Abstract
A sensitive enzyme immunoassay (EIA) specific for hepatitis B virus core antigen (HBcAg) and hepatitis B e antigen (HBeAg) was developed. We designated the precore/core gene products as hepatitis B virus (HBV) core-related antigens (HBcrAg). In order to detect HBcrAg even in anti-HBc/e antibody-positive specimens, the specimens were pretreated in detergents. The antibodies are inactivated by this pretreatment and, simultaneously, the antigens are released and the epitopes are exposed. The assay demonstrated 71 to 112% recovery using HBcrAg-positive sera. We observed no interference from the tested anticoagulants or blood components. When the cutoff value was tentatively set at 103 U/ml, all healthy control (HBsAg/HBV-DNA negative; n = 108) and anti-HCV antibody-positive (n = 59) sera were identified as negative. The assay showed a detection limit of 4 × 102 U/ml using recombinant antigen. Detection limits were compared in four serially diluted HBV high-titer sera. The HBcrAg assay demonstrated higher sensitivity than HBV-DNA transcription-mediated amplification (TMA) or HBeAg radio immunoassay (RIA) in the dilution test. HBcrAg concentrations correlated well with HBV-DNA TMA (r = 0.91, n = 29) and in-house real-time detection-PCR (r = 0.93, n = 47) in hepatitis B patients. On HBeAg/anti-HBe antibody seroconversion panels, the HBcrAg concentration changed in accordance with HBV-DNA levels. HBcrAg concentration provides a reflection of HBV virus load equivalent to HBV-DNA level, and the assay therefore offers a simple method for monitoring hepatitis B patients.
Many hepatitis B virus (HBV) markers are used for diagnosing and monitoring hepatitis B patients. HBV-DNA tests, such as the branched-chain DNA (b-DNA) signal amplification assay (7, 31), and transcription-mediated amplification (TMA)-based (11) or PCR-based (12, 14, 20) assays are used to diagnose and monitor the efficacy of treatment. However, these methods require cumbersome procedures and expensive equipment, thus requiring considerable skill and high costs. These gene amplification assays also present some limitations (22, 23, 35). The b-DNA assay provides quantitative results but requires a long incubation time and lacks adequate sensitivity. Amplification assays have adequate sensitivity but are less quantitative.
Immunoassays are generally easy and inexpensive. There have been a few reports of serum HBcAg assays with specimen pretreatment (4, 32). The concentration of HBcAg in these assays correlated with levels of HBV-associated DNA polymerase (4). Thus, HBcAg could be a marker for virus load. However, the use of these assays is limited because of relatively low sensitivity and complex procedures.
Serum HBeAg concentration reflects virus replication and hepatitis activity and is closely correlated with virus load in anti-HBe antibody-negative patients (8). Seroconversion of HBeAg to anti-HBe antibody reveals the inactive phase of infection (17, 25). However, after seroconversion, many patients may exhibit reactivation and high viral load (3, 10, 18). In these cases, HBeAg is usually negative due to masking by anti-HBe antibody (24), although the HBeAg/anti-HBe immune complex can be indirectly detected according to the levels of alanine aminotransferase (ALT) and HBV-DNA (6). Therefore, HBcAg and HBeAg could be expected to be efficient markers of virus load if antibodies were inactivated and the antigens released.
In the present study, for the purpose of developing a simple, sensitive, and inexpensive assay for determining HBV virus load, we targeted HBcrAg, which is comprised of HBcAg and HBeAg, products of precore/core gene and under the control of the same promoter. HBcAg and HBeAg share the first 149 amino acids (aa) encoded by the core gene (27). We developed a sensitive and specific enzyme immunoassay (EIA) for HBcrAg. The specimens were pretreated in order to inactivate antibodies and to denature antigen before the assay. This assay was able to detect HBcAg and HBeAg even in anti-HBc or anti-HBe antibody-positive specimens. The correlation between HBcrAg and HBV-DNA was assessed with sera of hepatitis B patients.
MATERIALS AND METHODS
Serum samples.
Hepatitis B sera panels were purchased from Boston Biomedica, Inc. (BBI) (West Bridgewater, Mass.), BioClinical Partners, Inc. (BCL) (Franklin, Mass.), or Nabi Diagnostics (Boca Raton, Fla.). Control samples negative for HBV were obtained from blood donors. Serum samples were collected from chronic hepatitis B patients and hepatitis C patients at the Shinshu University Hospital in 1997. All sera were stored at −80°C until tested.
Recombinant HBV core-related antigens and peptides.
Recombinant HBc antigen (rHBcAg; aa 1 to 183) was expressed in Escherichia coli, and HBcAg particles in soluble fraction were purified by gel filtration chromatography followed by centrifugation on a sucrose density gradient (33). Recombinant HBe antigen (rHBeAg; aa −10 to 149) was expressed and purified by a Yeast N-Terminal Expression System (SIGMA-ALDRICH KK, Tokyo, Japan). Recombinant ProHBe antigen (rProHBeAg; aa −10 to 183) was expressed as a TrpE-fusion protein in E. coli (16) and was solubilized and purified from inclusion bodies by gel filtration chromatography. The concentrations of these antigens were determined using the bicinchoninic acid protein assay kit (Pierce Chemical Co., Rockford, Ill.) and bovine serum albumin standards according to the manufacturer's instructions.
Twenty-residue-long peptides containing 10 aa overlapping the ProHBe antigen were chemically synthesized by the F-moc method and purified by reversed-phase chromatography at Asahi Techno Glass Corp. (Tokyo, Japan).
Monoclonal antibodies.
BALB/c mice were immunized with rHBcAg mixed with Freund's adjuvant (Wako Pure Chemical Industries, Osaka, Japan) five times intraperitoneally. Splenocytes from the immunized mice were fused with SP2/O Ag14 myeloma cells. The fused hybridoma cells were selected in RPMI 1640 medium supplemented with 10% fetal calf serum, hypoxanthine, aminopterin, and thymidine. Anti-HBcAg monoclonal antibody-producing hybridomas were selected by rHBcAg-coated wells in an enzyme-linked immunosorbent assay (ELISA) and cloned by limiting dilution. The subclasses of the monoclonal antibodies were determined using the mouse MonoAb-ID kit (Zymed, San Francisco, Calif.).
