Abstract
Candida samples were taken over a period of 2 years from 54 human immunodeficiency virus (HIV)-positive asymptomatic subjects to evaluate changes in yeast carriage, intensity of carriage, and genotype over time. Overall, we found that HIV-positive patients with CD4+-cell counts of between 200 and 400/μl had significantly more yeast colonization than healthy control subjects. Of the 54 patients, 11 developed thrush. We found that intensity of carriage in these 11 patients increased significantly in the progression from asymptomatic yeast carrier to an episode of oral thrush. Also, the most common yeast species isolated was Candida albicans; however, we did see a number of patients harboring multiple species at the same time. Using the C. albicans-specific probe Ca3, we found that 54% (n = 6) of the 11 patients who developed thrush maintained genetically similar strains throughout the study period, with minor genetic variations in all patients except one. Forty-six percent of these patients had either multiple strains throughout the study period (n = 2), strain replacement (n = 1), or species replacement (n = 2). Of the patients who had multiple strains, one (I4) was infected by two different strains of Candida dubliniensis distinguished by a recently developed species-specific probe. These results suggest that commensal strains colonizing HIV-positive individuals can undergo alterations prior to producing an episode of thrush.
Oropharyngeal Candida infections are the most common opportunistic diseases in human immunodeficiency virus (HIV)-infected individuals, occurring in up to 90% of patients during the course of their disease (15, 38, 39, 48). Studies have shown that symptom-free HIV-positive patients with CD4+-lymphocyte counts of less than 400 cells/μl have a 50% risk of progression to full-blown AIDS within 3 years. However, such patients who also have thrush have a 90% risk of progression in the same period (29).
The majority of candidal infections are caused by Candida albicans; however, reports of the emergence of other species of Candida have begun to appear (2, 25, 47). Barchiesi et al. described an increase in the frequency of isolation of non-C. albicans yeast species from 3 to 4% of isolates in 1988-1989 to 16 to 18% of isolates in 1990-1991 (2). Similarly, Morace et al. found that 25% of the yeast species isolated from persons with AIDS were non-C. albicans species (28). Masia Canuto et al. recently evaluated 153 HIV-positive patients and found that 21% of these patients had non-C. albicans species, the most common of which was Candida glabrata (23)
Asymptomatic oral C. albicans carriage has been demonstrated in HIV-positive patients (5), and an increased incidence of asymptomatic oral C. albicans carriage in HIV-positive patients compared to that in other at-risk groups has also been noted (4). Thus, a higher prevalence of oral C. albicans colonization may be a predisposing factor for the subsequent development of clinical thrush.
Despite the increased prevalence of C. albicans and the increased frequency of candidiasis in immunocompromised patient populations, little is known about the changes, if any, which may occur in C. albicans when HIV-positive patients develop AIDS, although several theories have been proposed. First, the infecting strain may simply represent the commensal strain colonizing the oral cavities prior to infection; second, the commensal strain and the infecting strain are not related; and third, the commensal strain and the infecting strain are similar but have undergone microevolution. The theory that the commensal strain and the infecting strain are the same has been assumed to be true for all types of Candida infections in HIV-negative, healthy individuals (33). With the development of methods for assessing genetic relatedness (e.g., DNA fingerprinting), it is possible to determine whether commensal strains or exogenously derived, more pathogenic strains are the disease-causing organisms in any immunocompromised individual (45, 46). Two studies have provided indirect evidence, suggesting that strain replacement can occur in some situations (7, 41). Schmid et al. compared oral isolates of C. albicans from 11 nonhospitalized AIDS patients suffering from recurrent episodes of oral thrush in Leicester, England, with oral isolates from a group of control individuals; they found that the genetic diversity among strains isolated from AIDS patients was significantly reduced compared with that among control strains (41).
Further evidence comes from the demonstration of C. albicans strain replacement in a DNA fingerprinting study of strains from male partners of women with vaginal candidiasis (42). It was found that the strains in the male partners were identical or highly related to the infecting strains from their female counterparts.
Similarly, Lockhart et al. assessed the genetic relatedness of C. albicans strains from 18 patients with recurrent infections; although they found strain maintenance to be the dominant scenario, 56% of these patients had minor genetic changes in the colonizing yeast strains (20).
