Abstract
16S ribosomal DNA (rDNA) and 16S-23S internal transcribed spacer rDNA sequence analyses were performed on Mycobacterium farcinogenes and M. senegalense strains and 26 strains of other rapidly growing mycobacteria to investigate the phylogenetic structure of bovine farcy mycobacteria within the M. fortuitum complex. M. farcinogenes and M. senegalense were indistinguishable in their 5"-end 16S rDNA but showed both considerable interspecies spacer sequence divergence and a high level of intraspecies sequence stability. A rapid detection assay using PCR and hybridization with species-specific probes was developed. The assay was specific among 46 species other than M. farcinogenes and M. senegalense and correctly identified all M. farcinogenes and M. senegalense strains. PCR- and 16S-23S rDNA sequence-based detection will be a valuable approach for diagnosis of the causal agents of African bovine farcy in cattle.
The chronic infectious disease of zebu cattle known as bovine farcy is endemic to East and Central Africa, notably, Chad, Nigeria, Senegal, Somalia, and Sudan (12, 14, 18). The disease, which causes great economic loss, was originally thought to be caused by Nocardia farcinica, but it is now clear that Mycobacterium farcinogenes and M. senegalense are the principal, if not sole, causal agents (3, 4, 6, 14). These closely related species form a distinct 16S ribosomal DNA (rDNA) clade, together with M. fortuitum and M. peregrinum, within the evolutionary radiation encompassed by fast-growing mycobacteria (9). Members of all four species have a mycolic acid pattern in common (22); M. farcinogenes and M. senegalense strains, together with the sorbitol-positive third biovariant of M. fortuitum, have identical 16S rDNA sequences (17, 20). DNA relatedness, chemotaxonomic, numerical taxonomic, and serotaxonomic data underpin the similarity of bovine farcy strains to M. fortuitum but also confirm the distinction between M. farcinogenes and M. senegalense (1, 2, 11, 14, 15, 21, 22). Despite the improvements in the classification of these taxa, better diagnostic methods are needed to distinguish between M. farcinogenes and M. senegalense. Strains isolated from animals with bovine farcy can be presumptively identified by using a small number of phenotypic properties based on biochemical and growth tests (14, 21). There are instances in which the sequences of 16S rDNA genes have been found to be very similar, if not identical, between different species in a genus, making it necessary to find alternative specific sequences. The intergenic 16S-23S internal transcribed spacer (ITS) region is considered to be less prone to selective pressure and consequently can be expected to have accumulated a higher percentage of mutations than the corresponding rDNA (5, 7, 10, 24). Sequencing of the ITS regions of diverse bacteria indicates that considerable length and primary sequence variation occurs, and this variability has been successfully used to distinguish between closely related mycobacteria, such as those assigned to the M. avium-M. intracellulare complex (5, 7) or to M. gastri and M. kansasii (24); representatives of the latter have identical 16S rDNA sequences (23). These initial investigations have recently been extended to the development of a method for the identification of a broad panel of mycobacterial species, including the closely related, rapidly growing species discussed above, by using either restriction enzymes or species-specific primers (19, 25). The aim of the present study was to sequence and examine the ITS regions of representative M. farcinogenes and M. senegalense strains in order to determine its potential for differentiating between members of these and related taxa, such as the M. fortuitum complex, and to evaluate probes for the rapid identification of the causal agents of bovine farcy.
