains of S. hominis.
ica operon, biofilm production, and clinical groups.
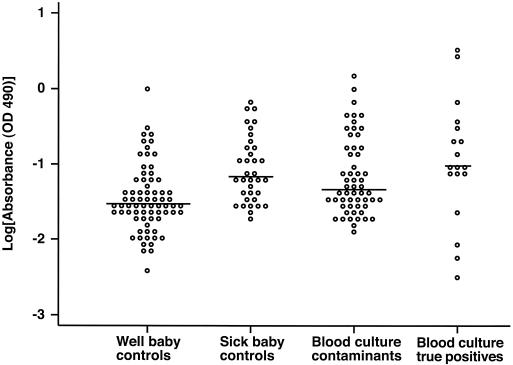
There were no significant differences in ica operon or CRA positivity among the four groups (invasive blood culture isolates, blood culture contaminants, sick baby skin isolates, and well baby [control] skin isolates) (Table 2). However, quantitative biofilm production was significantly higher in strains isolated from either the blood or the skin of neonates with CoNS bacteremia than in strains isolated from the skin of well (control) neonates (P < 0.001) (Fig. 2). This was true for the S. epidermidis subgroup but not for the other CoNS species when considered together as a “non-S. epidermidis” group.
TABLE 2.
Relationships among the presence of genes in the ica operon, qualitative (CRA) and quantitative (CRA) and quantitative (microtiter plate) assays for biofilm production, and clinical groups
| Sample | Results for:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| All species
|
S. epidermidis only
|
|||||||
| CRA test positivea | Biofilm productionb | Positive for:
|
CRA test positivea | Biofilm productionb | Positive for icaA, icaB, icaC, and icaDa | |||
| icaA, icaB, icaC, and icaDa | icaC and icaDa | |||||||
| Blood culture isolates | ||||||||
| Clinical positives | 7/18 (39) | 0.14 (0.03-0.32) | 5/18 (28) | 8/18 (44) | 3/12 (25) | 0.25 (0.07-0.56) | 5/12 (42) | |
| Clinical contaminants | 16/56 (29) | 0.11 (0.07-0.16) | 15/56 (27) | 17/56 (30) | 12/39 (31) | 0.13 (0.08-0.19) | 15/39 (38) | |
| All | 23/74 (31) | 0.12 (0.08-0.16) | 0/74 (27) | 22/74 (31) | 15/51 (29) | 0.15 (0.10-0.21) | 20/51 (39) | |
| Skin controls | ||||||||
| Sick babies | 7/35 (20) | 0.15 (0.10-0.21) | 12/35 (34) | 15/35 (43) | 5/25 (20) | 0.19 (0.13-0.27) | 12/25 (48) | |
| Well babies | 15/71 (21) | 0.03 (0.01-0.06) | 17/71 (24) | 22/71 (31) | 9/46 (20) | 0.04 (0.01-0.07) | 17/46 (37) | |
| Pc | 0.31 | 0.0001 | 0.53 | 0.46 | 0.49 | 0.0001 | 0.72 | |
Reported as number of strains positive/number tested (percent).
Reported as optical density at 490 nm (95% confidence interval).
P value is for the three-way comparison of results obtained with blood culture isolates, sick baby skin controls, and well baby skin controls.
FIG. 2.
Biofilm production by strains in the four clinical groups. As some of the microtiter plate assay absorbance values were negative after subtraction of background, 0.2 was added to all values prior to log transformation. The horizontal line represents the median value of each distribution. For statistical comparisons between groups, see the text. OD, optical density.
Relationships among ica operon, biofilm production, and PFGE types.
PFGE had previously been performed on all isolates (data not shown), and the results were used to evaluate whether the presence of the ica operon was conserved within the bacterial clones identified. The 122 strains of S. epidermidis were represented by 53 PFGE types. The ica operon either was found in all strains within each PFGE type or was universally absent. The three most frequent PFGE types, containing a total of 36 strains, were all negative for the ica operon and, consequently, negative by the CRA test. In contrast, all 29 S. capitis strains were of a single PFGE type, but the PCR and CRA test results were positive for less than half of these. All four S. hominis strains found positive by the CRA test belonged to the same PFGE type.
ica operon and IS256 in S. epidermidis.
