Abstract
Chlamydia pneumoniae and Mycoplasma pneumoniae were evaluated as agents of persistent cough in adolescents and adults (n = 491). Tests of 473 respiratory specimens by culture or PCR or both identified four episodes (0.8%) of M. pneumoniae-associated illness and no episodes of C. pneumoniae illness, suggesting that these bacteria do not frequently cause persistent cough.
In the United States, adults frequently seek medical attention for a persistent cough (9), a symptom often associated with Chlamydia pneumoniae and Mycoplasma pneumoniae infections. This study evaluated the frequency with which these bacteria cause persistent cough in adolescents and adults by using molecular, culture, and serologic techniques.
Healthy subjects (n = 491) were enrolled at seven centers in the United States between September 1997 and December 1998 as part of an acellular pertussis vaccine efficacy study approved by an internal review board. The subjects were monitored for up to 2 years for a nonimproving cough lasting 5 days or longer. The subjects had a median age of 35.4 years (range, 15 to 65 years); 64.4% were females, 66.4% were Caucasians, and 28.9% were African-Americans. Throat swab specimens were obtained at the baseline visit and at each visit during illness (illness visit) and a nasopharyngeal aspirate was obtained at each illness visit. The specimens were suspended in transport medium (6, 7, 12) and stored at −80°C. Cultures for M. pneumoniae and C. pneumoniae were done as described previously (6, 7, 12).
A multiplex PCR assay was designed using primers for amplification of a 438-bp target (internal control) in the β-actin gene of human DNA (16) and primer sets MP1-MP2 and P4A-P4E (1) for amplification of 124- and 230-bp targets, respectively, within the P1 cytadhesion gene of M. pneumoniae. Probes HAC7, MP7, and P4C are complementary to the respective targets. Primers CP10 and CP11 were designed to amplify a 323-bp target in the C. pneumoniae-specific 474-bp PstI restriction fragment (2); probe CP13 is complementary to this target. The oligonucleotides unique to this study and their 5" → 3" nucleotide sequences were as follows: HAC7, GCCATGTACGTTGCTATCCAGGCTGTGCTATCCCTG; MP1, GTGGGACACTTCACAAGTACC; MP2, GATACGTTCACGGGGTTAAGC; MP7, AATTTAGCTACACCCGCCCTGACGAGGTCGC; P4A, AGGCTCAGGTCAATCTGGCGTGGA; P4C, TGGGGCAGTTACCAAGCACGAGTGACGGAA; CP10, CAGATTACGAAACGGCATTAC; CP11, TGCTGCATAACCTACGGTGTG; and CP13, GGTGTCATTCGCCAAGGTTAAAGTCTACG. Sample lysates were prepared as described previously (11). DNA was amplified in 100-μl reaction mixtures containing a 200 μM concentration (each) of dATP, dUTP, dCTP, and dGTP, 2.0 U of AmpliTaq DNA polymerase, 1.0 U of uracil-N"-glycosylase, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, and 25 μl of sample. The reaction mixtures for the multiplex PCR assay contained 3.5 mM MgCl2, 1 ng of CEM cell DNA (16), a 0.25 μM concentration (each) of primers HAC3 and HAC5, and a 0.1 μM concentration (each) of primers MP1, MP2, P4A, and P4E. Reaction mixtures for amplification of C. pneumoniae DNA included 3.75 mM MgCl2 and a 0.25 μM concentration (each) of primers CP10 and CP11. The thermal cycling conditions used were described previously (14), except that a 55°C annealing temperature and 40 PCR cycles were used in the C. pneumoniae assay. PCR products were detected by a liquid hybridization gel retardation assay using 32P-labeled DNA probes, a highly sensitive platform (15) that avoids the use of cumbersome washing steps needed with colorimetric systems.
The PCR assays were initially evaluated by testing purified DNAs (16). Single copies of M. pneumoniae DNA (strain ATCC 15531) and C. pneumoniae DNA (strain ATCC 2282-VR [TW-183]) were detected in PCRs with 1 μg (approximately 300,000 copies) of human DNA (CEM cell line). Tests of the human DNA (i.e., 1 μg/reaction) did not yield any M. pneumoniae or C. pneumoniae PCR products or any nonspecific PCR products as determined by agarose gel electrophoretic analysis. None of the DNAs from two panels (n = 65 and n = 85) of nontarget microorganisms (Table 1), mostly isolated from the respiratory tract, yielded PCR products in the multiplex or C. pneumoniae assays, respectively. These results indicate that primers for M. pneumoniae and C. pneumoniae are sensitive, specific, and do not react with human DNA, results which are consistent with those of previous studies demonstrating high specificity of the C. pneumoniae PstI fragment (2).
TABLE 1.