All hybridoma cell lines were transplanted into the mouse abdominal cavity. From the mouse ascites, monoclonal antibodies were purified by protein A column chromatography (Amersham-Pharmacia). The epitopes of the monoclonal antibodies were analyzed as described previously (13) by an EIA system using 20-residue-long synthesized peptides containing 10 aa that overlapped the −10 to 183 amino acid region of the ProHBe antigen.
Preparation of AP-conjugated monoclonal antibody.
Monoclonal antibodies HB91 and HB110 were digested by pepsin (Worthington Biochemical Corp., Freehold, NJ.) in 100 mM acetate buffer (pH 4.0), and the F(ab)"2 fragment was isolated by gel filtration on Superdex 200HR (Amersham-Pharmacia). The Fab" fragment was prepared by reducing the F(ab)"2 fragment and was conjugated to alkaline phosphatase (AP) (Roche Diagnostics) by the maleimide hinge method (34). The conjugate was purified by gel filtration chromatography on a Superdex 200HR column.
Specimen pretreatment and EIA for HBcrAg.
Microtiter wells (FluoroNunc Black; NUNC, Roskilde, Denmark) were coated with 100 μl of a mixture of anti-HBcrAg monoclonal antibodies HB44, HB61, and HB114. Thereafter, the wells were washed twice with phosphate-buffered saline (PBS; pH 7.4) followed by incubation with a blocking reagent (3% sucrose, 0.5% casein-Na in PBS, pH 7.4) at room temperature for 2 h. After removing the blocking solution, the wells were vacuum dried and stored at 4°C.
A 100-μl specimen aliquot was mixed with 50 μl of pretreatment solution (15% sodium dodecyl sulfate [SDS], 2% Tween 60) and incubated for 30 min at 70°C. All assays were performed in duplicate. Pretreated specimen (50 μl) was then added to each well filled with 100 μl of assay buffer (pH 7.5). The wells were incubated for 2 h at room temperature with gentle agitation and then washed five times with washing buffer (0.05% Tween 20 in PBS, pH 7.4). A 100-μl aliquot of AP-conjugated HB91 and HB110 monoclonal antibodies solution was added to each well and incubated for 1 h at room temperature. After washing six times with the washing buffer, 100 μl of substrate solution (CDP-Star with Emerald II; Applied Biosystems, Bedford, Mass.) were added and incubated for 20 min at room temperature. The relative luminescence intensity (RLI) was measured using a microplate reader (LUMINOUS CT-9000D; DIA-IATRON, Tokyo, Japan).
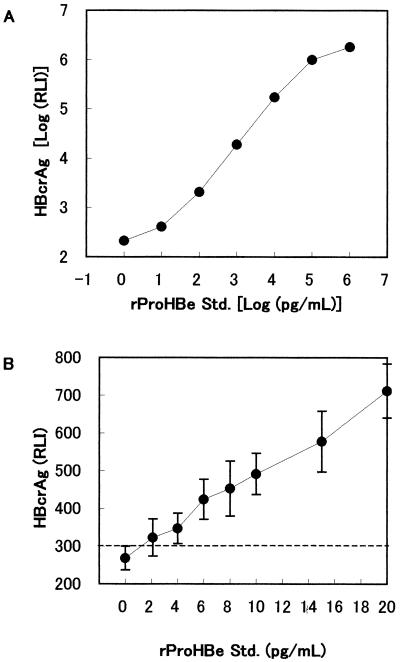
Recombinant ProHBeAg (aa −10 to 183) was used as a standard, and the immunoreactivity of rProHBeAg at 10 fg/ml was defined as 1 U/ml. This standard was serially diluted with normal human serum and assayed at the same time as the specimens. The standard log RLI was plotted versus log concentration (Fig. 1A), and HBcrAg concentrations in each specimen were calculated from the calibration curve.
FIG. 1.
Standard curve and detection limit of HBcrAg assay. Recombinant ProHBe standard was diluted in normal human serum and detected by the HBcrAg assay. The assay reactivity was shown as relative luminescence intensity (RLI). (A) Standard curve; (B) detection limit. Results are means of 10 assays, and the error bars show 2 SD.
HBs antigen, HBe antigen, anti-HBe antibody, and other HBV markers.
HBs antigen, HBe antigen, and anti-HBe antibody were measured by radioimmunoassay (RIA) (Dinabbott, Tokyo, Japan). Data for HBV markers in the BBI PHM 935B and BCP HBV6281 panels were provided by the supplier.
HBV-DNA measurement.
HBV-DNA was detected by TMA (Chugai Diagnostics Science Co., Ltd., Tokyo, Japan), which has a detection range between 3.7 and 8.7 log genome equivalent/ml (corresponding to 5 × 103 to 5 × 108 copies/ml).
The results for HBV-DNA in BBI PHM 935B or BCP HBV6281 panels were obtained using the supplier's data sheet and either the Roche Amplicor HBV Monitor test (detection range, between 4 × 102 and 4 × 107 copies/ml) or BCP PCR (detection limit, 100 copies/ml).
RTD-PCR assay was used for detecting HBV-DNA quantitatively. DNA was extracted from serum by the proteinase K method as follows. A 200-μl aliquot of serum was added to 200 μl of a freshly prepared working solution containing 100 mM Tris-HCl (pH 8.4), 20 mM EDTA, 4% SDS, and 400 mM NaCl supplemented with 10 μg of tRNA and 400 μg of proteinase K. After incubation for 30 min at 54°C, the mixture was extracted by phenol/chloroform and precipitated with isopropanol. Purified DNA was washed twice with 70% ethanol and resuspended in 20 μl of distilled water. A 5-μl aliquot of DNA solution was used for RTD-PCR, which was performed using the LightCycler System (Roche Diagnostics). The sequences of the primers for the HBV surface gene were as follows: forward primer, 5"-ACAACATCAGGATTCCTAGGAC-3" (nucleotides [nt] 166 to 187), and reverse primer, 5"-GGTTGGTGAGTGATTGGAGGTT-3" (nt 345 to 324). The HBV surface gene probe was a TaqMan probe, 5"-FAM-CAGAGTCTAGACTCGTGGTGGACTTC-TAMRA-3" (nt 244 to 269). For the preparation of an external standard, we subcloned the HBV genome (nt 20 to 1805) from the serum into a pUC vector. The recombinant plasmid was purified and subsequently quantified by measuring the optical density at 260 nm.