Insertion and excision of insertion sequence element IS256 have been reported to be associated with phase variation in the production of PIA in two laboratory isolates of S. epidermidis (49). We evaluated whether the presence of an insertion element was responsible for the failure of biofilm expression in S. epidermidis isolates that were found positive for the ica operon but negative by the CRA test. All PCR products from S. epidermidis (both CRA positive and negative) were of the predicted sizes. The presence of IS256 in the genome was confirmed by PCR for 25 of 27 ica operon-positive strains of S. epidermidis chosen at random.
ica operon, biofilm production, and antibiotic resistance.
With the exception of trimethoprim, antibiotic resistance was more frequent in isolates of S. epidermidis found positive for the ica operon, this difference reaching statistical significance for oxacillin, netilmicin, rifampin, and ciprofloxacin (Table 3).S. epidermidis strains with the ica operon were resistant to an average of 5.5 (95% confidence interval, 4.9 to 6.2) of the 12 antibiotics tested, compared with an average of 4.2 (3.8 to 4.6) for those without the operon (P = 0.0003). Similarly, for resistant isolates of all species, there was an overall trend for higher quantitative biofilm production which was significantly different for oxacillin, netilmicin, and amikacin. In addition, there was a significant correlation between the number of antibiotics to which a strain was resistant and biofilm production (value for Spearman's rho test, 0.28; P < 0.0001). In contrast, CRA positivity was not significantly associated with antibiotic resistance. The association between biofilm production and antibiotic resistance was mirrored by an association between the clinical source of the strains and antibiotic resistance. Strains derived from the blood or skin of patients with clinical CoNS sepsis were on average resistant to more antibiotics than those derived from the skin of healthy (control) neonates (5.3 [95% confidence interval, 4.8 to 5.9] versus 4.1 [3.8 to 4.4]; P = 0.0002). As there was an association between quantitative biofilm production and the clinical source of the strains, it is difficult to draw any conclusions concerning the etiology of the increase in antibiotic resistance. Antibiotics administered prior to bacterial isolation were matched between neonates with bacteremia and controls, as were the numbers of patients who left the hospital alive.
TABLE 3.
Relationships among the ica operon, biofilm production, and antibiotic resistancea
| Antibiotic | No. of strains positive/no. tested (%) with the following characteristics
|
OD490 (95% CI) in the microtiter plate assay for 176 strains of all species that wered:
|
|||||
|---|---|---|---|---|---|---|---|
|
ica operon presentb
|
CRA test positivec
|
||||||
| Resistant | Sensitive | Resistant | Sensitive | Resistant | Sensitive | ||
| Penicillin | 48/113 (42) | 0/7 (0) | 45/168 (27) | 0/8 (0) | 0.09 (0.06-0.11) | 0.05 (0.0-0.11) | |
| Oxacillin | 48/105 (46) | 0/15 (0) | 44/155 (28) | 1/21 (5) | 0.1 (0.07-0.22) | 0.01 (−0.02-0.05) | |
| Erythromycin | 23/52 (44) | 25/68 (37) | 11/64 (17) | 34/112 (30) | 0.07 (0.04-0.11) | 0.09 (0.07-0.13) | |
| Tetracycline | 19/33 (58) | 29/87 (33) | 13/49 (27) | 32/127 (25) | 0.05 (0.02-0.09) | 0.1 (0.07-0.13) | |
| Trimethoprim | 21/55 (38) | 27/65 (42) | 18/74 (24) | 27/102 (26) | 0.12 (0.08-0.16) | 0.07 (0.04-0.09) | |
| Gentamicin | 42/97 (43) | 6/23 (43) | 40/146 (27) | 5/30 (17) | 0.1 (0.07-0.13) | 0.03 (0.0-0.07) | |
| Netilmicin | 20/33 (61) | 28/87 (32) | 14/44 (32) | 31/132 (23) | 0.21 (0.13-0.30) | 0.05 (0.04-0.07) | |
| Amikacin | 13/23 (57) | 35/97 (36) | 8/30 (27) | 37/109 (25) | 0.23 (0.13-0.36) | 0.06 (0.04-0.08) | |
| Rifampin | 14/17 (82) | 34/103 (33) | 3/17 (18) | 42/159 (26) | 0.19 (0.1-0.3) | 0.08 (0.06-0.1) | |
| Fusidic acid | 9/20 (45) | 39/100 (39) | 4/22 (18) | 41/154 (27) | 0.13 (0.06-0.22) | 0.08 (0.06-0.1) | |
| Ciprofloxacin | 10/11 (91) | 38/109 (35) | 2/17 (12) | 43/159 (27) | 0.11 (0.03-0.23) | 0.08 (0.06-0.11) | |
Results shown in bold signify a P value of <0.01. The P value was calculated in a test of association between antibiotic sensitivity and the characteristic for which a result is shown.