Microorganisms used in the specificity studies
| Microorganism | Origina | No. of strains tested in assay
|
|
|---|---|---|---|
| Multiplex PCR | C. pneumoniae PCR | ||
| Acinetobacter baumannii | CI | 1 | |
| Candida albicans | CI | 5 | 7 |
| Chlamydia trachomatis | CI | 1 | |
| Corynebacterium species | CI | 5 | 7 |
| Enterobacter cloacae | CI | 1 | |
| Enterococcus species | CI | 1 | |
| Escherichia coli | CI | 1 | |
| Haemophilus influenzae | CI | 5 | 14 |
| Haemophilus parainfluenzae | CI | 3 | 8 |
| Klebsiella pneumoniae | CI | 1 | |
| Legionella pneumophila | CI | 1 | |
| Moraxella catarrhalis | CI | 5 | 6 |
| Mycoplasma fermentans | ATCC 19989 | 1 | 1 |
| Mycoplasma genitalium | ATCC 33530 | 1 | |
| Mycoplasma orale | ATCC 23714 | 1 | |
| Mycoplasma salivarium | ATCC 23064 | 1 | |
| Neisseria mucosa | CI | 2 | 2 |
| Neisseria species other than N. meningitidis and N. gonorrhoeae | CI | 3 | 4 |
| Proteus vulgaris | CI | 1 | |
| Pseudomonas aeruginosa | CI | 1 | |
| Salmonella enterica subsp. I | CI | 1 | |
| Staphylococcus aureus | CI | 5 | 6 |
| Staphylococcus species, coagulase negative | CI | 5 | 6 |
| Streptococcus pneumoniae | CI | 5 | 7 |
| Streptococcus pyogenes | CI | 5 | 7 |
| Streptococcus species, β-hemolytic group C | CI | 1 | |
| Streptococcus species, viridans group | CI | 5 | 7 |
CI, clinical isolate.
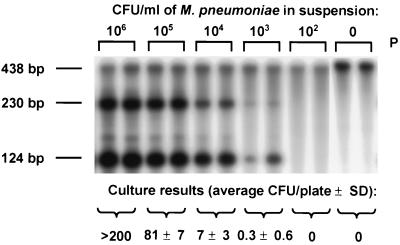
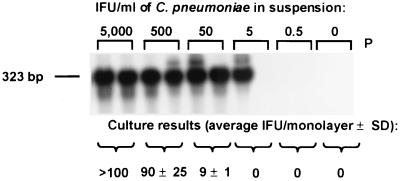
The detection limits of the PCR assays were compared to those of culture assays (6, 7, 12) by testing swabs which had been seeded with suspensions of M. pneumoniae and C. pneumoniae and processed as described previously (14). The detection limits of the two assays were nearly identical (Fig. 1 and 2). Because M. pneumoniae and C. pneumoniae survive poorly outside the human host, in situations where specimens are not processed promptly and carefully, the PCR assays may be more sensitive than the culture assays.
FIG. 1.
Comparison of the results of culture assay and the multiplex PCR assay for the detection of M. pneumoniae in seeded swab samples. The β-actin PCR product (i.e., internal control) (438 bp) and the M. pneumoniae PCR products (230 and 124 bp) are indicated. P, probe control; SD, standard deviation.
FIG. 2.
Comparison of the results of the PCR and culture assays for the detection of C. pneumoniae in seeded swab samples. IFU, inclusion-forming units. The C. pneumoniae PCR product (323 bp) is indicated. P, probe control; SD, standard deviation.
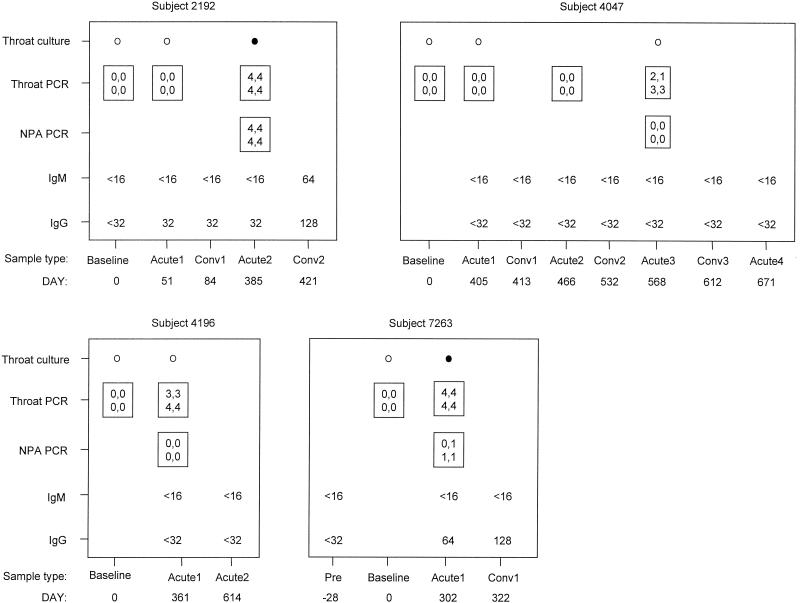
During the subject follow-up period, 516 episodes of persistent cough were identified (incidence rate = 0.75 episode/person year). M. pneumoniae was detected by PCR in 4 of 473 (0.8%) throat swab specimens collected at illness visits but not in any of 437 throat swab specimens collected at baseline visits and by culture in 2 of 350 (0.6%) throat swab specimens collected at illness visits but not in any of 457 throat swab specimens collected at baseline visits. Two of four subjects with an M. pneumoniae-associated cough illness had ≥4-fold increases in anti-M. pneumoniae immunoglobulin G antibody titer measured by microimmunofluorescence (3), and M. pneumoniae was detected in the nasopharynges of these two subjects by PCR (Fig. 3). Totals of 54 and 458 throat swab specimens obtained at baseline visits, 416 and 411 throat swab specimens obtained at illness visits, and 414 and 396 nasopharyngeal aspirate specimens obtained at illness visits were evaluated for C. pneumoniae by the PCR and culture assays, respectively. All of the specimens yielded negative results. A limitation in funding precluded the testing of all of the collected specimens by the different assays.