The sources, nucleotide sequence accession numbers, and spacer sequevar assignments of the representatives of the 12 rapidly growing mycobacterial species investigated in this study are listed in Table 1. In addition to these 52 strains that were used for sequencing, a further 174 mycobacterial strains were examined to test for the specificity of the probes described below: (i) 3 additional M. farcinogenes strains (M15, M16, and M39); (ii) the type strains of 34 species, namely, M. asiaticum, M. aurum, M. avium, M. bohemicum, M. celatum, M. chlorophenolicum, M. conspicuum, M. duvalii, M. gastri, M. gordonae, M. heckeshornense, M. hassiacum, M. hodleri, M. interjectum, M. intermedium, M. intracellulare, M. kansasii, M. lentiflavum, M. malmoense, M. marinum, M. mucogenicum, M. neoaurum, M. nonchromogenicum, M. obuense, M. parafortuitum, M. rhodesiae, M. scrofulaceum, M. shimoidei, M. simiae, M. szulgai, M. terrae, M. triviale, M. ulcerans, and M. xenopi; and (iii) strains randomly selected from the strain collection used in a previous study (25), i.e., 30 M. avium, 4 M. celatum, 2 M. flavescens, 5 M. fortuitum, 4 M. gastri, 4 M. genavense, 8 M. gordonae, 1 M. haemophilum, 22 M. intracellulare, 2 M. lentiflavum, 2 M. malmoense, 4 M. marinum, 4 M. simiae, 3 M. szulgai, 1 M. triplex, 26 M. tuberculosis, and 15 M. xenopi strains. Details of the strain histories and sources of the 21 representative M. farcinogenes and 8 representative M. senegalense strains (Table 1) have been given elsewhere (2, 14, 22). The position of two bovine farcy isolates received as M. farcinogenes (strains M280 and M555) was considered equivocal in the light of data from earlier studies (2, 14).
TABLE 1.
Strains and sequences used for analysis of 16S-23S spacer sequences
| Species | Strain(s); referencea | 16S-23S spacer
|
EMBL accession no. | |
|---|---|---|---|---|
| Sequevarb | Size (bp) | |||
| M. abscessus | DSM 44196T, S321, S323 | Mab-A | 297 | AJ291580 |
| S322 | Mab-B | 297 | AJ291581 | |
| Unknown; 19 | Mab-C | 297 | AF163815 | |
| M. chelonae | DSM 43804T, K52 | Mch-A | 294 | AJ291582 |
| DSM 43484, DSM 43488 | Mch-B | 294 | AJ291583 | |
| DSM 43276 | Mch-C | 296 | AJ291584 | |
| M. diernhoferi | Unknown; 19 | 396 | AF186463 | |
| M. farcinogenes | M262T (DSM 43637T), M9, M57, M191, M217, M269, M274, M275, M276, M281, M285, M687, N710, N725, N785 | Mfa-A | 381 | Y10384 |
| M52 | Mfa-B | 381 | AJ291585 | |
| M280, M555 | Mse | See below | ||
| M. flavescens | DSM 43991T | 359 | AJ291586 | |
| M. fortuitum | DSM 46621T | Mfo-A | 342 | AJ291587 |
| DSM 44220T | Mfo-B | 363 | AJ291588 | |
| M368 | Mfo-C | 342 | AJ291589 | |
| S336 | Mfo-E | 364 | AJ291591 | |
| S312 | Mfo-F | 380 | AJ291592 | |
| S358 | Mfo-G | 362 | AJ291593 | |
| M. peregrinum | DSM 43271T | Mpe-A | 359 | AJ291594 |
| S254 | Mpe-B | 360 | AJ291595 | |
| M. phlei | DSM 43239T, S270 | Mph-A | 363 | AJ291596 |
| S271 | Mph-B | 368 | AJ291597 | |
| R82; 27 | Mph-C | 363 | X74493 | |
| M. porcinum | DSM 44242T | 380 | AJ291598 | |
| M. senegalense | M263T, (DSM 43656T), M266, M283, M728, N714, N717, N718, N721 | Mse | 354 | Y10385 |
| M. smegmatis | DSM 43756T | Msm-A | 365 | AJ291599 |
| ATCC 607 | Msm-B | 376 | U07955 | |
| M. tuberculosis | ATCC 27294T | 275 | L15623 | |
| M. vaccae | DSM 43292T, DSM 43229, S345 | 409 | AJ291600 | |
ATCC, American Type Culture Collection, Manassas, Va; DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany; M and N numbers, strain collection of the Department of Agricultural and Environmental Science, University of Newcastle, Newcastle, United Kingdom; S and K numbers, strain collection of the Institut für Mikrobiologie, Lungenklinik Heckeshorn, Berlin, Germany; T, type strain.