S. epidemidis strains only (n = 120).
Strains of all species (n = 176).
See Table 1, footnote a.
DISCUSSION
Production of biofilm by CoNS is widely considered to be an important determinant of prosthetic-device-related infections. Such an effect is likely to be mediated through the ability to colonize and persist on prosthetic material, resist the effects of antibiotics, and evade the immune system. Biofilm production has also been shown to be associated with virulence in the absence of prosthetic material in an animal model (11). The production of PIA is an important component in the process of biofilm formation, suggesting that the ica operon plays an important role in disease pathogenesis. Evidence for this comes from both animal models and clinical studies. Inactivation of icaA was reported to be associated with a decrease in the pathogenicity of a strain of S. epidermidis in two animal models of foreign-body infection (40, 41). The presence of the ica operon was also found to be more common in S. epidermidis strains associated with disease than in carriage strains in four studies of human disease (1, 16, 19, 48).
Our results lead us to conclude that there is no association between the presence of the ica operon and CoNS bacteremia in our unit. However, we found that quantitative biofilm production was higher in S. epidermidis strains isolated from the blood and skin of neonates with CoNS bacteremia than in strains isolated from the skin of well (control) neonates. The biological significance of the magnitude of this difference is a matter for speculation, but there are several possible explanations for the finding. One possibility is that infants with CoNS sepsis represented a subgroup who had a poorer prognosis and who had also received more antibiotics before bacterial isolation. Given this case scenario, antibiotics could select for high biofilm producers, and CoNS sepsis might be a marker of a less fit host. However, we examined antibiotic usage before bacterial isolation together with the number of neonates who left the hospital alive for cases and controls and found that these characteristics did not differ. The alternative explanation is that colonizing strains which produce more biofilm have enhanced pathogenic potential.
Our findings suggest that it may be gene regulation rather than the presence or absence of the ica operon that is involved in bacterial virulence. Using transposon mutagenesis, three gene loci have been shown to have a regulatory influence on the expression of the synthetic genes for PIA (34). The genetic defect has been described for one of these loci, the transposon insertion site being located in rsbU (30). This is the first gene of an operon that is highly homologous to the sigB operons of S. aureus and Bacillus subtilis and that acts as a positive regulator of alternative sigma factor σB (30). Biofilm is also known to be phase variable, and a study demonstrated that phase variation of PIA produced by S. epidermidis strains RP62A and 229 under laboratory conditions was due to alternating insertion and excision of insertion sequence element IS256 (49). This element is localized at the termini of aminoglycoside resistance-mediating transposon Tn4001 and is present in multiple copies on the chromosome of aminoglycoside-resistant staphylococci (15). Our study found that of the 49 S. epidermidis strains positive for the ica operon, only 29 were found to be biofilm producers by the CRA method. We confirmed the presence of IS256 in a subset of ica-positive S. epidermidis strains, but there was no evidence for insertion of this element into the ica operon, the PCR products of the ica genes being of the predicted sizes. These results are consistent with those of a study which found no evidence for an insertion sequence associated with a 2.8-kb PCR product from the ica operon of 10 S. epidermidis isolates that were ica positive but biofilm negative (19). These data suggest that the variability in expression of the ica operon in natural bacterial populations cannot be explained by IS256 insertion in this region.