On the other hand, Miyasaki et al. (27) used restriction fragment polymorphism analysis to compare the genotypic relatedness between strains of C. albicans isolated from HIV-infected patients with or without oral candidiasis and strains isolated from their sexual partners. They found that recurrent oral candidiasis in HIV-positive patients was usually caused by a single persistent strain unique to each patient (27).
Using the C. albicans repetitive element 2 (CARE-2) DNA fragment, Lischewski et al. found no “signature genotype” associated with samples of C. albicans isolated from AIDS patients in Wurzburg, Germany (16).
From the evidence presented, the most frequent scenarios of genetic relatedness of yeast strains isolated from HIV-positive individuals are strain maintenance with no genetic variation and strain replacement. These studies, however, evaluated patients who already had candidiasis or developed thrush very shortly after the first sample was obtained. Therefore, the purpose of this study was to evaluate Candida samples isolated longitudinally from asymptomatic HIV-positive individuals with no prior history of thrush. The results of this study demonstrate that for this population, the intensity of oral yeast carriage and the genetic relatedness of oral Candida strains varied over time.
MATERIALS AND METHODS
Collection of isolates.
Prior to initiation of this study, approval was obtained from the Human Subjects Committee at the University of Iowa. A total of 54 HIV-positive individuals voluntarily enrolled in the study between July 1994 and July 1996: 48 from the Infectious Diseases Clinic at The University of Iowa Hospitals and Clinics, Iowa City; 4 from The William Beaumont Army Hospital, Fort Bliss, El Paso, Tex.; and 2 from the Bering Dental Clinic, Houston, Tex. For comparison, 38 volunteer subjects who had been recruited from the general population of Iowa City, Iowa, were used as control subjects (13). All subjects were initially free of signs or symptoms of oral candidiasis or other mucosal diseases, and all HIV-positive subjects had starting CD4+-lymphocyte counts of greater than 200 cells/μl. A sample was collected from each of three oral locales: the buccal mucosa, the floor of the mouth, and the dorsal surface of the tongue.
Samples were coded according to location of collection (FB [The William Beaumont Army Hospital, Fort Bliss, El Paso, Tex.]; H [Bering Dental Clinic, Houston, Tex.]; or I [Infectious Diseases Clinic at The University of Iowa Hospitals and Clinics, Iowa City]), test individual number (1, 2, 3, and so forth), sample location (B [buccal mucosa], F [floor of the mouth], or T [dorsal surface of the tongue]), and whether or not the sample was obtained prior to oral thrush, during an episode of oral thrush, or after antifungal therapy (a, b, and c, respectively). Therefore, FB1aF represents a sample obtained from the floor of the mouth of patient 1 at The William Beaumont Army Hospital, Fort Bliss, El Paso, Tex., prior to an episode of thrush.
Samples were collected by only one investigator (K.G.V.) using methodology previously described (50). Briefly, each sample was collected by passing a sterile cotton swab (Culturette; Becton Dickinson Microbiology Systems, Cockeysville, Md.) several times across the particular oral surface. Immediately after sampling, each swab was replaced in its sterile containment tube and was moistened with sterile salt solution by crushing the glass ampoule in the tube. The containment tubes were transported within 2 h of sampling from the place of collection to the laboratory. The cotton end of each swab was inserted into 0.5 ml of sterile water in a polypropylene test tube and vigorously mixed for 30 s. A 0.15-ml sample was spread on each of two agar plates containing supplemented Lee's medium (15a). This agar is very useful for evaluating phenotypic switching but is not appropriate for the growth of certain yeast species, such as C. glabrata (18, 19). A 0.15-ml sample was spread on a CHROMagar plate (Hardy Diagnostics, Santa Monica, Calif.) for primary species typing (32). This procedure resulted in three plates from each oral location. The Lee's agar plates were incubated for 7 days at 25°C, and the CHROMagar plate was incubated for 48 h at 37°C.
After incubation, the total number of yeast colonies was considered the relative intensity of carriage per oral site, and the total number of yeast colonies on the nine plates was considered the relative intensity of oral carriage.
To assess genetic homogeneity within a clone, multiple colonies with the same colony morphologies from selected primary plates were individually streaked onto agar slants and stored for subsequent analysis.
Typing of the yeast species was performed with the IDS rapid yeast identification system (Remel) or the Vitek YBC automated yeast identification system (Vitek, BioMerieux, Inc.). For both systems, the manufacturer's instructions were followed.