A nested PCR assay was used for highly sensitive and qualitative detection of HBV-DNA. Five microliters of DNA solution was mixed with 45 μl of PCR mixture containing 250 μM concentrations of each deoxynucleoside triphosphate (dNTP), 1× PCR buffer, 0.25 U of Taq DNA polymerase (Takara, Kyoto, Japan), and 0.25 μM concentrations of each primer pair. The primer sets for the HBV core region were 5"-TTTTTCACCTCTGCCTAATCATCT-3" (nt 1823 to 1846) and 5"-GTAGAAGAATAAAGCCCAGTAA-3" (nt 2505 to 2484) for the first round of PCR and 5"-TGTTCATGTCCTACTGTTCAAG-3" (nt 1849 to 1870) and 5"-AGTTTCCCACCTTATGAGTCCA-3" (nt 2483 to 2462) for the second round of PCR. PCR was started using the hot-start technique. The first round of PCR was performed using an outer primer set for 35 cycles (94°C for 1 min, 55°C for 1 min, and 72°C for 1 min) followed by an extension step at 72°C for 7 min. The second round was performed using an inner primer set for 30 cycles followed by the extension reaction. Nested PCR products were subjected to electrophoresis on a 1.2% agarose gel, stained by ethidium bromide, and visualized using a UV transilluminator.
RESULTS
Selection and characterization of monoclonal antibodies for HBcrAg assay.
A total of 54 hybridoma cell lines that produced anti-HBc antibody were established from four fusions. We used some of these monoclonal antibodies for the assay to detect both HBcAg and HBeAg. These monoclonal antibodies were selected for reactivity to denatured recombinant HBcAg and HBeAg. Many combinations of capture and detector monoclonal antibodies were evaluated for sandwich EIA to detect HBcrAg in SDS-pretreated HBeAg-positive serum. Consequently, we selected HB44, HB61, and HB114 as the immobilized monoclonal antibodies for capturing HBcrAg, while HB91 and HB110 were selected as detector monoclonal antibodies. All five of the monoclonal antibodies were immunoglobulin G1 (IgG1), kappa. The epitope of each monoclonal antibody was determined using 20-residue-long peptides containing 10 aa overlapping the precore region. The epitopes of HB44, HB61, HB91, and HB110 were located at amino acid residues 31 to 49, 131 to 140, 1 to 19, and 21 to 40, respectively. HB114 reacted to recombinant HBc (aa 1 to 81) antigen, but not to any of the peptides tested. The reactivity to recombinant HBc (aa 1 to 183) was strengthened about fourfold when antigen was pretreated with SDS (data not shown). HB114 would therefore recognize a nonlinear epitope on aa 1 to 81 formed after denaturing.
Detection range and linearity of HBcrAg assay.
In the HBcrAg assay, the calibration curve was determined using the rProHBeAg standard serially diluted in normal human serum (Fig. 1). The upper detection limit of this assay was approximately 107 U/ml (Fig. 1A). The analytical lower detection limit was 4 × 102 U/ml, the concentration at which the mean minus two standard deviations (SD) of the RLI did not overlap with the mean plus 2 SD of the zero calibrator (n = 10; Fig. 1B). This assay displayed a broad dynamic range, from 4 × 102 to 1 × 107 U/ml.
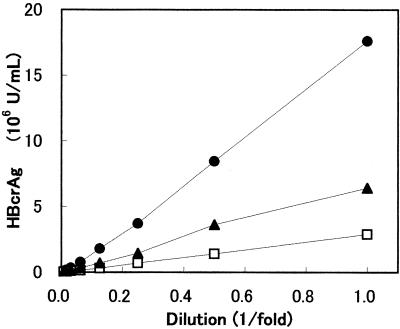
The linearity of the assay was examined by serially diluting three HBV-positive sera, all of which were HBeAg positive and anti-HBe antibody negative (Fig. 2). The quantity of HBcrAg decreased linearly with dilution on a straight line through the zero point.
FIG. 2.
Dilution linearity of hepatitis B sera. Three HBV-DNA and HBeAg positive sera were serially diluted in normal human serum and detected by the HBcrAg assay.
Reproducibility of HBcrAg assay.
Intra-assay reproducibility was assessed from 10 measurements of four specimens. The mean HBcrAg values of the specimens were 6.7 × 103, 2.1 × 104, 3.1 × 106, and 8.2 × 106 U/ml, and the coefficients of variation (CV) were 9.8, 11.3, 12.1, and 10.1%, respectively. Interassay reproducibility was assessed from 11 assays of three specimens. The mean HBcrAg values of the specimens were 3.4 × 104, 8.0 × 104, and 3.2 × 106 U/ml, and the CV were 12.7, 11.8, and 4.3%, respectively.
Recovery, interference, and stability of HBcrAg assay.
Aliquots of 10 μl of HBcrAg- and HBV-DNA-positive serum were added to 90 μl of reference serum. Five different reference sera were utilized, including three anti-HBc/anti-HBe antibody (+/+) hepatitis B sera, anti-HBc/anti-HBe antibody (−/−) serum, and normal human serum. No HBcrAg was detected in any reference sera by this method. Antigen measured in these samples was designated recovered HBcrAg, and recovery rates were calculated as follows: (recovered HBcrAg)/(added HBcrAg) × 100% (Table 1). Recovery rates ranged from 71 to 112%, and the mean recovery rates were 81, 97, or 101% in the case of anti-HBc/anti-HBe antibody-positive reference sera, whereas rates of 97 or 91% were achieved with anti-HBc/anti-HBe antibody-negative or control reference sera. The recovery rates did not demonstrate any significant differences between anti-HBc/anti-HBe antibody positivity and negativity. These data indicated that anti-HBc and anti-HBe antibodies did not interfere with the HBcrAg assay.