Given that several previous investigators found a link between the ica operon and invasive S. epidermidis disease in other clinical settings, is it possible that the lack of an association in this study is a false-negative finding arising from an aspect of the study design? Possible factors that could have influenced our results include clinical definitions and the populations sampled. Distinguishing between true CoNS bacteremia and blood culture contamination is notoriously difficult; this is particularly true for neonates, since it is often difficult to take a blood sample in a sterile manner. Furthermore, defining sepsis in these individuals is complex. However, we believe that our definitions are robust and were applied in a rigorous fashion by the primary physicians at the bedside during this prospective study. A second factor is that comparisons between CoNS associated with nosocomial disease and carriage flora of either healthy volunteers or inpatients from a different hospital or unit may be influenced by bacterial clonality. Endemic clones of CoNS were shown to be present in several hospital settings, including an NICU and a hematology-oncology unit (3, 36, 45, 46), and appeared to be unit specific. Such a scenario may lead to the finding that a given determinant is either common or rare in a given setting, depending on its presence in the most common genotypes. Thus, comparison between predominant clones present in different hospital settings may be misleading. The genetic population structure of CoNS colonizing healthy volunteers in the community is unknown, but these strains are under less antibiotic pressure, and there may be little overlap with those found in the hospital setting. To address such issues, we chose a case-control study design. This design was used in preference to a cohort study, as in a cohort study a very large number of infants would have to be recruited for a reasonable number of cases to be prospectively identified.
Our finding that the production of biofilm was associated with antibiotic resistance is consistent with the findings of several previous studies (12, 32). Under laboratory conditions, variants of S. epidermidis RP62A with diminished slime production became susceptible to oxacillin, penicillin, and ampicillin, suggesting a direct association between slime and resistance (4). This is not always the case, however (37, 38), and it is also possible that biofilm production and antibiotic resistance have been independently selected as determinants that confer a selective advantage for colonization and survival in the NICU environment.
S. epidermidis is the most common cause of nosocomial infections in neonates, and putative virulence determinants have been studied most intensively for this species of CoNS. However, slime production has been reported for S. hominis and S. haemolyticus (5, 22, 29, 38, 43), S. warneri (22), S. saprophyticus (12, 29), and S. simulans (12). Cross-species hybridization with DNA blots and simultaneous probes for S. epidermidis and S. aureus icaA has also been demonstrated for S. auricularis and S. capitis but not S. haemolyticus, S. hominis, or S. warneri (8). The difference in results for hybridization and slime production may reflect the fact that only a single strain of each species was tested by hybridization for genes that are variably present. We found that 12 of 29 S. capitis isolates were positive by PCR for icaC and icaD, indicating the presence of an ica homolog. However, all 29 isolates appeared to belong to a single clone on the basis of PFGE, suggesting either that the ica genes have undergone changes at the DNA level resulting in a lack of primer annealing or that genes have been gained or lost in about half of the clone. The findings that PCR positive S. capitis isolates were positive for biofilm production by the CRA test but that ica negative strains did not produce biofilm suggest that the latter explanation is more likely.
In conclusion, this study has demonstrated an association between quantitative biofilm production and CoNS associated with disease but a lack of an association for the ica operon. These results suggest that regulation of biofilm expression plays a central role in the disease process and represents a potentially important area for further research.
Acknowledgments
Funding was provided by SPARKS (Sport Aiding Medical Research for Kids), the Oxford Sick Newborn Education and Research Trust, and the Oxfordshire Health Services Research Fund. G.D.I.D.S. was funded by a Commonwealth Scholarship, and N.P.J.D. was a Wellcome Trust career development fellow during the study period.
We thank the medical and nursing staff of the NICU, John Radcliffe Hospital. We thank J. Buttery and M. Herbert for assistance in assigning the clinical significance of positive blood cultures. Grateful thanks go to Wilma Ziebuhr for bacterial strains and technical advice.