Bacterial lysates were obtained and used for PCR amplification of three different ribosomal regions as described elsewhere (24, 25). i.e., (i) the 5" region of 16S rDNA (24), (ii) the complete ITS (7, 24), and (iii) part of the ITS, with genus-specific primers Sp1 and Sp2 (25). Both strands of the amplicons obtained from the first two protocols were sequenced as described earlier (24). ITS amplicons were cloned by using standard procedures provided by a commercial company (Genexpress, Berlin, Germany) for strains that showed interoperon spacer sequence variabilities after a first direct sequencing approach (M. fortuitum strains S336, S312, and S358 and M. peregrinum DSM 43271T). Thereafter, one clone for each strain was sequenced. The ITS sequences were aligned by computer and corrected manually by using DNASIS software (version 2.5; Hitachi Software Engineering Co., Ltd., San Bruno, Calif.). Distance estimation and tree topology determination were done by using the neighbor-joining algorithm (26), which was applied to distances in accordance with Kimura's two-parameter model (16) with the aid of the TREECON for Windows software (version 3.1b; University of Antwerp, Antwerp, Belgium). Digoxigenin-11-UTP, which was incorporated into amplicons during PCR with primers Sp1 and Sp2, was detected with the PCR-enzyme-linked immunosorbent assay system (DIG-Labeling and DIG-Detection kits; Roche Molecular Biochemicals, Mannheim, Germany). The success of amplification with these genus-specific primers was verified by ethidium bromide-stained agarose gel electrophoresis prior to the detection step. For hybridization, two probes designed on the basis of all of the available spacer sequences were evaluated for specificity, namely, biotin-3"-TCAGCCAGCATCTGTAG and biotin-3"-AGGAGTCTGTGCGCTGT, as probes for the detection of M. farcinogenes or M. senegalense, respectively. Hybridization was performed at 52°C by using 1 μl of the PCR products and following the recommendations of the manufacturer.
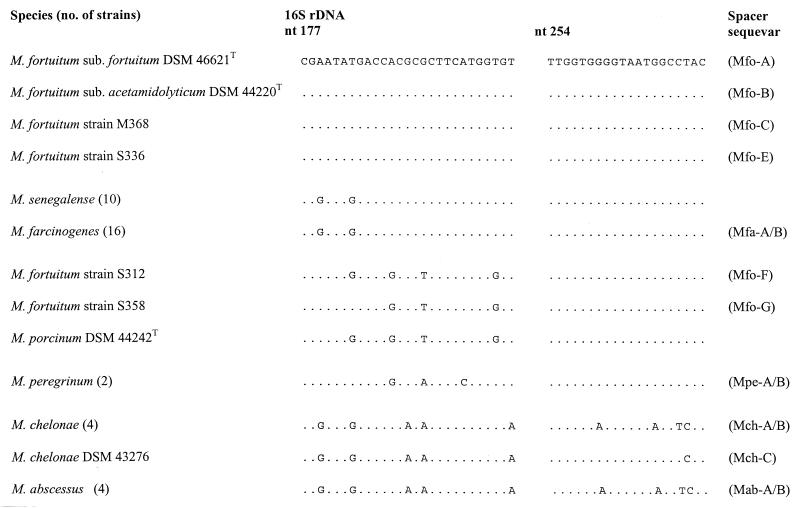
The results presented in Fig. 1 show that M. farcinogenes and M. senegalense have identical partial 16S rDNA sequences that distinguish them from representatives of closely related taxa, with the exception of the sorbitol-positive third biovariant of M. fortuitum (strain ATCC 49403); this variant was not included in the present study (the 16S rDNA sequence is shown in reference 17, and its ITS sequence, which differs from that of M. farcinogenes and M. senegalense, has been recently submitted to the RIDOM database [http://www.ridom.de]). In contrast, the interspecies ITS sequence variability of strains of these two species was considerable (sequence similarity of less than 75%), a finding that was already evident from the significant difference in spacer size (Table 1) and which is in good agreement with previous PCR-restriction fragment length polymorphism data (25). The complete ITS sequences of 15 M. farcinogenes strains were identical (sequevar Mfa-A, Table 1), although one organism, strain M52, showed a minor intraspecies sequence polymorphism (one base substitution [G224T] at position 95; sequevar Mfa-B). No base differences were found in the ITS sequences of the eight M. senegalense isolates. Hence, these data suggest that ITS sequences can be used as a genetic target with which to differentiate between members of the causal agents of bovine farcy. It was interesting that strains M280 and M555, which had been received as M. farcinogenes, showed ITS sequences characteristic of M. senegalense. Strain M280, which was isolated from a case of bovine farcy in Chad, is known to contain glycolipids that characterize it as a strain of M. senegalense (2, 13). It is clear from both the present and earlier studies that strain M280 is a bona fide M. senegalense strain. It is also apparent from the present study that strain M255, an isolate from a case of bovine farcy in Somalia, was misclassified as M. farcinogenes.