TABLE 1.
Recovery of HBcrAg
| Reference serum | Antibody resultsa
|
Amt of recovered HBcrAg from serum containingb:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PHJ201-11
|
PHJ201-02
|
PHJ201-03
|
|||||||||
| Anti-HBc (s/co) | Anti-HBe (s/co) | U/ml | % Recovery | U/ml | % Recovery | U/ml | % Recovery | Mean % recovery | |||
| P0339/2-01 | 0.051 | 0.241 | 4.11 × 103 | 71 | 2.32 × 104 | 89 | 5.53 × 105 | 81 | 81 | ||
| P0339/2-03 | 0.024 | 0.011 | 6.26 × 103 | 109 | 2.31 × 104 | 89 | 6.35 × 105 | 93 | 97 | ||
| P0339/2-04 | 0.018 | 0.010 | 6.16 × 103 | 107 | 2.53 × 104 | 98 | 6.63 × 105 | 97 | 101 | ||
| P0339/2-08 | 1.113 | 1.805 | 5.22 × 103 | 91 | 2.92 × 104 | 112 | 5.88 × 105 | 86 | 97 | ||
| Normal | — | — | 5.06 × 103 | 88 | 2.59 × 104 | 100 | 5.90 × 105 | 87 | 91 | ||
| Mean | 5.36 × 103 | 93 | 2.53 × 104 | 98 | 6.06 × 105 | 89 | 93 | ||||
| SD | 8.84 × 102 | 2.47 × 103 | 4.31 × 104 | ||||||||
| CV | 16.5% | 9.7% | 7.1% | ||||||||
Anti-HBc and anti-HBe antibody results are expressed as signal/cutoff (s/co). Ratios of <1.0 are considered positive. —, serum was negative.
Aliquots of 10 μl of HBcrAg- and HBV-DNA-positive serum were added to 90 μl of reference serum. The amount of recovered HBcrAg is the amount of antigen in these samples. % Recovery = (amount of recovered HBcrAg)/(amount of added HBcrAg) × 100%. The amounts of HBcrAg in the sera were as follows: PHJ201-11, 5.75 × 103 U/ml; PHJ201-02, 2.60 × 104 U/ml; PHJ201-03, 6.81 × 105 U/ml.
The influence of blood elements was assessed using an interference check kit (International Reagents Corp., Kobe, Japan). Five concentrations of each blood element were added to two samples of HBcrAg-positive sera, and HBcrAg was measured. Unconjugated bilirubin (<19 mg/dl), conjugated bilirubin (<20 mg/dl), hemoglobin (<440 mg/dl), chyle (<2,350 formazin turbidity units), and IgM rheumatoid factor (<50 IU/ml) did not interfere with this assay (data not shown).
Antigen stability in HBcrAg-positive specimens was examined. From the BBI PHJ201 HBV performance panels, 13 HBcrAg-positive specimens were divided into three aliquots and stored at −20, 4, or 37°C for 7 days. Except for two specimens stored at 37°C, which showed 29 and 28% lower concentrations of HBcrAg than samples at −20°C, no significant changes (−13 to +18%) were observed in HBcrAg amounts, regardless of storage temperature.
HBcrAg in healthy and hepatitis C sera.
HBcrAg was examined using healthy control (HBsAg/HBV-DNA negative; n = 108) and hepatitis C (anti-HCV antibody positive; n = 59) sera. HBcrAg levels were below 8.9 × 102 (mean, 4.5 × 102; SD, 1.3 × 102) and 6.2 × 102 (mean, 4.5 × 102; SD, 1.0 × 102) U/ml, respectively. Based on these data, we set a tentative cutoff for HBcrAg positivity at 1.0 × 103 U/ml. The immunoreactivities of all healthy control and hepatitis C sera were under this cutoff, indicating a high specificity of this assay for HBcrAg.
Detection limits of HBV markers in hepatitis B sera dilutions.
The detection limits of HBcrAg EIA, HBsAg RIA, HBeAg RIA, and HBV-DNA TMA for hepatitis B sera were compared by a limiting-dilution assay. Four hepatitis B panel (BBI, PHJ201) sera were serially diluted 10-fold, and HBV markers were measured. HBsAg and HBeAg values were evaluated by a cutoff index (COI): a COI of over 1.0 indicated a positive response. The results are summarized in Table 2. The assay was able to detect HBcrAg even in anti-HBe and anti-HBc antibody-positive sera, such as PHJ201-13, which yield very poor results with current HBeAg assays. The HBcrAg assay demonstrated higher sensitivity than HBV-DNA TMA or the HBeAg test, and equivalent sensitivity to HBsAg test in these specimens.
TABLE 2.
Dilution tests of hepatitis B seraa
| Specimens (anti-HBe/anti-HBc) | Dilution | Detection limit
|
|||
|---|---|---|---|---|---|
| HBcrAg EIA (U/ml) | DNA TMA (LGE/ml) | HBsAg RIA (COI) | HBeAg RIA (COI) | ||
| PHJ201-04 (−/+) | 102 | 5.3 × 106 | 6.4 | 62.7 | 157.5 |
| 103 | 4.5 × 105 | 5.7 | 63.1 | 38.0 | |
| 104 | 4.8 × 104 | 4.7 | 31.5 | 5.3 | |
| 105 | 5.4 × 103 | 4.0 | 5.4 | — | |
| 106 | — | — | — | — | |
| PHJ201-07 (−/+) | 102 | 1.4 × 106 | 5.7 | 73.3 | 96.5 |
| 103 | 1.2 × 105 | 5.2 | 56.1 | 15.4 | |
| 104 | 1.2 × 104 | 3.9 | 22.4 | 2.0 | |
| 105 | 1.1 × 103 | — | 3.7 | — | |
| 106 | — | — | — | — | |
| PHJ201-08 (−/+) | 102 | 7.4 × 105 | 5.0 | 52.3 | 76.1 |
| 103 | 7.8 × 104 | 3.9 | 19.6 | 10.5 | |
| 104 | 8.6 × 103 | — | 2.6 | 1.6 | |
| 105 | — | — | — | — | |
| 106 | — | — | — | — | |
| PHJ201-13 (+/+) | 102 | 1.3 × 106 | 5.7 | 62.2 | 1.8 |
| 103 | 1.3 × 105 | 4.8 | 42.4 | 1.0 | |
| 104 | 1.4 × 104 | 4.0 | 11.1 | — | |
| 105 | 1.3 × 103 | — | 1.6 | — | |
| 106 | — | — | — | — | |
| Cutoff | 1.0 × 103 | 3.7 | 1.0 | 1.0 | |
Detection limits of serially diluted hepatitis B sera are compared.