FIG. 3.
Detection of M. pneumoniae by the PCR and culture assays and serologic assessment of infections in the four subjects with an M. pneumoniae-associated illness. Results are categorized by study day relative to the baseline (day 0) visit and by the following sample types: acute, sample obtained at a visit during an acute illness; conv, sample obtained at a visit during convalescence. The number following the sample type refers to the number of the consecutive illness visit for each subject. NPA, nasopharyngeal aspirate; ○, negative culture result; •, positive culture result. Boxes with four numbers are intensity results for the PCR-based assay: the values in the upper row correspond to the intensities of the 230-bp M. pneumoniae band in the duplicate assays, and the values in the lower row correspond to the intensities of the 124-bp M. pneumoniae band in the duplicate assays. Band intensity ratings: 0, not visible; 1, barely visible; 2, dim but definite; 3, dark (approximately 75% of maximum intensity); and 4, maximum intensity.
Initially, 17% of specimens (n = 910) tested in the multiplex PCR yielded 438-bp control bands with 50 to 75% reductions in intensity. However, repeat testing yielded bands with maximum intensities. Because earlier studies in our laboratory showed that the β-actin amplification control system had an adverse effect on amplification of the C. pneumoniae DNA target, this internal control was not included in the C. pneumoniae PCR assay. Instead, inhibition was assessed by random testing of 150 specimens in PCR mixtures containing 1,000 copies of C. pneumoniae DNA. Only one of these samples was negative, but repeat testing yielded a C. pneumoniae band of maximum intensity. Technical variability in sample processing or in probe hybridization may have accounted for the different results observed in the initial and repeat assays. Overall, these studies confirmed that the specimen processing procedure (11) provides material suitable for PCR studies.
Paired sera from the first cough illness visits of 99 subjects with negative results from PCR and culture studies were evaluated by microimmunofluorescent-antibody tests (3, 8). There were no increases in anti-M. pneumoniae IgG antibody titers of ≥4-fold or any positive anti-M. pneumoniae IgM titers. There were no increases in anti-C. pneumoniae IgA or IgG antibody titers of ≥4-fold or any detectable anti-C. pneumoniae IgM antibody titers. The seroprevalence rates of anti-M. pneumoniae IgG antibody (i.e., titer of ≥32) and anti-C. pneumoniae IgG antibody (i.e., titer of ≥8) were 38 and 74%, respectively, and the coseroprevalence rate of the two antibodies together was 27%.
Sensitive, specific, and robust PCR assays, as well as culture and serologic techniques, were utilized to assess M. pneumoniae and C. pneumoniae infections in adolescents and adults. The frequency of M. pneumoniae-associated cough illness was 0.8%, and no episodes of C. pneumoniae-associated illness were detected. The high seroprevalence rates of IgG antibodies to M. pneumoniae and C. pneumoniae identified in a subgroup of the study subjects suggest that many of the subjects had acquired some measure of protective immunity prior to the development of their first episode of persistent cough (10, 13). Accordingly, differences in M. pneumoniae and C. pneumoniae infection rates may be expected in other populations where the seroprevalence rates are lower or at times when the agents are more common in the community (4, 5).
In conclusion, the results of this study suggest that M. pneumoniae is an infrequent agent of cough illnesses that are present for 5 days or longer in adolescents and adults in the United States and that C. pneumoniae is of even less importance.
Acknowledgments
This work was supported by a National Institutes of Health subcontract with Harbor UCLA (AI-45249).
