Abstract
A rapid (time to completion, <4 h, including DNA extraction) and quantitative touch-down (QTD) real-time diagnostic Pneumocystis carinii PCR assay with an associated internal control was developed, using fluorescence resonance energy transfer (FRET) probes for detection. The touch-down procedure significantly increased the sensitivity of the assay compared to a non-touch-down procedure. Tenfold serial dilutions of a cloned target were used as standards for quantification. P. carinii DNA has been detected in respiratory specimens from patients with P. carinii pneumonia (PCP) and from patients without clinical evidence of PCP. The latter probably represents colonization or subclinical infection. It is logical to hypothesize that quantification might prove helpful in distinguishing between infected and colonized patients: the latter group would have lower copy numbers than PCP patients. A blinded retrospective study of 98 respiratory samples (49 lower respiratory tract specimens and 49 oral washes), from 51 patients with 24 episodes of PCP and 34 episodes of other respiratory disease, was conducted. PCR-positive samples from colonized patients contained a lower concentration of P. carinii DNA than samples from PCP patients: lower respiratory tract samples from PCP and non-PCP patients contained a median of 938 (range, 2.4 to 1,040,000) and 2.6 (range, 0.3 to 248) (P < 0.0004) copies per tube, respectively. Oral washes from PCP and non-PCP patients contained a median of 49 (range, 2.1 to 2,595) and 6.5 (range, 2.2 to 10) (P < 0.03) copies per tube, respectively. These data suggest that this QTD PCR assay can be used to determine if P. carinii is present in respiratory samples and to distinguish between colonization and infection.
The opportunistic fungus Pneumocystis carinii f. sp. hominis is an important cause of morbidity and mortality, causing P. carinii pneumonia (PCP) in AIDS and other immunocompromised patients. The standard method for diagnosis of PCP is microscopic examination of stained (immunofluorescent or conventional tinctorial) invasive lower respiratory tract specimens: bronchoalveolar lavage (BAL), lung biopsy, or induced sputum specimens, the latter being the least sensitive, with reports of sensitivity varying from <50% to >90% (4-6, 9, 17, 18, 21, 23). Molecular detection systems have the potential to provide a higher degree of sensitivity than microscopic examination. PCR methods have been applied to lower respiratory tract specimens, and recently to non-invasive oral washes as well (1-5, 10, 12-14, 19-36, 38-42). However, some of these techniques are cumbersome, often requiring several steps in order to increase sensitivity and leaving them open to possible contamination.
A single-round, nonnested, PCR assay, with no manipulations of amplicons required, would significantly reduce risks of contamination problems and be ideal for use in clinical diagnostic laboratories. A rapid PCR assay for detection of PCP, with a turnaround time comparable to smears, would enhance the clinical utility of molecular testing for P. carinii.
Due to the high sensitivity of molecular methods, P. carinii DNA has been detected in respiratory samples from patients without PCP, probably representing either colonization or subclinical infection (3, 10, 19-21, 23, 25, 26, 29-33, 36, 38, 41, 42). These clinical false-positive but biological true-positive results lower the clinical specificity and positive predictive value of the qualitative test. It is logical to hypothesize that quantification might prove helpful in distinguishing between clinical true- and false-positive P. carinii PCR results: PCP patients would have a higher organism burden than colonized or subclinically infected patients, reflected by a higher amount of P. carinii DNA present in the extracted specimens from PCP patients.
The principal aim of this study was to develop and validate a rapid, sensitive and quantitative P. carinii PCR assay, with internal and external controls, for routine use in a clinical laboratory setting.
MATERIALS AND METHODS
The laboratory in which this study was conducted is practicing strict physical separation of all the various steps involved in PCR, and unidirectional workflow is employed to reduce risk of carryover contamination.
Standards.
A plasmid containing a P. carinii major surface glycoprotein (MSG) gene insert was generated by cloning HuMSG14 (3,083 bp) (GenBank accession no. AF033209) into the pCR2.1 vector. After propagation and purification of the plasmid, concentration (number of MSG copies per microliter) was derived by the A260 optical density measurement and the plasmid molecular weight. Tenfold serial dilutions (10−2 to 105 MSG copies/μl) of the linearized plasmid in water were prepared with glycogen (33.3 μg/ml) as the DNA carrier.
Patient specimens.