—, data are below the cutoff.
Correlation with HBV-DNA.
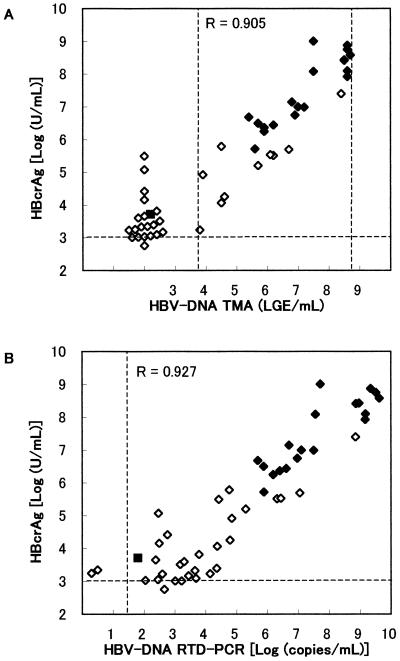
HBcrAg was examined in 50 sera of hepatitis B patients (Fig. 3). Sera with over 107 U of HBcrAg/ml were remeasured after 10- or 100-fold dilution in normal human serum. HBV-DNA was also measured by TMA and by in-house RTD-PCR (detection limit of 20 copies/ml). HBV-DNA and HBcrAg levels were relatively low, but still detectable in most anti-HBe antibody-positive sera. When the cutoff was set at 1.0× 103 U/ml, 49 of 50 samples were HBcrAg positive, whereas TMA identified only 29 samples as positive for HBV-DNA and RTD-PCR identified 48 positives. The positivity rate of HBcrAg was significantly higher than that of HBV-DNA TMA (P < 0.0001; McNemar test). The correlation between the log concentration of HBcrAg and that of HBV-DNA is shown in Fig. 3. The correlation coefficients for HBV-DNA- and HBcrAg-positive sera were 0.905 (n = 29, by TMA) or 0.927 (n = 47 by RTD-PCR), respectively, indicating the results of the HBcrAg assays were well correlated with HBV-DNA.
FIG. 3.
Correlation between concentrations of HBcrAg and HBV-DNA in serum of hepatitis B patients. ⧫, HBeAg positive; ◊, anti-HBe positive; ▪, HBeAg and anti-HBe antibody negative. The HBV-DNA level was measured by TMA (detection limit, 3.7 to 8.7 LGE/ml) (A) or RTD-PCR (detection limit, 20 copies/ml) (B).
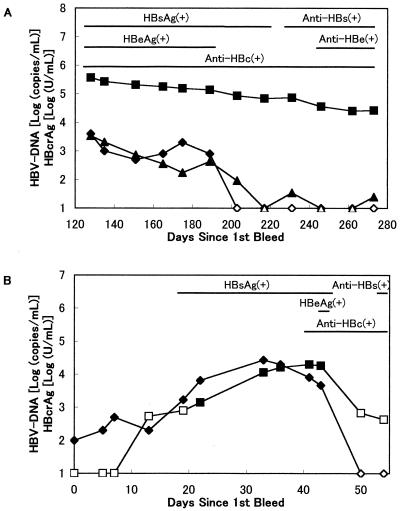
HBcrAg in HBeAg/anti-HBe antibody seroconversion panels.
HBcrAg was examined in commercially available seroconversion panel sera (Fig. 4). These panels are sets of serial bleeds taken from a donor. On the BBI PHM935B panel (Fig. 4A), which was a case of seroconversion of HBeAg to anti-HBe antibody, HBcrAg were detectable throughout the panel and the HBcrAg levels gradually decreased. We also examined HBV-DNA by in-house RTD-PCR and nested PCR. RTD-PCR revealed that HBV-DNA levels gradually decreased and became very low or negative, results similar to those of the Roche Amplicor Monitor test, whereas nested PCR was positive between days 203 and 273. During days 128 to 273, the HBcrAg levels decreased from 5.57 log (U/ml) to 4.43 log, while HBV-DNA RTD-PCR did from 3.53 log (copies/ml) to 1.39 log. Although the decrease was smaller, the variation in HBcrAg levels reflected HBV-DNA levels throughout the panel.
FIG. 4.
HBcrAg and HBV-DNA patterns in seroconversion panels. The data on HBsAg, HBeAg, and anti-HBs, anti-HBe, and anti-HBc antibodies were obtained from the supplier's data sheet. Closed symbols, positive; open symbols, under the cutoff. (A) BBI PHM935B panel. ▪ and □ HBcrAg [log(units/milliliter)]; ⧫ and ◊, HBV-DNA detected by the Roche Amplicor HBV Monitor test (detection limit, 4 × 102 to 4 × 107 copies/ml); ▴ and ▵, in-house RTD-PCR (detection limit 20 copies/ml). (B) BCP HBV6281 panel. ▪ and □, HBcrAg [log (units/milliliter)]; ⧫ and ◊, HBV-DNA detected by BCP PCR (detection limit, 102 copies/ml).
The BCP HBV6281 panel (Fig. 4B) is an acute and recovered hepatitis B seroconversion panel. HBcrAg was positive between days 22 and 43, and the changes in concentration were similar to those of HBV-DNA levels, although there was slightly lower sensitivity in this panel.