FIG. 1.
Comparison of 16S rDNA signature sequences with the respective spacer sequevars. The alignment comprises the two variable regions found in the 5" 16S rRNA genes of selected members of species closely related to M. farcinogenes and M. senegalense. Dots indicate identity, and the corresponding positions of the Escherichia coli 16S rDNA are also shown. nt, nucleotide.
The high ITS sequence divergence previously found among representatives of rapidly growing mycobacteria (25) was confirmed and extended in the present study. It is particularly interesting that the representatives of M. abscessus and M. chelonae, which are difficult to distinguish in routine clinical practice because of their phenotypic similarity and which have the same 16S rDNA sequence within the variable 5" region normally used for identification (Fig. 1), can be distinguished on the basis of their ITS sequences.
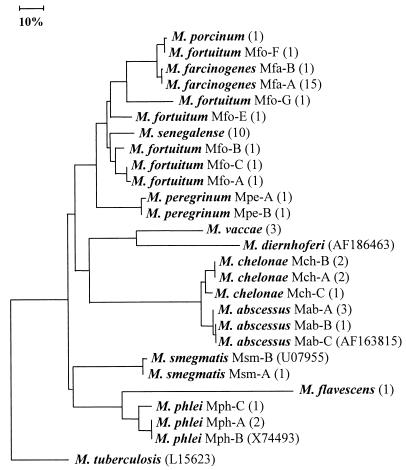
The relationships found between all of the test strains based on their ITS sequences are shown in Fig. 2. The high degree of ITS variability is reflected by the great length of some of the branches, which suggests that the rate of base changes in the ITS may not allow comparison of distantly positioned groups. Strains known to show significant interoperon heterogeneity, such as M. peregrinum, were not sequenced in all of the operons in this study. Hence, the impact of interoperon variability on tree topology remains unknown. Despite these limitations, the association of M. flavescens, M. phlei, and M. smegmatis; M. abscessus and M. chelonae; and M. peregrinum, M. fortuitum, M. farcinogenes, and M. senegalense is in good agreement with the results of 16S rDNA sequencing studies (10, 20). M. fortuitum strains can be assigned to three groups based on three 16S rDNA signature sequences (Fig. 1). Thus, the type strains of the two subspecies of M. fortuitum have a sequence in common with one another and with strains S336 and M368, strain S312 has the sequence described for the sorbitol-negative third biovariant M. fortuitum reference strain ATCC 49404 (17), and strain S358 has a novel sequence that is characterized by one base substitution compared to the sequence of strain S312. The latter two 16S genotypes show the greatest similarity to the sequence of M. porcinum. Strains DSM 46621T, DSM 44220 T, and M368 from the first genotype formed a tight ITS cluster (Mfo-A to -C), while the members of the two other 16S genotypes showed distinct spacer sequences (Mfo-F and Mfo-G, respectively), which showed a high level of similarity to the spacer sequence of M. porcinum. The remaining organism, strain S336, showed a spacer sequence that occupied an intermediate position between the other two ITS subclusters. These data are also in good agreement with those of previous studies (8, 24) that show that ITS sequences give greater resolution than corresponding 16S rDNA data. The two probes recognized all of the representatives of the respective target species, that is, either M. farcinogenes or M. senegalense, but gave negative results for the representatives of the remaining mycobacterial species (Table 1 and the strains listed in Materials and Methods). These results are very encouraging, as they show that the rapid PCR protocol combined with the specific hybridization is suitable for distinguishing between M. farcinogenes and M. senegalense. This opens up the possibility of detecting the causal agents of bovine farcy directly in clinical material and determining the primary environmental reservoir of these organisms.