Fingerprinting of C. albicans isolates with probe Ca3
The complex DNA fingerprinting probe Ca3 (1, 12, 17, 37) was used to assess the genetic relatedness of C. albicans isolates by methods previously described (21, 35, 43). In brief, cells from agar storage slants were streaked onto a YPD (2% glucose-2% Bacto-Peptone-1% yeast extract) agar plate and allowed to incubate for 48 h at 25°C. DNA was prepared from each clone by the method of Scherer and Stevens (40), and the concentrations were measured with a Sequoia-Turner 45 fluorometer. DNA was digested with EcoRI and electrophoresed in an 0.8% (wt/vol) gel overnight at 35 V. DNA from reference strain 3153A was run in the outer lanes of each gel. The gel was stained with ethidium bromide to compare loading between lanes. DNA was then transferred by capillary blotting to a nylon Hybond-N+ membrane (Amersham, Piscataway, N.J.) and hybridized with a random-primer-labeled ([32P]dCTP) probe. The membrane was washed at 45°C and exposed to XAR-S film (Eastman Kodak, Rochester, N.Y.) with a Cronex Lightning-Plus intensifying screen (Du Pont Co., Wilmington, Del.). DNA hybridization patterns were digitized into the DENDRON software program (version 2.0; Solltech, Inc., Oakdale, Iowa). The methods used for automatic processing and analysis of Southern blot hybridization patterns were described in detail recently (46). Similarity coefficients (SAB) were computed with a formula based on band positions only, and dendrograms were generated by the unweighted pair-group method with arithmetic averages (44).
Statistical methods.
Significance was determined by nonparametric one-way analysis of variance (ANOVA), Student's t test, and Fisher's exact test.
RESULTS
Patient demographics and CD4 status.
A total of 54 HIV-positive individuals with no signs or symptoms of oral candidiasis were recruited into this study between 1994 and 1996. Because complete histories from the patients monitored in El Paso and Houston could not be obtained, most of the demographic information pertains to the 48 subjects recruited at The University of Iowa Hospitals and Clinics, unless noted otherwise.
Of the 48 Iowa patients, 25 (52%) had CD4+-cell counts of greater than 400 cells/μl, 23 (48%) had counts of between 200 and 400 cells/μl, and none had counts of less than 200 cells/μl. The mean CD4+-T-cell count was 364 cells/μl, with a range of 203 to 1,000. Thirty-eight subjects were male, and 10 were female. All 48 were HIV positive and asymptomatic upon entering the study, had not had an episode of oral or esophageal candidiasis prior to enrollment, and had not been previously exposed to an antifungal agent. For comparison, 38 HIV-negative, healthy individuals were used as control subjects, with a mean age of 38 years (range, 30 to 45 years). Nineteen subjects were male, and 19 were female.
At various times during the 2 years of the study, samples from the HIV-positive subjects were taken from the following three regions of the oral cavity: the buccal mucosa, the floor of mouth, and the dorsum of the tongue. During the course of the study, 11 patients (5 from Iowa City, 4 from El Paso, and 2 from Houston) developed oral thrush (pseudomembranous). The average CD4+-cell count for these individuals was 332, with a range of 203 to 560. All of the patients from Iowa were routinely taking the antiretroviral medications zidovudine, dideoxyinosine, and stavudine alone or in combination.
Yeast carriage and CD4+-cell counts.
HIV-positive subjects were evaluated for yeast carrier status by computing the percentage of individuals whose samples grew one or more yeast colonies on the nine culture plates. Samples were taken on the day on which the subject enrolled in the study and at every subsequent visit to the Infectious Diseases Clinic at The University of Iowa Hospitals and Clinics until completion of the follow-up period. Sixty-five percent (15) of subjects with CD4+-cell counts of 200 to 399/μl had yeasts isolated at each visit (persistent carriers), 22% (5) of the same group had yeasts isolated occasionally (intermittent carriers), and 12% (3) had no yeast colonization throughout the study period (noncarriers). In subjects with CD4+-cell counts of greater than 400/μl, 56% (14) were persistent carriers, 20% (3) were intermittent carriers, and 32% (8) were noncarriers (Table 1). With Fisher's exact test, no relationship was found between yeast carrier status and CD4+-cell count. Although the healthy control subjects were not monitored longitudinally to evaluate intermittent carrier status, we did find that 55% (21) were yeast carriers and that 45% (17) were noncarriers (Table 1). When persistent and intermittent carriers were combined, 87% (P > 0.05) of patients with CD4+-cell counts of 200 to 399/μl and 68% of patients with counts of greater than 400/μl had yeasts present at some point during the study, compared to only 55% of the control population. Regardless of CD4 status, a total of 37 HIV-positive patients (77%) had yeasts during the study period, a rate which also differed significantly from the carriage rate seen in healthy control subjects (55%) (P < 0.05).