DISCUSSION
Recently, HBV-DNA TMA and PCR have been widely used to monitor virus load (11, 12, 14, 20, 22, 23, 35). However, conventional EIA has some advantages over nucleic acid amplification assays. EIA is a relatively simple method and provides a low-cost and quantitative analysis with high reproducibility. HBsAg assays are highly sensitive immunoassays that indicate the presence of HBV. However, HBsAg concentration shows little or no correlation with HBV-DNA (9). Therefore, HBsAg cannot be used as virus load marker.
Measurement of the HCV core antigen after specimen pretreatment has been reported useful for the diagnosis and monitoring of hepatitis C (13, 15, 19, 21, 28, 30). We previously developed a highly sensitive EIA for HCV core antigen (1, 29). In this assay, HCV core antigen was released from virus virion by SDS pretreatment prior to detection. In the present study, we applied this detection system to HBV core and e antigen. We developed a sensitive and specific EIA for HBcrAg, which includes a single-step pretreatment with SDS. High concentrations of SDS generally denature proteins and destroy their epitopes. We used monoclonal antibodies reactive to denatured HBcAg and HBeAg. Moreover, we selected monoclonal antibodies that can bind to antigen in SDS-containing solutions. The ingredients in the pretreatment buffer, reaction buffer, and labeled antibody solution were optimized for sensitive HBcrAg measurement by a specific set of monoclonal antibodies. Consequently, the immobilized monoclonal antibodies could react to HBcrAg in 1.7% SDS solution. Three monoclonal antibodies were used as capture antibodies and two were used as detector antibodies, which enabled the assay to detect HBcrAg with mutations at one or more epitopes.
There are shared strong linear specific epitopes for HBcAg and HBeAg near aa 80 (26). However, none of our 54 monoclonal antibodies to denatured HBcAg reacted with peptides comprised of aa 61 to 80, 71 to 90, or 81 to 100. The linear epitopes of natural anti-HBc/e antibody might differ from mouse antibodies to denatured HBc/e antigens.
The SDS and heat treatment in the present assay efficiently inactivated anti-HBc and anti-HBe antibodies in specimens, and therefore HBcrAg could be measured even in anti-HBc/e antibody-positive specimens. This improvement was confirmed by the recovery test using anti-HBc/e antibody-positive sera (Table 1).
Observed recovery rates (Table 1) displayed a wider variation (CV, 7.1 to 16.5% in recovered HBcrAg) than reproducibility tests (4.3 to 12.7%) in cases where PHJ201-11 was added. This was indicative of low HBcrAg levels. We attributed the wider variation to factors such as low HBcrAg levels near the cutoff, smaller volumes (10 μl) of added sample than other tests (100 μl), and smaller sample numbers (5, compared to the 10 or 11 in the reproducibility tests). In addition, it should be noted that the values for added HBcrAg also displayed variation; thus, the recovery rates contained variations for both recovered HBcrAg and added HBcrAg. The mean recovery rate was lowest at 81% in the case of P0339/1-01 as reference sera. On the other hand, recovery rates were 97 and 101% for P0339/2-03 and -04, which showed stronger anti-HBc/anti-HBe antibody responses. Recovery rates displayed considerable variation but were independent of anti-HBc/anti-HBe antibody positivity or strength.
The HBcrAg assay showed higher sensitivity than the HBV-DNA TMA test and comparable sensitivity to the HBV-DNA PCR test (Table 2; Fig. 3 and 4). This assay exhibited better quantitative linearity than HBV-DNA TMA, HBsAg RIA, or HBeAg RIA over a wide range of concentrations (Table 2; Fig. 2). The broad dynamic range of the HBcrAg assay is appropriate for monitoring HBV virus loads that vary over a wide range.
Due to the stability of HBcrAg concentration in serum, the HBcrAg assay is expected to have a lower risk of false results due to target degradation during storage of specimens than HBV-DNA assay.
HBcrAg concentrations were well correlated with HBV-DNA levels (Fig. 3). Thus, the HBcrAg assay could be a viable alternative to HBV-DNA assays. As HBeAg is a secreting protein that exists in serum at much higher concentrations than HBcAg (roughly 100:1; unpublished data), HBcrAg concentrations mainly reflect levels of HBeAg and HBeAg/anti-HBe antibody complex. The high correlation between HBcrAg concentrations and HBV-DNA levels suggests that HBeAg is produced proportionately to virus load. HBeAg expression may be correlated to HBcAg and thus to the HBV virus load, because HBcAg and HBeAg are controlled by the same promoter.
Current commercial HBeAg assays do not detect HBeAg/anti-HBe complex, because the epitopes are masked by anti-HBe antibody (24). The results of our HBcrAg assay, which mainly reflect HBeAg and HBeAg/anti-HBe antibody complexes, were not necessarily affected by HBeAg (Table 2, PHJ201-13; Fig. 3, anti-HBe antibody positives; Fig. 4A, days 203 to 273). These results support previous studies that demonstrated that anti-HBe antibody seroconversion does not necessarily indicate subsequent HBeAg clearance (24). Anti-HBe antibody-positive, HBV-DNA positive sera like PHJ201-03 would contain high concentrations of HBeAg, while the results of commercial HBeAg assay were either only slightly positive or negative.
However, HBeAg are not accurate markers in cases of precore mutant HBV infections (2, 5). Thus, one potential drawback of using HBcrAg as an HBV virus load marker is that HBcrAg would not reflect the HBV virus load of precore mutants. Nevertheless, one of the advantages of the HBcrAg assay is the potential to detect not only HBeAg but also HBcAg. Therefore, we believe this assay could detect HBcAg even in HBV-infected specimens with 100% precore mutation, although with lower sensitivity.
In the seroconversion panels, decreases in HBcrAg seemed to lag behind and to be smaller than the changes seen for HBV-DNA (Fig. 4). We hypothesized that in these cases, HBV virions were undergoing a rapid decline in the blood while a considerable amount of virus remained in tissues, such as liver. HBeAg would therefore be secreted from these tissues and detected by the HBcrAg assay. If this hypothesis is true, HBcrAg levels could be a marker of HBV activity in liver. It has been reported that the presence of HBeAg/anti-HBe antibody immune complexes was restricted to patients suffering a reactivation of HBV replication in HBsAg and anti-HBe antibody chronic carriers (6). HBcrAg levels might therefore reflect a risk of reactivation. At present, we have no other evidence to support this hypothesis, and further study is needed to clarify the mechanisms behind this observation.