Paired samples of oral washes with either induced sputum or BAL fluid were obtained from patients with respiratory symptoms, who were felt to be susceptible to PCP while being seen at the Clinical Center, National Institutes of Health (NIH), Washington Hospital Center, Washington, D.C., or the Johns Hopkins Hospital, Baltimore, Md. Patients agreed to provide the oral washes, induced sputum, and BAL fluid for research under a protocol approved by the respective institutional review boards. Oral wash samples were collected within 24 h prior to either induction of sputum or BAL fluid by having patients rinse the oral cavity vigorously for 10 to 30 s with 50 ml of sterile saline. One hundred respiratory samples (51 oral washes and 49 lower respiratory tract specimens: 30 BAL and 19 induced sputum samples), previously examined by conventional qualitative PCR, from 51 patients with 58 episodes of respiratory disease were selected for inclusion in the study. Of the 51 patients, 24 were human immunodeficiency virus positive, 10 had hematological malignancies, 6 had inflammatory rheumatological diseases, 4 had solid-organ malignancies, and 7 had other underlying conditions, including rare congenital immunodeficiency disorders. Samples had been tested previously (7a) by two conventional qualitative PCR methods targeting MSG and mitochondrial rRNA genes (14, 38). Since the objective of this study was to investigate the utility of quantification, the selected samples were deliberately biased toward a higher frequency of expected false positive results as compared to a random sample series: all available samples which had previously tested positive by PCR (n = 54) as well as randomly selected samples previously tested negative by PCR (n = 46) were included. The investigator performing the assays and interpreting the results was blinded to all earlier data, including the expected proportion of positive and negative samples.
Samples obtained within 3 days were regarded as being from the same episode of respiratory disease. P. carinii was detected by microscopic examination using a direct immunofluorescent assay (DFA), Monofluo Test Kit (Bio-Rad/Sanofi Diagnostics, Hercules, Calif.), at the NIH or conventional tinctorial stains at the other two hospitals. Positive BAL or induced sputum smears were considered diagnostic for PCP. If the BAL smear was negative, the patient was regarded as not having PCP. If an induced sputum was negative by smear for PCP, the collection of BAL specimen was encouraged. In 13 episodes, a smear-negative induced sputum was not followed by a diagnostic BAL sample. Review of all these patients' charts confirmed that none had clinical evidence of PCP, and all recovered without receiving treatment for PCP during the episodes in question.
Control specimens.
Twenty-five oral washes from 25 healthy control subjects were obtained as described above.
External controls.
P. carinii-positive and -negative BAL samples, as determined by previous DFA smear and PCR, were included with each DNA extraction and in each PCR run. In addition, a water control and dilutions of cloned standards for quantification were included in each PCR run.
Internal controls.
In order to detect inhibitors in the patient specimens, a mimic amplifiable by the MSG primers was constructed by amplifying a sequence of the tetracycline resistance gene of pBR322 with primers tailed with the corresponding MSG primer sequences. This PCR product was cloned, and the plasmid was propagated and purified. Concentration was calculated as described above.
DNA extraction.
DNA was extracted from patient and control specimens and external controls using a NucliSens kit (Organon Teknika) according to the manufacturer's recommendations. Specimens were stored at −70°C in NucliSens lysis buffer. A 200-μl volume of lower respiratory tract specimen (induced sputum or BAL fluid) was extracted directly, whereas the entire volume of oral wash fluid was centrifuged at 2,800 × g for 10 min. The oral wash pellet was resuspended in 1 ml of original fluid, and 200 μl of this concentrate was used for DNA extraction.
DNA amplification.
A PCR method previously described (7a 15), targeting the multicopy MSG gene of P. carinii, was modified for the real-time assay. Primers commercially synthesized (Midland Certified Reagent Company, Midland, Tex.), JKK14/15 (5"-GAA TGC AAA TCY TTA CAG ACA ACA G-3") and JKK17 (5"-AAA TCA TGA ACG AAA TAA CCA TTG C-3"), amplify a 250-bp segment of the multicopy MSG gene family. The MSG primers also amplify a 295-bp fragment of the mimic, designed to be longer than the MSG amplicon to give the MSG target a competitive advantage during simultaneous amplification.
PCR conditions.