TABLE 1.
Yeast carriage and relationship to CD4+-lymphocyte counts
| CD4 count (cells/mm3) | No. of patients tested | No. (%) of patients with the following carrier status:
|
|||
|---|---|---|---|---|---|
| Persistent | Intermittent | Total | Noncarrier | ||
| 200-400 | 23 | 15 (65) | 5 (22) | 20 (87)a | 3 (13) |
| >400 | 25 | 14 (56) | 3 (12) | 17 (68) | 8 (32) |
| Control | 38 | 21 (55) | NAb | 21 (55)a | 17 (45) |
With Fisher's exact test, a significant difference was found at a P value of <0.05.
NA, not applicable.
Intensity of carriage varies over time.
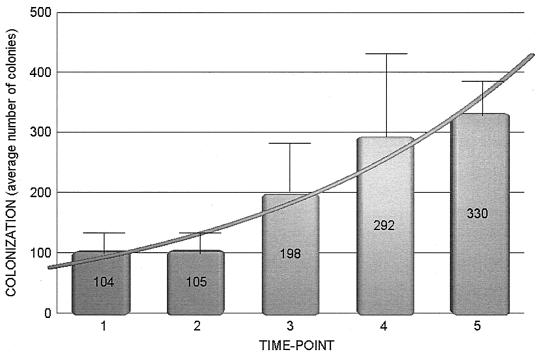
The intensity of yeast carriage for each individual was computed as the combined number of colonies on the nine agar plates containing samples derived from the oral cavity. Of the HIV-positive subjects positive for yeast carriage (37), the average intensities of carriage were 104 colonies/nine plates at the first swab, 105 colonies at the second, 198 at the third, 282 at the fourth, and 330 at the fifth (Fig. 1). With ANOVA, statistically significant differences were found between the average intensities obtained at the first and second swabs and the average intensity found at the fifth swab (P < 0.05).
FIG. 1.
Quantitative evaluation of oral yeast carriage over time in 48 HIV-positive individuals at The University of Iowa of Hospitals and Clinics. Time points refer to the first, second, third, fourth, or fifth time a sample was taken from a patient; the actual time varied from patient to patient. Error bars indicate standard errors.
Relationship among yeast carriage, CD4+-cell counts, and development of thrush.
Of the 48 subjects recruited in Iowa, 5 (10%) developed oral thrush over the 2-year study period. All five of these individuals had CD4+-cell counts of 200 to 400/μl (Table 2). None of the subjects with CD4+-cell counts of greater than 400 cells/μl had any manifestation of thrush during the study period. The median intensity of yeast carriage for the five Iowa subjects who developed thrush was 4,351 colonies (P < 0.05). Of the remaining subjects with CD4+-cell counts of 200 to 400/μl, 15 (65%) carried yeasts as commensals, with a median carriage intensity of 1,090 colonies (Table 2). On the other hand, the median intensity of yeast carriage for the 17 subjects (68%) who had CD4+-cell counts of greater than 400 cells/μl was 309 colonies/nine plates. This difference was significant, at P < 0.05 (Table 2).
TABLE 2.
Median yeast concentrations over the 2-year time period and development of thrush in subjects from lowa City
| CD4 count (cells/mm3) and thrush status | No. of patients positive/total no. tested (%) | Median CFU |
|---|---|---|
| 200-400 | ||
| No thrush | 15/23 (65) | 1,090a |
| Thrush | 5/23 (22) | 4,351a |
| >400 | 17/25 (68) | 309 |
The median CFU for patients with CD4 counts of 200-400 cells/mm3 (thrush or no thrush) were significantly (P < 0.05) different from the median CFU for patients with CD4 counts of >400 (these patients did not have thrush).
The onset of clinical thrush for the five Iowa subjects occurred on day 320 for subject I1 (Fig. 2), day 483 for subject I2 (Fig. 2), day 500 for subject I3 (Fig. 2), day 350 for subject I4, and day 527 for subject I5 (data not shown). For subjects from El Paso and Houston, the onset of clinical thrush occurred on day 74 for subject FB1, day 216 for subject FB3, day 119 for subject FB4, day 106 for subject H1, and day 60 for subject H2; the day of onset for subject FB2 was not determined (data not shown).