In this study, we developed a sensitive EIA specific for HBcAg and HBeAg. The specimens are pretreated in order to inactivate antibodies and to denature antigen before assay. This assay was able to detect HBcrAg even in anti-HBc or anti-HBe antibody-positive specimens. HBcrAg concentrations well correlated with HBV virus load. The HBcrAg concentration varied in accordance to HBV-DNA level. The HBcrAg assay could be used to monitor hepatitis B patients or carriers, especially during lamivudine or interferon therapy. Also, highly sensitive HBcrAg assays would contribute not only to monitoring hepatitis B patients but also to the early diagnosis of HBV infection. To confirm these preliminary observations, additional clinical and diagnostic studies of much larger populations are required.
Acknowledgments
We are grateful for the technical assistance of Sanae Nakatsuka.
REFERENCES
- 1.Aoyagi, K., C. Ohue, K. Iida, T. Kimura, E. Tanaka, K. Kiyosawa, and S. Yagi. 1999. Development of a simple and highly sensitive enzyme immunoassay for hepatitis C virus core antigen. J. Clin. Microbiol. 37:1802-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, B. L., A. M. D. Stecle, I. Peenze, and M. J. Mphahlele. 1995. Detection and quantitation of hepatitis B virus DNA: comparison of two commercial hybridization assays with polymerase chain reaction. J. Viral Hepatitis 2:107-111. [DOI] [PubMed] [Google Scholar]
- 3.Bonino, F., F. Rosina, M. Rizzetto, R. Rizzi, E. Chiaberge, R. Tardanico, F. Callea, and G. Verme. 1986. Chronic hepatitis in HBsAg carriers with serum HBV-DNA and anti-HBe. Gastroenterology 90:1268-1273. [DOI] [PubMed] [Google Scholar]
- 4.Bredehorst, R., H. von Wulffen, and C. Granato. 1985. Quantitation of hepatitis B virus (HBV) core antigen in serum in the presence of antibodies to HBV core antigen: comparison with assays of serum HBV DNA, DNA polymerase, and HBV e antigen. J. Clin. Microbiol. 21:593-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carman, W. F., and H. C. Thomas. 1992. Genetic variation in hepatitis B virus. Gastroenterology 102:711-719. [DOI] [PubMed] [Google Scholar]
- 6.Castillo, I., J. Bartolome, J. A. Quiroga, J. C. Porres, and V. Carreno. 1990. Detection of HBeAg/anti-HBe immune complexes in the reactivation of hepatitis B virus replication among anti-HBe chronic carriers. Liver 10:79-84. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C.-H., J.-T. Wang, C.-Z. Lee, J.-C. Sheu, T.-H. Wang, and D.-S. Chen. 1995. Quantitative detection of hepatitis B virus DNA in human sera by branched-DNA signal amplification. J. Virol. Methods 53:131-137. [DOI] [PubMed] [Google Scholar]
- 8.Gowans, E. J., 1986. Relationship between HBeAg and HBV DNA in patients with acute and persistent hepatitis B infection. Med. J. Aust. 145:439-441. [DOI] [PubMed] [Google Scholar]
- 9.Guillou, D. B., J. C. Duclos-Vallee, F. Eberle, F. Capel, and M. A. Petit. 2000. Evaluation of enzyme-linked immunosorbent assay for detection and quantification of hepatitis B virus PreS1 envelope antigen in serum samples: comparison with two commercial assays for monitoring hepatitis B virus DNA. J. Viral Hepat. 7:387-392. [DOI] [PubMed] [Google Scholar]
- 10.Hadziyannis, S. J., H. M. Lieberman, G. G. Karvountzis, and D. A. Shafritz. 1983. Analysis of liver disease, nuclear HBcAg, viral replication, and hepatitis B virus DNA in liver and serum of HBeAg vs. anti-HBe positive carriers of hepatitis B virus. Hepatology 3:656-662. [DOI] [PubMed] [Google Scholar]
- 11.Kamisango, K., C. Kamogawa, M. Sumi, S. Goto, A. Hirao, F. Gonzales, K. Yasuda, and S. Iino. 1999. Quantitative detection of hepatitis B virus by transcription-mediated amplification and hybridization protection assay. J. Clin. Microbiol. 37:310-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaneko, S., S. M. Feinstone, and R. H. Miller. 1989. Rapid and sensitive method for detection of serum hepatitis B virus DNA using the polymerase chain reaction technique. J. Clin. Microbiol. 27:1930-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kashiwakuma, T., A. Hasegawa, T. Kajita, A. Takata, H. Mori, Y. Ohta, E. Tanaka, K. Kiyosawa, T. Tanaka, S. Tanaka, N. Hattori, and M. Kohara. 1996. Detection of hepatitis C virus specific core protein in serum of patients by a sensitive fluorescence enzyme immunoassay (FEIA). J. Immunol. Methods 190:79-89. [DOI] [PubMed] [Google Scholar]
- 14.Kessler, H., H. S. Preininger, E. Stelzl, E. Daghofer, B. I. Santner, E. Marth, H. Lackner, and R. E. Stauber. 2000. Identification of different states of hepatitis B virus infection with a quantitative PCR assay. Clin. Diagn. Lab. Immunol. 7:298-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi, M., E. Tanaka, A. Matsumoto, K. Yoshizawa, H. Imai, T. Sodeyama, and K. Kiyosawa. 1998. Clinical application of hepatitis C virus core protein in early diagnosis of acute hepatitis C. J. Gastroenterol. 33:508-511. [DOI] [PubMed] [Google Scholar]
- 16.Kohara, K. T., K. Yamaguchi, N. Maki, Y. Ohta, K. Miki, M. Mizokami, K. Ohba, S. Tanaka, N. Hattori, A. Nomoto, and M. Kohara. 1993. Antigenicities of group I and II hepatitis C virus polypeptides—molecular basis of diagnosis. Virology 192:430-437. [DOI] [PubMed] [Google Scholar]