All reactions were performed in glass capillaries (Roche) with a final reaction volume of 20 μl of 1× LightCycler-FastStart DNA Master Hybridization Probes reaction mixture (Roche) containing FastStart Taq, reaction buffer, and deoxynucleoside triphosphate and final concentrations of 1.0 μM for each primer, 0.2 μM for each MSG fluorescence resonance energy transfer (FRET) probe, 5.0 mM MgCl2, and 1 U of uracil-DNA glycosylase (UNG), heat-labile (from BMTU 3346; Roche), per tube. The FastStart Taq ensures hot start. In the reaction mixture dUTP is substituted for dTTP. Tubes with internal control contained, in addition to 400 copies of mimic, 4 pmol (final concentration, 0.2 μM) of each mimic FRET probe. Initial assay runs performed with standards only used 4 μl of a standard dilution added to each tube. Later experiments, including all patient samples, utilized 5 μl of patient specimen or standard dilution per tube. All samples were kept on ice during preparation. Tubes were incubated at room temperature (20.5 to 25.0°C) for 10 min for UNG activity to take place. Thermocycling and detection were performed on the LightCycler (Roche). An initial preheating step of 10 min at 95°C was used to activate the DNA polymerase, inactivate UNG, and melt double-stranded DNA. Then, a touch-down procedure followed, consisting of 5 s at 95°C, annealing for 10 s at temperatures decreasing from 65 to 50°C during the first 11 cycles (with 1°C decremental steps in cycles 2 to 6 and 2°C decremental steps in cycles 7 to 11), and ending with an extension step at 72°C for 20 s. A total of 46 cycles were performed. All patient samples were analyzed in triplicate, with one of three tubes containing the internal control.
Amplicon detection.
FRET detection probes were designed for both the MSG and the mimic. Each pair of FRET probes was designed to anneal with a one-base gap between the probes. For MSG detection the following probes were commercially synthesized (Genset, La Jolla, Calif.) and labeled with Red 640 as the reporter: PCMSGFRET1U (5"-CAA AAA TAA CAY TSA CAT CAA CRA GGC G-fluorescein-3") and PCMSGFRET1D (5"-Red 640-TGC AAA CCA ACC AAG TGT ACG ACA GG-3"). For mimic detection the following probes labeled with Red 705 as the reporter were utilized: PCMIM1U (5"-G ATA TCG TCC ATT CCG ACA GCA TC-fluorescein-3") and PCMIM1D (5"-Red 705- C CAG TCA CTA TGG CGT GCT GCT AG-3").
Data analysis.
All acquired fluorescence data were analyzed using LightCycler software. In theory, true quantification is achieved with real-time PCR, since the cycle number in which amplicons become detectable is proportional to the logarithm of initial number of templates. In each experiment at least three standards of cloned template (105, 103, and 10 copies/μl of specimen) were included to generate a standard curve for quantification of positive patient samples. For quantification of PCR-positive patient specimens, the means of tubes not containing mimic were recorded, since the concurrent mimic-amplification might interfere with the accuracy of the quantification.
Fisher's exact test was used to compare results obtained with and without the touch-down protocol. Wilcoxon's two-sample test was used to compare quantitative results (copies/tube) from clinical false positive to true positive samples. A two-sided P value of <0.05 was considered significant.
Interpretation.
A sample was regarded as positive if at least two of three tubes were positive by this assay. A positive result in only one tube warranted a retest of a new aliquot of the sample. If the retest had at least one of three tubes positive, the sample was considered positive for P. carinii. A negative MSG result had to have a positive result for the mimic to be considered valid, to ensure absence of inhibitors in the specimen. Presence of PCR inhibitors warranted retesting (reextraction and reamplification) of an additional aliquot of the original sample.
RESULTS
All negative controls (water and processed BAL samples) and positive controls (processed BAL samples) were appropriate. All 25 oral washes from healthy control subjects were negative. Inhibition detected by lack of mimic amplification occurred in 4 of 100 patient samples (4%). Two inhibited samples (both oral washes) had to be excluded from the analysis, since no additional specimen was available for retesting. Inhibition could be eliminated in the remaining two inhibited samples by processing and assaying new aliquots of the original specimens.
Touch-down PCR.
In developing the assay, a standard (non-touch-down) protocol using exactly the same conditions and materials as described, except that the annealing temperature was 50°C through all cycles, was initially investigated. However, the assay was less sensitive than desired. Therefore, the described touch-down protocol was designed and evaluated through a side-by-side comparison to the standard non-touch-down protocol. Twenty-four tubes with five copies (1 copy/μl of specimen) of cloned MSG templates per tube were analyzed in six separate experiments. All 12 tubes analyzed by using the touch-down protocol were positive, whereas only 7 of 12 tubes were positive by using the standard protocol ([two-sided] P < 0.04).
Quantification.
Performance of the touch-down assay for quantification was evaluated by analyzing 10-fold serial dilutions of the cloned MSG template. Standard curves (cycle number versus log concentration) were constructed by linear regression, and calculated values for standards and clinical specimens were recorded. Quantitative results for all 261 standards analyzed in this study are shown as means ± 1.96 standard deviations (SD) corresponding to 95% confidence intervals (Table 1). All standards with 0.01 copies/μl were negative, and all except 1 of 33 standards with 1 copy/μl were positive, indicating a sensitivity of one to five copies per tube. All standards with ≥10 copies/μl of specimen (n = 195) were positive, and the calculated concentration deviated 15% or less from the expected absolute value for 193 (99%) of them, as reflected by the means and SD (Table 1).