FIG. 2.
Neighbor-joining tree based on sequences derived from 16S-23S rDNA ITS regions showing relationships between M. farcinogenes and M. senegalense and between organisms and representatives of other rapidly growing mycobacterial species. The horizontal lengths represent genetic distances, and the scale bar represents 10% sequence divergence. The values in parentheses are the numbers of strains analyzed (for details, see Table 1). The tree was rooted by using M. tuberculosis as the outgroup.
Nucleotide sequence accession numbers. The sequences determined in this study have been deposited in the EMBL database under accession numbers Y10384, Y10385, and AJ291580 to AJ291600. The partial 16S rDNA sequences of the M. farcinogenes and M. senegalense strains were deposited under accession numbers Y11581 and Y11582, respectively.
Acknowledgments
We are indebted to Sabine Michalke for technical assistance.
M. E. Hamid was supported by a fellowship from the Alexander Humboldt Foundation and by a Wellcome Trust Research Development Award in Tropical Medicine (grant 043935/Z/95/A).
REFERENCES
- 1.Baess, I. 1982. Deoxyribonucleic acid relatedness among species of rapidly growing mycobacteria. Acta Pathol. Microbiol. Immunol. Scand. Sect. B 90:371-375. [DOI] [PubMed] [Google Scholar]
- 2.Besra, G. S., S. S. Gurcha, K.-H. Khoo, H. R. Morris, A. Dell, M. E. Hamid, D. E. Minnikin, M. Goodfellow, and P. J. Brennan. 1994. Characterization of the specific antigenicity of representatives of M. senegalense and related bacteria. Zentbl. Bakteriol. 281:415-432. [DOI] [PubMed] [Google Scholar]
- 3.Chamoiseau, G. 1973. Mycobacterium farcinogenes agent causal du farcin du boeuf en Afrique. Ann. Microbiol. 124A:215-222. [PubMed]
- 4.Chamoiseau, G. 1979. Etiology of farcy in African bovines: nomenclature of the causal organisms Mycobacterium farcinogenes Chamoiseau and Mycobacterium senegalense (Chamoiseau) comb. nov. Int. J. Syst. Bacteriol. 29:407-410. [Google Scholar]
- 5.De Smet, A. L., I. N. Brown, M. Yates, and J. Ivanyi. 1995. Ribosomal internal transcribed spacers are identical among Mycobacterium avium-intracellulare complex isolates from AIDS patients, but vary among isolates from elderly pulmonary disease patients. Microbiology 141:2739-2747. [DOI] [PubMed] [Google Scholar]
- 6.El-Sanousi, S. M., and M. A. M. Salih. 1979. Miliary bovine farcy experimentally induced in a zebu calf. Vet. Pathol. 16:372-373. [DOI] [PubMed] [Google Scholar]
- 7.Frothingham, R., and K. H. Wilson. 1993. Sequence-based differentiation of strains in the Mycobacterium avium complex. J. Bacteriol. 175:2818-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frothingham, R., and K. H. Wilson. 1994. Molecular phylogeny of the Mycobacterium avium complex demonstrates clinically meaningful divisions. J. Infect. Dis. 169:305-312. [DOI] [PubMed] [Google Scholar]
- 9.Goodfellow, M., and J. G. Magee. 1997. Taxonomy of mycobacteria, p. 1-71. In P. R. J. Gangadharam and P. A. Jenkins (ed.), Mycobacteria. I. Basic aspects. Chapman & Hall, New York, N.Y.
- 10.Gürtler, V., and V. A. Stanisch. 1996. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology 142:3-16. [DOI] [PubMed] [Google Scholar]
- 11.Hall, R. M., and C. Ratledge. 1985. Equivalence of mycobactins from Mycobacterium senegalense, Mycobacterium farcinogenes and Mycobacterium fortuitum. J. Gen. Microbiol. 131:1691-1696. [DOI] [PubMed] [Google Scholar]
- 12.Hamid, M. E. 1994. Classification and identification of actinomycetes associated with bovine farcy. Ph.D. thesis. University of Newcastle, Newcastle upon Tyne, United Kingdom.