REFERENCES
- 1.Arciola, C. R., L. Baldassarri, and L. Montanaro. 2001. Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J. Clin. Microbiol. 39:2151-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayston, R., and S. R. Penny. 1972. Excessive production of mucoid substance in staphylococcus SIIA: a possible factor in colonisation of Holter shunts. Dev. Med. Child. Neurol. Suppl. 27:25-28. [DOI] [PubMed] [Google Scholar]
- 3.Burnie, J. P., M. Naderi-Nasab, K. W. Loudon, and R. C. Matthews. 1997. An epidemiological study of blood culture isolates of coagulase-negative staphylococci demonstrating hospital-acquired infection. J. Clin. Microbiol. 35:1746-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen, G. D., L. M. Baddour, B. M. Madison, J. T. Parisi, S. N. Abraham, D. L. Hasty, J. H. Lowrance, J. A. Josephs, and W. A. Simpson. 1990. Colonial morphology of staphylococci on Memphis agar: phase variation of slime production, resistance to beta-lactam antibiotics, and virulence. J. Infect. Dis. 161:1153-1169. [DOI] [PubMed] [Google Scholar]
- 5.Christensen, G. D., J. T. Parisi, A. L. Bisno, W. A. Simpson, and E. H. Beachey. 1983. Characterization of clinically significant strains of coagulase-negative staphylococci. J. Clin. Microbiol. 18:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen, G. D., W. A. Simpson, A. L. Bisno, and E. H. Beachey. 1982. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect. Immun. 37:318-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen, G. D., W. A. Simpson, J. J. Younger, L. M. Baddour, F. F. Barrett, D. M. Melton, and E. H. Beachey. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Gotz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Angio, C. T., K. L. McGowan, S. Baumgart, J. St Geme, and M. C. Harris. 1989. Surface colonization with coagulase-negative staphylococci in premature neonates. J. Pediatr. 114:1029-1034. [DOI] [PubMed] [Google Scholar]
- 10.Davenport, D. S., R. M. Massanari, M. A. Pfaller, M. J. Bale, S. A. Streed, and W. J. Hierholzer, Jr. 1986. Usefulness of a test for slime production as a marker for clinically significant infections with coagulase-negative staphylococci. J. Infect. Dis. 153:332-339. [DOI] [PubMed] [Google Scholar]
- 11.Deighton, M. A., R. Borland, and J. A. Capstick. 1996. Virulence of Staphylococcus epidermidis in a mouse model: significance of extracellular slime. Epidemiol. Infect. 117:267-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deighton, M. A., J. C. Franklin, W. J. Spicer, and B. Balkau. 1988. Species identification, antibiotic sensitivity and slime production of coagulase-negative staphylococci isolated from clinical specimens. Epidemiol. Infect. 101:99-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Silva, G. D. I., A. Justice, A. R. Wilkinson, J. Buttery, M. Herbert, N. P. J. Day, and S. J. Peacock. 2001. Genetic population structure of coagulase-negative staphylococci associated with carriage and disease in preterm infants. Clin. Infect. Dis. 33:1520-1528. [DOI] [PubMed]
- 14.Dunne, W. M., Jr., D. B. Nelson, and M. J. Chusid. 1987. Epidemiologic markers of pediatric infections caused by coagulase-negative staphylococci. Pediatr. Infect. Dis. J. 6:1031-1035. [PubMed] [Google Scholar]
- 15.Dyke, K. G., S. Aubert, and N. el Solh. 1992. Multiple copies of IS256 in staphylococci. Plasmid 28:235-246. [DOI] [PubMed] [Google Scholar]
- 16.Frebourg, N. B., S. Lefebvre, S. Baert, and J. F. Lemeland. 2000. PCR-based assay for discrimination between invasive and contaminating Staphylococcus epidermidis strains. J. Clin. Microbiol. 38:877-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freeman, D. J., F. R. Falkiner, and C. T. Keane. 1989. New method for detecting slime production by coagulase-negative staphylococci. J. Clin. Pathol. 42:872-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freeman, J., M. F. Epstein, N. E. Smith, R. Platt, D. G. Sidebottom, and D. A. Goldmann. 1990. Extra hospital stay and antibiotic usage with nosocomial coagulase-negative staphylococcal bacteremia in two neonatal intensive care unit populations. Am. J. Dis. Child. 144:324-329. [DOI] [PubMed] [Google Scholar]