TABLE 1.
Quantitative results of all 261 standards analyzed
| Standards (copies/μl) | n | No. of positive reactions (%) | Mean calculated concna (1.96 SD) (copies/μl) |
|---|---|---|---|
| 0.01 | 12 | 0 (0) | |
| 0.1 | 21 | 3 (14) | 0.8 |
| 1 | 33 | 32 (97) | 1.6 (1.8) |
| 10 | 60 | 60 (100) | 10.0 (1.2) |
| 100 | 11 | 11 (100) | 101 (9) |
| 1,000 | 58 | 58 (100) | 994 (126) |
| 10,000 | 8 | 8 (100) | 10,800 (900) |
| 100,000 | 58 | 58 (100) | 99,700 (8,200) |
Mean calculated concentration for each known standard concentration.
Patient specimens.
Forty-nine oral washes and 49 lower respiratory tract specimens (30 BAL and 19 induced sputa), from 51 patients with 24 episodes of PCP and 34 episodes of other respiratory disease, were analyzed. Quantitative, touch-down (QTD) PCR results with and without a retrospective arbitrary cutoff of 10 MSG copies/tube applied are presented in Table 2 with conventional qualitative MSG PCR results as references. There was concordance with regard to the QTD PCR assay without cutoff and the conventional qualitative MSG PCR assay in 92 of 98 samples (94%). All tubes that tested positive by this QTD PCR assay had evidence of being biological true positives, since independently processed samples from the same episodes had previously tested positive by conventional qualitative PCR and/or by smear.
TABLE 2.
QTD PCR assay results with and without cutoff value of 10 MSG copies/tube applieda
| Specimen and assay | No. of PCR-positive specimens/total no. of PCP samples | No. of PCR-positive specimens/total no. of non-PCP samples |
|---|---|---|
| Lower respiratory tract (n = 49) | ||
| QTD PCR | 19/19 | 10/30 |
| QTD PCR > 10 copies | 16/19 | 2/30 |
| Conventional MSG PCR | 18/19 | 11/30 |
| Oral wash (n = 49) | ||
| QTD PCR | 17/18 | 3/31 |
| QTD PCR > 10 copies | 15/18 | 0/31 |
| Conventional MSG PCR | 18/18 | 6/31 |
Conventional qualitative MSG PCR assay results are given for reference. Concordance between the QTD PCR assay without cutoff and the conventional qualitative MSG PCR assay was 94% (92 of 98 samples).
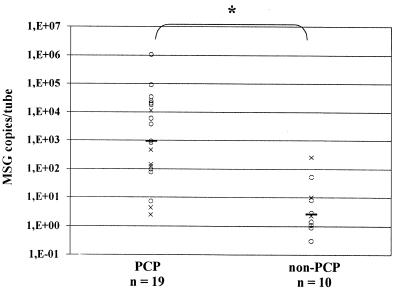
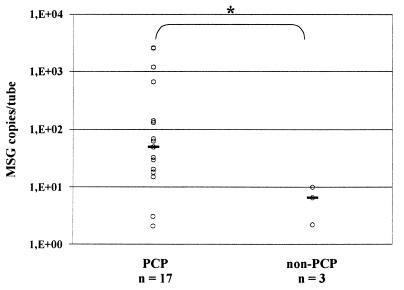
Quantification of the P. carinii positive samples revealed that lower respiratory tract samples and oral washes from non-PCP patients contained significantly lower concentrations of P. carinii DNA than samples from PCP patients (Fig. 1 and 2). PCR-positive lower respiratory tract samples from PCP and non-PCP patients contained 938 (2.4 to 1,040,000) and 2.6 (0.3 to 248) (P < 0.0004) copies/tube (values presented are medians, with ranges shown in parentheses), respectively. PCR positive oral washes from PCP and non-PCP patients contained 49 (2.1 to 2,595) and 6.5 (2.2 to 10) (P < 0.03) copies/tube, respectively. A total of only nine induced sputum samples were PCR positive, and analyzing differences between PCP and non-PCP patients in quantitative results from PCR positive induced sputa separately did not reach the level of significance (Fig. 1).
FIG. 1.
QTD PCR-positive lower respiratory samples from PCP and non-PCP patients. Symbols: ○, BAL fluid; ×, induced sputum; —, median; ∗, two-sided P < 0.0004.
FIG. 2.
QTD PCR-positive oral wash samples from PCP and non-PCP patients. Symbols: ○, oral washes; —, median; ∗, two-sided P < 0.03.