- 17.Liaw, Y. F., C. M. Chu, I. J. Su, M. J. Huang, D. Y. Lin, and C. S. Chang-Chien. 1983. Clinical and histological events preceding hepatitis B e antigen seroconversion in chronic type B hepatitis. Gastroenterol. 84:216-219. [PubMed] [Google Scholar]
- 18.Matsuyama, Y., M. Omata, O. Yokosuka, F. Imazeki, Y. Ito, and K. Okuda. 1985. Discordance of hepatitis B e antigen/antibody and hepatitis B virus deoxyribonucleic acid in serum. Analysis of 1063 specimens. Gastroenterology 89:1104-1108. [DOI] [PubMed] [Google Scholar]
- 19.Moriya, T., F. Sasaki, J. Tanaka, M.-A. Mizui, T. Nakanishi, K. Takahashi, and H. Yoshizawa. 1994. Comparison of HCV core antigen activity by ELISA and amount of HCV RNA by branched DNA assay. Int. Hepatol. Commun. 2:175-177. [Google Scholar]
- 20.Noborg, U., A. Gusdal, E. K. Pisa, A. Hedrum, and M. Lindh. 1999. Automated quantitative analysis of hepatitis B virus DNA by using the Cobas Amplicor HBV Monitor Test. J. Clin. Microbiol. 37:2793-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orito, E., M. Mizokami, T. Tanaka, J. Y. Lau, K. Suzuki, M. Yamauchi, Y. Ohta, A. Hasegawa, S. Tanaka, and M. Kohara. 1996. Quantification of serum hepatitis C virus core protein level in patients chronically infected with different hepatitis C virus genotypes. Gut 39:876-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pawlowsky, J. M., A. Bastie, I. Lonjon, J. Remire, F. Darthuy, C. J. Soussy, and D. Dhumeaux. 1997. What technique should be used for routine detection and quantification of HBV DNA in clinical samples? J. Virol. Methods 65:245-253. [DOI] [PubMed] [Google Scholar]
- 23.Quint, W. G. V., R. A. Heijtink, J. Schirm, W. H. Gerlich, and H. G. M. Niesters. 1995. Reliability of methods for hepatitis B virus DNA detection. J. Clin. Microbiol. 33:225-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raimondo, G., S. Recchia, C. Lavarini, O. Crivelli, and M. Rizzetto. 1982. Dane particle-associated hepatitis B e antigen in patients with chronic hepatitis B virus infection and hepatitis B e antibody. Hepatology 2:449-454. [DOI] [PubMed] [Google Scholar]
- 25.Realdi, G., A. Alberti, M. Rugge, et al. 1980. Seroconversion from hepatitis B e antigen to anti-HBe in chronic hepatitis B virus infection. Gastroenterology 79:195-199. [PubMed] [Google Scholar]
- 26.Salfeld, J., E. Pfaff, M. Noah, and H. Schaller. 1989. Antigenic determinants and functional domains in core antigen and e antigen from hepatitis B virus. J. Virol. 63:798-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi, K., A. Machida, G. Funatsu, M. Nomura, S. Usuda, S. Aoyagi, K. Tachibana, H. Miyamoto, M. Imai, T. Nakamura, Y. Miyakawa, and M. Mayumi. 1983. Immunochemical structure of hepatitis B e antigen in the serum. J. Immunol. 130:2903-2907. [PubMed] [Google Scholar]
- 28.Tanaka, E., K. Kiyosawa, A. Matsumoto, T. Kashiwakuma, A. Hasegawa, H. Mori, O. Yanagihara, and Y. Ohta. 1996. Serum levels of hepatitis C virus core protein in patients with chronic hepatitis C treated with interferon alfa. Hepatology 23:1330-1333. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka, E., C. Ohue, K. Aoyagi, K. Yamaguchi, S. Yagi, K. Kiyasawa, and H. J. Alter. 2000. Evaluation of a new enzyme immunoassay for hepatitis C virus (HCV) core antigen with clinical sensitivity approximating that of genomic amplification of HCV RNA. Hepatology 32:388-393. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka, T., J. Y. Lau, M. Mizokami, E. Orito, E. Tanaka, K. Kiyosawa, K. Yasui, Y. Ohta, A. Hasegawa, S. Tanaka, and M. Kohara. 1995. Simple fluorescent enzyme immunoassay for detection and quantification of hepatitis C viremia. J. Hepatol. 23:742-745. [DOI] [PubMed] [Google Scholar]
- 31.Urdea, M. S. 1993. Synthesis and characterization of branched DNA (bDNA) for the direct and quantitative detection of CMV, HBV, HCV, and HIV. Clin. Chem. 39:725-726. [Google Scholar]
- 32.Usuda, S., H. Okamoto, F. Tsuda, T. Tanaka, Y. Miyakawa, and M. Mayumi. 1998. An enzyme-linked immunosorbent assay with monoclonal antibodies for the determination of phosphorylated hepatitis B core protein (p21c) in serum. J. Virol. Methods 72:95-103. [PubMed] [Google Scholar]
- 33.Wingfield, P. T., S. J. Stahl, R. W. Williams, and A. C. Steven. 1995. Hepatitis core antigen produced in Escherichia coli: Subunit composition, comformational analysis, and in vitro capsid assembly. Biochemistry 34:4919-4932. [DOI] [PubMed] [Google Scholar]
- 34.Yoshitake, S., M. Imagawa, E. Ishikawa, Y. Niitsu, I. Urushizaki, M. Nishiura, R. Kanazawa, H. Kurosaki, S. Tachibana, N. Nakazawa, and H. Ogawa. 1982. Mild and efficient conjugation of rabbit Fab" and horseradish peroxidase using a maleimide compound and its use for enzyme immunoassay. J. Biochem. 92:1413-1424. [DOI] [PubMed] [Google Scholar]
- 35.Zaaijer, H. L., F. ter Borg, H. T. M. Cuypers, M. C. A. H. Hermus, and P. N. Lelie. 1994. Comparison of methods for detection of hepatitis B virus DNA. J. Clin. Microbiol. 32:2088-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]