- 13.Hamid, M. E., D. E. Minnikin, M. Goodfellow, and M. Ridell. 1993. Thin-layer chromatographic analysis of glycolipids and mycolic acids from Mycobacterium farcinogenes and Mycobacterium senegalense and related taxa. Zentbl. Bakteriol. 279:354-367. [DOI] [PubMed] [Google Scholar]
- 14.Hamid, M. E., G. E. Mohamed, M. T. Abu-Samra, S. M. El-Sanousi, and M. E. Barri. 1991. Bovine farcy: a clinico-pathological study of the disease and its aetiological agent. J. Comp. Pathol. 105:287-301. [DOI] [PubMed] [Google Scholar]
- 15.Hamid, M. E., M. Ridell, D. E. Minnikin, and M. Goodfellow. 1998. Serotaxonomic analysis of glycolipids from the Mycobacterium chelonae-Mycobacterium fortuitum complex and bovine farcy strains. Zentbl. Bakteriol. 288:23-34. [DOI] [PubMed] [Google Scholar]
- 16.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 17.Kirschner, P., M. Kiekenbeck, D. Meissner, J. Wolters, and E. C. Böttger. 1992. Genetic heterogeneity within Mycobacterium fortuitum complex species: genotypic criteria for identification. J. Clin. Microbiol. 30:2772-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mostafa, E. I. 1966. Bovine nocardiosis (cattle farcy): a review. Vet. Bull. 36:189-193. [Google Scholar]
- 19.Park, H., H. Jang, C. Kim, B. Chung, C. L. Chang, S. K. Park, and S. Song. 2000. Detection and identification of mycobacteria by amplification of the internal transcribed spacer regions with genus- and species-specific PCR primers. J. Clin. Microbiol. 38:4080-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pitulle, C., M. Dorsche, J. Kazda, J. Wolters, and E. Stackebrandt. 1992. Phylogeny of rapidly growing members of the genus Mycobacterium. Int. J. Syst. Bacteriol. 42:337-343. [DOI] [PubMed] [Google Scholar]
- 21.Ridell, M., and M. Goodfellow. 1983. Numerical classification of Mycobacterium farcinogenes, Mycobacterium senegalense and related taxa. J. Gen. Microbiol. 129:599-611. [DOI] [PubMed] [Google Scholar]
- 22.Ridell, M., M. Goodfellow, D. E. Minnikin, S. M. Minnikin, and I. G. Hutchinson. 1982. Classification of Mycobacterium farcinogenes and Mycobacterium senegalense by immunodiffusion and thin-layer chromatography of long-chain components. J. Gen. Microbiol. 128:1299-1307. [DOI] [PubMed] [Google Scholar]
- 23.Rogall, T., J. Wolters, T. Floher, and E. C. Böttger. 1990. Towards a phylogeny and definition of the species at the molecular level within the genus Mycobacterium. Int. J. Syst. Bacteriol. 40:323-330. [DOI] [PubMed] [Google Scholar]
- 24.Roth, A., M. Fischer, H. E. Hamid, W. Ludwig, S. Michalke, and H. Mauch. 1998. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J. Clin. Microbiol. 36:139-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roth, A., U. Reischl, A. Streubel, L. Naumann, R. M. Kroppenstedt, M. Habicht, M. Fischer, and H. Mauch. 2000. Novel diagnostic algorithm for identification of mycobacteria using genus-specific amplification of the 16S-23S rRNA gene spacer and restriction endonucleases. J. Clin. Microbiol. 38:1094-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 27.Van der Giessen, J. W. B., R. M. Haring, and, B. A. M. van der Zeijst. 1994. Comparison of the 23S ribosomal RNA and the spacer region between the 16S and 23S rRNA genes of the closely related Mycobacterium avium and Mycobacterium paratuberculosis and the fast-growing Mycobacterium phlei. Microbiology 140:1103-1108. [DOI] [PubMed] [Google Scholar]