DISCUSSION
A rapid, sensitive and QTD real-time PCR assay for P. carinii, with an internal control for detecting PCR inhibitors and applicable for routine clinical testing was developed. This QTD PCR assay has the ability to detect P. carinii in oral washes or lower respiratory tract specimens. The results suggest that quantitative PCR with application of cutoff values could improve the specificity of previously described qualitative PCR, by distinguishing between colonization and infection.
It has been shown that touch-down protocols increase both the specificity and sensitivity of PCR assays (11). At relatively low annealing temperatures the formation of spurious PCR products might interfere with amplification of a specific target in low-copy-number samples. On the other hand, the FRET signal is suboptimal at higher annealing temperatures. Therefore, this touch-down procedure was designed to achieve highly stringent conditions in the first few cycles, where simultaneous FRET detection is not needed, to ensure efficient and specific amplification in low-copy-number samples. The annealing temperature is gradually lowered to a level that increases yield and is optimal for FRET detection. A comparison of the described touch-down procedure to the non-touch-down protocol for detection of low-copy-number samples (five MSG copies/tube) demonstrated a significant increase in sensitivity with touch down. The sensitivity of the assay was comparable to an optimized conventional qualitative in-house MSG assay (data not shown), and was determined, by serial dilutions of cloned target, to be five or fewer copies of the MSG gene of which there are multiple copies per genome (8, 16).
Inclusion of an internal PCR control has been suggested as necessary, in assays such as these, in order to detect false-negative results due to the presence of PCR inhibitors in the specimens (12, 23, 24, 37). Simultaneous amplification and detection, with separate FRET probes, of a mimic internal control and the P. carinii target were implemented in addition to traditional positive and negative external controls.
No manipulations of amplicons are required at any step during real-time PCR. This is desirable for routine testing in clinical laboratories, since it lessens the possibility of amplicon contamination problems. To reduce further the contamination risk, UNG and dUTP were used for amplicon carryover prevention (39). Use of a simple commercial DNA extraction kit also enhances the utility of the assay for clinical laboratories (7). The entire assay, including DNA extraction, has a turnaround time of <4 h, further increasing the potential clinical utility of this procedure.
It was determined, that quantification by this assay is reliable and reproducible, especially for tubes containing ≥50 MSG gene copies (i.e., ≥10 MSG copies/μl) (Table 1). In addition to assays for other infectious agents or host mutations in clinical specimens, the enhanced performance of this QTD real-time assay design could be useful in some research settings where precise quantification is warranted (15). The traditional quantification method for P. carinii is enumeration of organisms by microscopic examination, which is difficult due to the organism’s growth in clusters.
There are multiple reports of detection of P. carinii DNA in respiratory specimens from immunocompromised patients without clinical PCP, suggesting colonization or subclinical infection (3, 10, 19-21, 23, 25, 26, 29-33, 36, 38, 41, 42). Although intriguing from an epidemiological point of view, this fact impairs the diagnostic value of the test, since the positive predictive value for PCP is lowered. Other investigators have previously theorized that a quantitative approach could solve this problem (28).
To evaluate the performance of the assay and the utility of quantification, a blinded study on selected respiratory samples previously examined by conventional qualitative PCR was conducted. QTD PCR detection of P. carinii DNA in respiratory samples was compared to microscopic detection of P. carinii in induced sputum and BAL fluid as the diagnostic standard.
P. carinii DNA was detected in all 19 lower respiratory tract specimens and 17 of 18 oral washes from PCP episodes by this assay. Only 16 of the lower respiratory tract samples were positive by smear. For the three smear false-negative results, PCP was diagnosed on the basis of a smear of another specimen from the same episode. P. carinii DNA was detected in 10 of 30 lower respiratory tract specimens and 3 of 31 oral washes from non-PCP episodes. This study was designed to evaluate the assay and the utility of quantification. Therefore, calculation of sensitivity, specificity, and positive and negative predictive values in this selected series is not meaningful. However, QTD PCR and QTD PCR of >10 is more sensitive than DFA smears, since oral washes from PCP patients are negative by DFA (7a). The amount of DNA detected in both lower respiratory tract samples and oral washes, was significantly lower in the clinical false-positive group than in the PCP group in this series (Fig. 1 and 2). This is in concordance with the hypothesis of lower organism burden in colonized or subclinically infected patients as compared to patients with PCP. Applying a retrospective arbitrary cutoff value of 10 MSG copies per tube reduces the number of false-positive results of lower respiratory tract samples and oral washes from 10 and 3 to 2 and 0, respectively, while the number of false-negative results increases slightly from 0 and 1 to 3 and 3, respectively (Table 2; Fig. 1 and 2).