We thank Eduardo Yunis and Ronald Jaffe for encouragement and Thomas Grayston for serologic testing for C. pneumoniae.
REFERENCES
- 1.Cadieux, N., P. Lebel, and R. Brousseau. 1993. Use of a triplex polymerase chain reaction for the detection and differentiation of Mycoplasma pneumoniae and Mycoplasma genitalium in the presence of human DNA. J. Gen. Microbiol. 139:2431-2437. [DOI] [PubMed] [Google Scholar]
- 2.Campbell, L. A., M. P. Melgosa, D. J. Hamilton, C.-C. Kuo, and J. T. Grayston. 1992. Detection of Chlamydia pneumoniae by polymerase chain reaction. J. Clin. Microbiol. 30:434-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fedorko, D. P., D. D. Emery, S. M. Franklin, and D. D. Congdon. 1995. Evaluation of a rapid enzyme immunoassay system for serologic diagnosis of Mycoplasma pneumoniae infection. Diagn. Microbiol. Infect. Dis. 23:85-88. [DOI] [PubMed] [Google Scholar]
- 4.Foy, H. M. 1993. Infections caused by Mycoplasma pneumoniae and possible carrier state in different populations of patients. Clin. Infect. Dis. 17(Suppl. 1):S37-S46. [DOI] [PubMed] [Google Scholar]
- 5.Grayston, J. T., L. A. Campbell, C. C. Kuo, C. H. Mordhorst, P. Saikku, D. H. Thom, and S. Wang. 1990. A new respiratory tract pathogen: Chlamydia pneumoniae strain TWAR. J. Infect. Dis. 161:618-625. [DOI] [PubMed] [Google Scholar]
- 6.Kenny, G. E. 1991. Mycoplasmas, p. 478-482. In A. Balows, W. J. Hausler, K. L. Herrmann, H. D. Isenberg, and H. J. Shadomy (ed.), Manual of clinical microbiology, 5th ed. American Society for Microbiology, Washington, D.C.
- 7.Kenny, G. E., G. G. Kaiser, M. K. Cooney, and H. M. Foy. 1990. Diagnosis of Mycoplasma pneumoniae pneumonia: sensitivities and specificities of serology with lipid antigen and isolation of the organism on soy peptone medium for identification of infections. J. Clin. Microbiol. 28:2087-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuo, C.-C., L. A. Jackson, L. A. Campbell, and J. T. Grayston. 1995. Chlamydia pneumoniae (TWAR). Clin. Microbiol. Rev. 8:451-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metlay, J. P., R. S. Stafford, and D. E. Singer. 1998. National trends in the use of antibiotics by primary care physicians for adult patients with cough. Arch. Intern. Med. 158:1813-1818. [DOI] [PubMed] [Google Scholar]
- 10.Mogabgab, W. J. 1973. Protective efficacy of killed Mycoplasma pneumoniae vaccine measured in large-scale studies in a military population. Am. Rev. Respir. Dis. 108:899-908. [DOI] [PubMed] [Google Scholar]
- 11.Reznikov, M., T. K. Blackmore, J. J. Finlay-Jones, and D. L. Gordon. 1995. Comparison of nasopharyngeal aspirates and throat swab specimens in a polymerase chain reaction-based test for Mycoplasma pneumoniae. Eur. J. Clin. Microbiol. Infect. Dis. 14:58-61. [DOI] [PubMed] [Google Scholar]
- 12.Roblin, P. M., W. Dumornay, and M. R. Hammerschlag. 1992. Use of HEp-2 cells for improved isolation and passage of Chlamydia pneumoniae. J. Clin. Microbiol. 30:1968-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Surcel, H. M., H. Syrjälä, M. Leinonen, P. Saikku, and E. Herva. 1993. Cell-mediated immunity to Chlamydia pneumoniae measured as lymphocyte blast transformation in vitro. Infect. Immun. 61:2196-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wadowsky, R. M., S. Laus, T. Libert, S. J. States, and G. D. Ehrlich. 1994. Inhibition of PCR-based assay for Bordetella pertussis by using calcium alginate fiber and aluminum shaft components of a nasopharyngeal swab. J. Clin. Microbiol. 32:1054-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wadowsky, R. M., T. Libert, and G. D. Ehrlich. 1994. Detection of Bordetella pertussis using PCR, p. 621-632. In G. D. Ehrlich and S. J. Greenberg (ed.), PCR-based diagnostics in infectious disease. Blackwell Scientific Publications, Boston, Mass.
- 16.Wadowsky, R. M., R. H. Michaels, T. Libert, L. A. Kingsley, and G. D. Ehrlich. 1996. Multiplex PCR-based assay for detection of Bordetella pertussis in nasopharyngeal swab specimens. J. Clin. Microbiol. 34:2645-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]