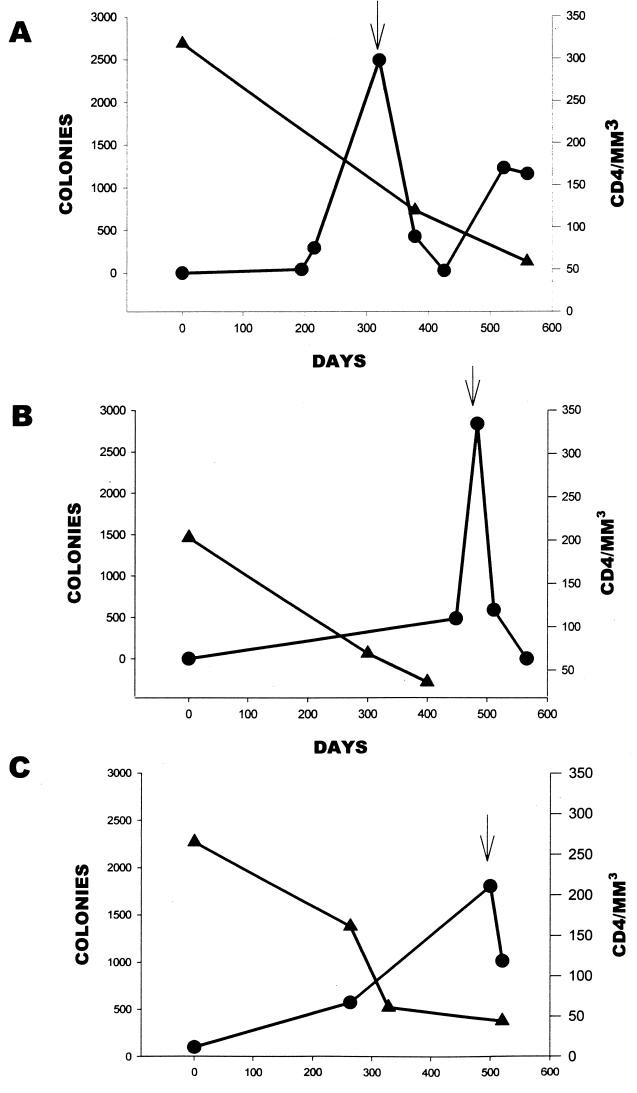
FIG. 2.
Relationship of CD4+-cell count to Candida colonization in patients I1(A), I2 (B), and I3 (C). The left y axis corresponds to number of colonies over time and is represented by the line graph with the circles. The right y axis corresponds to CD4+-cell count over time and is represented by the line graph with the triangles. The arrow represents the development of oral thrush for each patient.
To show the relationship between CD4+-cell count and manifestation of oral thrush, the intensity of colonization was plotted against CD4+-cell count over time for three of the patients from Iowa who developed thrush (Fig. 2). An inverse relationship was seen as the CD4+-cell count decreased and the intensity of colonization increased, culminating in the clinical manifestation of oral candidiasis at day 320 for patient I1 (Fig. 2A), day 483 for patient I2 (Fig. 2B), and day 500 for patient I3 (Fig. 2C).
After averaging the carriage rates for the 11 HIV-positive individuals who developed thrush, we found that the average carriage rates were 105 colonies (standard error, ±20) before thrush, 1,480 colonies (standard error, ±630) (P < 0.05) during an episode of thrush, and 415 colonies (standard error, ±150) after antifungal therapy.
The distribution of oral yeast isolates was also examined for the five Iowa patients who developed thrush. At some point during the 2-year study period, all five subjects had colonization with yeast isolates at multiple locations. The intensity of yeast carriage was highest on the buccal mucosa and dorsum of the tongue and lowest on the floor of the mouth, except for patient I4, who had carriage primarily on the dorsum of the tongue (data not shown). With ANOVA, significant differences were found for the intensity of yeast carriage on the buccal mucosa, the dorsum of the tongue, and the floor of the mouth (P < 0.05).
Multiple yeast species are present in HIV-positive individuals.