In conclusion, the preliminary results discussed here indicate that this assay has a high sensitivity for diagnosing PCP, even when applied to noninvasive oral wash specimens. Furthermore, the results of this study suggest that quantification and application of a cutoff value could have a role in distinguishing between colonized or subclinically infected patients and PCP patients. Further investigation and prospective studies are needed to confirm these findings, to establish an interpretative cutoff level, and to evaluate the specificity and sensitivity of the assay in different patient populations.
Acknowledgments
Hans Henrik Larsen was supported by the Danish Medical Research Council (grant 9900484).
We acknowledge Ellen Townley for her critical role in providing the patient specimens from Washington Hospital Center.
REFERENCES
- 1.Aztori, C., F. Agostini, E. Angeli, A. Mainini, G. Orlando, and A. Cargnel. 1998. Combined use of blood and oropharyngeal samples for noninvasive diagnosis of Pneumocystis carinii pneumonia using the polymerase chain reaction. Eur. J. Clin. Microbiol. Infect. Dis. 17:241-246. [DOI] [PubMed] [Google Scholar]
- 2.Aztori, C., E. Angeli, F. Agostini, A. Mainini, V. Micheli, and A. Cargnel. 1999. Biomolecular techniques to detect Pneumocystis carinii f. sp. hominis pneumonia in patients with acquired immunodeficiency syndrome. Int. J. Infect. Dis. 3:76-81. [DOI] [PubMed] [Google Scholar]
- 3.Calderon, E. J., C. Regordan, F. J. Medrano, M. Ollero, and J. M. Varela. 1996. Pneumocystis carinii infection in patients with chronic bronchial disease. Lancet 347:977.. [DOI] [PubMed] [Google Scholar]
- 4.Cartwright, C. P., N. A. Nelson, and V. J. Gill. 1994. Development and evaluation of a rapid and simple procedure for detection of Pneumocystis carinii by PCR. J. Clin. Microbiol. 32:1634-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chouaid, C., P. Roux, I. Lavard, J. L. Poirot, and B. Housset. 1995. Use of the polymerase chain reaction technique on induced-sputum samples for the diagnosis of Pneumocystis carinii pneumonia in HIV-infected patients. Am. J. Clin. Pathol. 104:72-75. [DOI] [PubMed] [Google Scholar]
- 6.Elvin, K. M., A. Bjorkman, E. Linder, N. Heurlin, and A. Hjerpe. 1988. Pneumocystis carinii pneumonia: detection of parasites in sputum and bronchoalveolar lavage fluid by monoclonal antibodies. Br. Med. J. 297:381-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fahle, G., and S. H. Fischer. 2000. Comparison of six commercial DNA extraction kits for recovery of cytomegalovirus DNA from spiked human specimens. J. Clin. Microbiol. 38:3860-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Fischer, S., V. J. Gill, J. Kovacs, P. Miele, J. Keary, V. Silcott, S. Huang, L. Borio, F. Stock, G. Fahle, D. Brown, B. Hahn, E. Townley, D. Lucey, and H. Masur. 2001. The use of oral washes to diagnose Pneumocystis carinii pneumonia: a blinded prospective study using a polymerase chain reaction-based detection system. J. Infect. Dis. 184:1485-1488. [DOI] [PubMed] [Google Scholar]
- 8.Garbe, T. R., and J. R. Stringer. 1994. Molecular characterization of clustered variants of genes encoding major surface antigens of human Pneumocystis carinii. Infect. Immun. 62:3092-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gill, V. J., G. Evans, F. Stock, J. E. Parrillo, H. Masur, and J. A. Kovacs. 1987. Detection of Pneumocystis carinii by fluorescent-antibody stain using a combination of three monoclonal antibodies. J. Clin. Microbiol. 25:1837-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauser, P. M., D. S. Blanc, J. Bille, A. Nahimana, and P. Francoli. 2000. Carriage of Pneumocystis carinii by immunosuppressed patients and molecular typing of the organisms. AIDS 14:461-463. [DOI] [PubMed] [Google Scholar]
- 11.Hecker, K. H., and K. H. Roux. 1996. High and low annealing temperatures increase both specificity and yield in touchdown and stepdown PCR. BioTechniques 20:478-485. [DOI] [PubMed] [Google Scholar]