- 19.Galdbart, J. O., J. Allignet, H. S. Tung, C. Ryden, and N. El Solh. 2000. Screening for Staphylococcus epidermidis markers discriminating between skin-flora strains and those responsible for infections of joint prostheses. J. Infect. Dis. 182:351-355. [DOI] [PubMed] [Google Scholar]
- 20.Gosden, P. E., J. M. Andrews, K. E. Bowker, H. A. Holt, A. P. MacGowan, D. S. Reeves, J. Sunderland, and R. Wise. 1998. Comparison of the modified Stokes' method of susceptibility testing with results obtained using MIC methods and British Society of Antimicrobial Chemotherapy breakpoints. J. Antimicrob. Chemother. 42:161-169. [DOI] [PubMed] [Google Scholar]
- 21.Gray, J. E., D. K. Richardson, M. C. McCormick, and D. A. Goldmann. 1995. Coagulase-negative staphylococcal bacteremia among very low birth weight infants: relation to admission illness severity, resource use, and outcome. Pediatrics 95:225-230. [PubMed] [Google Scholar]
- 22.Hall, R. T., S. L. Hall, W. G. Barnes, J. Izuegbu, M. Rogolsky, and I. Zorbas. 1987. Characteristics of coagulase-negative staphylococci from infants with bacteremia. Pediatr. Infect. Dis. J. 6:377-383. [DOI] [PubMed] [Google Scholar]
- 23.Hall, S. L., R. T. Hall, W. G. Barnes, and S. Riddell. 1988. Colonization with slime-positive coagulase-negative staphylococci as a risk factor for invasive coagulase-negative staphylococci infections in neonates. J. Perinatol. 8:215-221. [PubMed] [Google Scholar]
- 24.Hall, S. L., S. W. Riddell, W. G. Barnes, L. Meng, and R. T. Hall. 1990. Evaluation of coagulase-negative staphylococcal isolates from serial nasopharyngeal cultures of premature infants. Diagn. Microbiol. Infect. Dis. 13:17-23. [DOI] [PubMed] [Google Scholar]
- 25.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 26.Huebner, J., G. B. Pier, J. N. Maslow, E. Muller, H. Shiro, M. Parent, A. Kropec, R. D. Arbeit, and D. A. Goldmann. 1994. Endemic nosocomial transmission of Staphylococcus epidermidis bacteremia isolates in a neonatal intensive care unit over 10 years. J. Infect. Dis. 169:526-531. [DOI] [PubMed] [Google Scholar]
- 27.Isaacs, D., C. Barfield, T. Clothier, B. Darlow, R. Diplock, J. Ehrlich, K. Grimwood, I. Humphrey, H. Jeffery, R. Kohan, R. McNeil, A. McPhee, C. Minutillo, F. Morey, D. Tudehope, and M. Wong. 1996. Late-onset infections of infants in neonatal units. J. Paediatr. Child Health 32:158-161. [DOI] [PubMed] [Google Scholar]
- 28.Ishak, M. A., D. H. Groschel, G. L. Mandell, and R. P. Wenzel. 1985. Association of slime with pathogenicity of coagulase-negative staphylococci causing nosocomial septicemia. J. Clin. Microbiol. 22:1025-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones, J. W., R. J. Scott, J. Morgan, and J. V. Pether. 1992. A study of coagulase-negative staphylococci with reference to slime production, adherence, antibiotic resistance patterns and clinical significance. J. Hosp. Infect. 22:217-227. [DOI] [PubMed] [Google Scholar]
- 30.Knobloch, J. K., K. Bartscht, A. Sabottke, H. Rohde, H. H. Feucht, and D. Mack. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183:2624-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kotilainen, P. 1990. Association of coagulase-negative staphylococcal slime production and adherence with the development and outcome of adult septicemias. J. Clin. Microbiol. 28:2779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotilainen, P., J. Nikoskelainen, and P. Huovinen. 1991. Antibiotic susceptibility of coagulase-negative staphylococcal blood isolates with special reference to adherent, slime-producing Staphylococcus epidermidis strains. Scand. J. Infect. Dis. 23:325-332. [DOI] [PubMed] [Google Scholar]
- 33.Mack, D. 1999. Molecular mechanisms of Staphylococcus epidermidis biofilm formation. J. Hosp. Infect. 43(Suppl.):S113-S25. [DOI] [PubMed] [Google Scholar]