- 12.Helweg-Larsen, J., J. S. Jensen, T. Benfield, U. G. Svendsen, J. D. Lundgren, and B. Lundgren. 1998. Diagnostic use of PCR for detection of Pneumocystis carinii in oral wash samples. J. Clin. Microbiol. 36:2068-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helweg-Larsen, J., J. S. Jensen, and B. Lundgren. 1997. Non-invasive diagnosis of Pneumocystis carinii pneumonia by PCR on oral washes. Lancet 350:1363. [DOI] [PubMed] [Google Scholar]
- 14.Huang, S. N., S. H. Fischer, E. O'Shaugnessy, V. J. Gill, H. Masur, and J. A. Kovacs. 1999. Development of a PCR assay for diagnosis of Pneumocystis carinii pneumonia based on amplification of the multicopy major surface glycoprotein gene family. Diagn. Microbiol. Infect. Dis. 35:27-32. [DOI] [PubMed] [Google Scholar]
- 15.Kaiser, K., M. Rabodonirina, and S. Picot. 2001. Real time quantitative PCR and RT-PCR for analysis of Pneumocystis carinii hominis. J. Microbiol. Methods 45:113-118. [DOI] [PubMed] [Google Scholar]
- 16.Kovacs, J. A., F. Powell, J. C. Edman, B. Lundgren, A. Martinez, B. Drew, and C. W. Angus. 1993. Multiple genes encode the major surface glycoprotein of Pneumocystis carinii. J. Biol. Chem. 268:6034-6040. [PubMed] [Google Scholar]
- 17.Kovacs, J. A., V. Gill, J. C. Swan, F. Ognibene, J. Shelhamer, J. E. Parillo, and H. Masur. 1986. Prospective evaluation of a monoclonal antibody in diagnosis of Pneumocystis carinii pneumonia. Lancet ii:1-3. [DOI] [PubMed] [Google Scholar]
- 18.Kovacs, J. A., V. L. Ng, H. Masur, G. Leoung, W. K. Hadley, G. Evans, C. Lane, F. P. Ognibene, J. Shelhamer, J. E. Parillo, and V. J. Gill. 1988. Diagnosis of Pneumocystis carinii pneumonia: improved detection in sputum with use of monoclonal antibodies. N. Engl. J. Med. 318:589-593. [DOI] [PubMed] [Google Scholar]
- 19.Leigh, T. R., A. E. Wakefield, S. E. Peters, J. M. Hopkin, and J. V. Collins. 1992. Comparison of DNA amplification and immunofluorescence for detecting Pneumocystis carinii in patients receiving immunosuppressive therapy. Transplantation 54:468-470. [DOI] [PubMed] [Google Scholar]
- 20.Leigh, T. R., H. O. Kangro, B. G. Gazzard, D. F. Jeffries, and J. V. Collins. 1993. DNA amplification by the polymerase chain reaction to detect sub-clinical Pneumocystis carinii colonization in HIV-positive and HIV-negative male homosexuals with and without respiratory symptoms. Respir. Med. 87:525-529. [DOI] [PubMed] [Google Scholar]
- 21.Lipschik, G., V. J. Gill, J. D. Lundgren, V. A. Andrawis, N. A. Nelson, J. O. Nielsen, F. P. Ognibene, and J. A. Kovacs. 1992. Improved diagnosis of Pneumocystis carinii infection by polymerase chain reaction on induced sputum and blood. Lancet 340:203-206. [DOI] [PubMed] [Google Scholar]
- 22.Lu, J.-J., C.-H. Chen, M. S. Bartlett, J. W. Smith, and C.-H. Lee. 1995. Comparison of six different PCR methods for detection of Pneumocystis carinii. J. Clin. Microbiol. 33:2785-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lundgren, B., and A. E. Wakefield. 1998. PCR for detecting diagnosing Pneumocystis carinii in clinical or environmental samples. FEMS Immunol. Med. Microbiol. 22:97-101. [DOI] [PubMed] [Google Scholar]
- 24.Mathis, A., R. Weber, H. Kuster, and R. Speich. 1997. Simplified sample processing combined with a sensitive one-tube nested PCR assay for detection of Pneumocystis carinii in respiratory specimens. J. Clin. Microbiol. 35:1691-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nevez, G., C. Raccurt, V. Jounieaux, E. Dei-Cas, and E. Mazars. 1999. Pneumocystosis versus pulmonary Pneumocystis carinii colonization in HIV-negative and HIV-positive patients. AIDS 13:535-536. [DOI] [PubMed] [Google Scholar]
- 26.Olsson, M., K. Elvin, S. Lofdahl, and E. Linder. 1993. Detection of Pneumocystis carinii DNA in sputum and bronchoalveolar lavage samples by polymerase chain reaction. J. Clin. Microbiol. 31:221-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oz, H. S., and W. T. Hughes. 2000. Search for Pneumocystis carinii DNA in upper and lower respiratory tract of humans. Diagn. Microbiol. Infect. Dis. 37:161-164. [DOI] [PubMed] [Google Scholar]