- 34.Mack, D., H. Rohde, S. Dobinsky, J. Riedewald, M. Nedelmann, J. K. Knobloch, H. A. Elsner, and H. H. Feucht. 2000. Identification of three essential regulatory gene loci governing expression of Staphylococcus epidermidis polysaccharide intercellular adhesin and biofilm formation. Infect. Immun. 68:3799-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mehta, G., S. Singh, and S. Kumari. 1991. Observations on coagulase-negative staphylococci in a neonatal unit in India. J. Hosp. Infect. 19:273-281. [DOI] [PubMed] [Google Scholar]
- 36.Nouwen, J. L., A. van Belkum, S. de Marie, J. Sluijs, J. J. Wielenga, J. A. Kluytmans, and H. A. Verbrugh. 1998. Clonal expansion of Staphylococcus epidermidis strains causing Hickman catheter-related infections in a hemato-oncologic department. J. Clin. Microbiol. 36:2696-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oto, S., P. Aydin, N. Ciftcioglu, and D. Dursun. 1998. Slime production by coagulase-negative staphylococci isolated in chronic blepharitis. Eur. J. Ophthalmol. 8:1-3. [DOI] [PubMed] [Google Scholar]
- 38.Ponce de Leon, S., S. H. Guenthner, and R. P. Wenzel. 1986. Microbiologic studies of coagulase-negative staphylococci isolated from patients with nosocomial bacteraemias. J. Hosp. Infect. 7:121-129. [DOI] [PubMed] [Google Scholar]
- 39.Ronnestad, A., T. G. Abrahamsen, P. Gaustad, and P. H. Finne. 1998. Blood culture isolates during 6 years in a tertiary neonatal intensive care unit. Scand. J. Infect. Dis. 30:245-251. [DOI] [PubMed] [Google Scholar]
- 40.Rupp, M. E., J. S. Ulphani, P. D. Fey, K. Bartscht, and D. Mack. 1999. Characterization of the importance of polysaccharide intercellular adhesin-hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect. Immun. 67:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rupp, M. E., J. S. Ulphani, P. D. Fey, and D. Mack. 1999. Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin-hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect. Immun. 67:2656-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanghvi, K. P., and D. I. Tudehope. 1996. Neonatal bacterial sepsis in a neonatal intensive care unit: a 5 year analysis. J. Paediatr. Child Health 32:333-338. [DOI] [PubMed] [Google Scholar]
- 43.Schumacher-Perdreau, F., B. Jansen, G. Peters, and G. Pulverer. 1988. Typing of coagulase-negative staphylococci isolated from foreign body infections. Eur. J. Clin. Microbiol. Infect. Dis. 7:270-273. [DOI] [PubMed] [Google Scholar]
- 44.Stoll, B. J., T. Gordon, S. B. Korones, S. Shankaran, J. E. Tyson, C. R. Bauer, A. A. Fanaroff, J. A. Lemons, E. F. Donovan, W. Oh, D. K. Stevenson, R. A. Ehrenkranz, L. A. Papile, J. Verter, and L. L. Wright. 1996. Late-onset sepsis in very low birth weight neonates: a report from the National Institute of Child Health and Human Development Neonatal Research Network. J. Pediatr. 129:63-71. [DOI] [PubMed] [Google Scholar]
- 45.Vermont, C. L., N. G. Hartwig, A. Fleer, P. de Man, H. Verbrugh, J. van den Anker, R. de Groot, and A. van Belkum. 1998. Persistence of clones of coagulase-negative staphylococci among premature neonates in neonatal intensive care units: two-center study of bacterial genotyping and patient risk factors. J. Clin. Microbiol. 36:2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villari, P., C. Sarnataro, and L. Iacuzio. 2000. Molecular epidemiology of Staphylococcus epidermidis in a neonatal intensive care unit over a 3-year period. J. Clin. Microbiol. 38:1740-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Eiff, C., C. Heilmann, and G. Peters. 1999. New aspects in the molecular basis of polymer-associated infections due to staphylococci. Eur. J. Clin. Microbiol. Infect. Dis. 18:843-846. [DOI] [PubMed] [Google Scholar]
- 48.Ziebuhr, W., C. Heilmann, F. Gotz, P. Meyer, K. Wilms, E. Straube, and J. Hacker. 1997. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect. Immun. 65:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ziebuhr, W., V. Krimmer, S. Rachid, I. Lossner, F. Gotz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345-356. [DOI] [PubMed] [Google Scholar]