- 28.Peters, S. E., A. E. Wakefield, S. Banerji, and J. M. Hopkin. 1992. Quantification of the detection of Pneumocystis carinii by DNA amplification. Mol. Cell. Probes 6:115-117. [DOI] [PubMed] [Google Scholar]
- 29.Probst, M., H. Ries, T. Schmidt-Wieland, and A. Serr. 2000. Detection of Pneumocystis carinii DNA in patients with chronic lung diseases. Eur. J. Clin. Microbiol. Infect. Dis. 19:644-645. [DOI] [PubMed] [Google Scholar]
- 30.Rabodonirina, M., D. Raffenot, L. Cotte, A. Boibieux, M. Mayencon, G. Bayle, F. Persat, F. Rabatel, C. Trepo, D. Peyramond, and M.-A. Piens. 1997. Rapid detection of Pneumocystis carinii in bronchoalveolar lavage specimens from human immunodeficiency virus-infected patients: use of a simple DNA extraction procedure and nested PCR. J. Clin. Microbiol. 35:2748-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ribes, J. A., A. H. Limper, M. J. Espy, and T. F. Smith. 1997. PCR detection of Pneumocystis carinii in bronchoalveolar lavage specimens: analysis of sensitivity and specificity. J. Clin. Microbiol. 35:830-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sing, A., A. Roggenkamp, I. B. Autenrieth, and J. Heesemann. 1999. Pneumocystis carinii carriage in immunocompetent patients with primary pulmonary disorders as detected by single or nested PCR. J. Clin. Microbiol. 37:3409-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sing, A., K. Trebesius, A. Roggenkamp, H. Russmann, K. Tybus, F. Pfaff, J. R. Bogner, C. Emminger, and J. Heesemann. 2000. Evaluation of diagnostic value and epidemiological implications of PCR for Pneumocystis carinii in different immunosuppressed and immunocompetent patient groups. J. Clin. Microbiol. 38:1461-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skøt, J., A. G. Lerche, H. J. Kolmos, J. O. Nielsen, L. R. Mathiesen, and J. D. Lundgren. 1995. Pneumocystis carinii in bronchoalveolar lavage and induced sputum: detection with a nested polymerase chain reaction. Scand. J. Infect. Dis. 27:363-367. [DOI] [PubMed] [Google Scholar]
- 35.Sobck, H., M. Schmidt, B. Frey, and K. Kaluza. 1996. Heat-labile uracil-DNA glycosylase: purification and characterization. FEBS Lett. 388:1-4. [DOI] [PubMed] [Google Scholar]
- 36.Torres, J., M. Goldman, L. J. Wheat, X. Tang, M. S. Bartlett, J. W. Smith, S. D. Allen, and C.-H. Lee. 2000. Diagnosis of Pneumocystis carinii pneumonia in human immunodeficiency virus-infected patients with polymerase chain reaction: a blinded comparison to standard methods. Clin. Infect. Dis. 30:141-145. [DOI] [PubMed] [Google Scholar]
- 37.Ursi, J.-P., D. Ursi, M. Ieven, and S. R. Pattyn. 1992. Utility of an internal control for the polymerase chain reaction: application to detection of Mycoplasma pneumoniae in clinical specimens. APMIS 100:635-639. [DOI] [PubMed] [Google Scholar]
- 38.Wakefield, A. E., F. J. Pixley, S. Banerji, K. Sinclair, R. F. Miller, E. R. Moxon, and J. M. Hopkin. 1990. Detection of Pneumocystis carinii with DNA amplification. Lancet 336:451-453. [DOI] [PubMed] [Google Scholar]
- 39.Wakefield, A. E., L. A. Guiver, R. F. Miller, and J. M. Hopkin. 1991. DNA amplification on induced sputum samples for diagnosis of Pneumocystis carinii pneumonia. Lancet 337:1378-1379. [DOI] [PubMed] [Google Scholar]
- 40.Wakefield, A. E., R. F. Miller, L. A. Guiver, and J. M. Hopkin. 1993. Oropharyngeal samples for detection of Pneumocystis carinii by DNA amplification. Q. J. Med. 86:401-406. [PubMed] [Google Scholar]
- 41.Weig, M., H. Klinker, B. H. Bogner, A. Meier, and U. Gross. 1997. Usefulness of PCR for diagnosis of Pneumocystis carinii pneumonia in different patient groups. J. Clin. Microbiol. 35:1445-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weig, M., H. Klinker, M. Wilhelm, K. Lemmer, and U. Gross. 1996. Correlation of Pneumocystis carinii PCR with clinical diagnosis in immunocompromised patients. Lancet 347:1266.. [DOI] [PubMed] [Google Scholar]