From the 11 HIV-positive individuals who developed thrush, 443 isolates were analyzed for species identification: 51 from patient I1, 57 from patient I2, 45 from patient I3, 20 from patient I4, 33 from patient I5, 30 from patient FB1, 21 from patient FB2, 49 from patient FB3, 11 from patient FB4, 30 from patient H1, and 66 from patient H2. Of these, 374 (84%) were found to be C. albicans, 29 (7%) were Candida tropicalis, and 20 (5%) were Candida dubliniensis. Although 5% of the isolates were C. dubliniensis, they all came from only one patient (I4). The remaining 20 isolates (4%) were Candida parapsilosis, Candida humicola, C. glabrata, Candida guillermondii, and Saccharomyces cerevisiae (Table 3). In comparison, 75% of the isolates from the 38 control subjects were C. albicans, 5% were C. parapsilosis, and 20% were other species (Candida zelanoides, Trichosporan beigelii, and Candida famata). It is of interest that only 0.2% of the isolates were C. glabrata. The low proportion of this yeast was probably due to the fact that platings were done on modified Lee's agar plates, which do not support the growth of this species.
TABLE 3.
Proportions of yeast species in isolates collected from 11 HIV-positive subjects who developed thrush and healthy control subjects
| Species | % of subjects
|
|
|---|---|---|
| HIV positive | Control | |
| Candida albicans | 84 | 75 |
| Candida tropicalis | 7 | 0 |
| Candida dubliniensis | 5 | 0 |
| Candida parapsilosis | 2 | 5 |
| Candida guillermondii | 1 | 0 |
| Saccharomyces cerevisiae | 1 | 0 |
| Candida humicola | 0.2a | 0 |
| Candida glabrata | 0.2a | 0 |
| Other | 0 | 20 |
| Total | 100 | 100 |
Isolation of this species was minimal and is not reflected in the total percentage.
Of the 11 HIV-positive subjects, 3 had multiple species or species other than C. albicans. This group represents 27% of the persons who developed thrush. The combinations were C. albicans plus C. tropicalis for individual FB1; C. albicans, C. guillermondii, C. tropicalis, C. glabrata, and S. cerevisiae for individual I2; and C. dubliniensis for individual I4. None of the healthy control subjects had multiple species. It is of interest that patient I2 had the lowest CD4+-cell count upon entry into the study (205) and had the most morbidity.
Multiple scenarios exist in infecting populations.
To test strain heterogeneity, several individual colonies from primary cultures for each of the 11 individuals with thrush were fingerprinted with the complex DNA fingerprinting probe Ca3. Ca3 is specific for C. albicans and has been demonstrated to be effective in distinguishing between completely unrelated strains, in identifying the same strain in different samples, and in identifying microevolution within a strain (35, 46). DNA fingerprints from the 11 HIV-positive patients who developed thrush showed four different scenarios: single strain maintenance, multiple strain maintenance, strain replacement, and species replacement.
(i) Single strain maintenance.
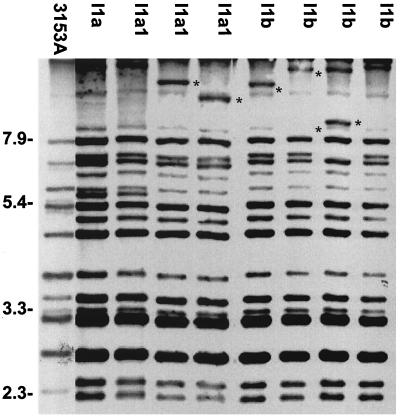
Based on the SAB calculations, dendrograms were generated which provided a means for quantifying the genetic relatedness of test strains. We found that 5 of the 11 HIV-positive subjects (FB2, FB3, FB4, I1, and I3) who developed thrush had an average SAB among all their strains of 0.85, indicating that the strains were closely related but not identical and underwent microevolution throughout the study period. An example of the fingerprinting patterns of the collection of isolates from patient I1 is presented in Fig. 3; lanes 1 to 4 represent isolates obtained at two different times before an episode of thrush, and lanes 5 to 8 represent isolates obtained during an episode of thrush. Pattern variations can be seen in the high-molecular-weight bands of the isolates in Fig. 3, lanes 3, 4, 5, 6, and 7, indicating a high degree of microevolution over time.
FIG. 3.
Southern blot hybridization patterns generated with probe Ca3 for colonies isolated from patient I1. Molecular sizes in kilobases are denoted to the left of the Southern blot. Asterisks indicate bands representative of microevolution.
(ii) Multiple strain maintenance.
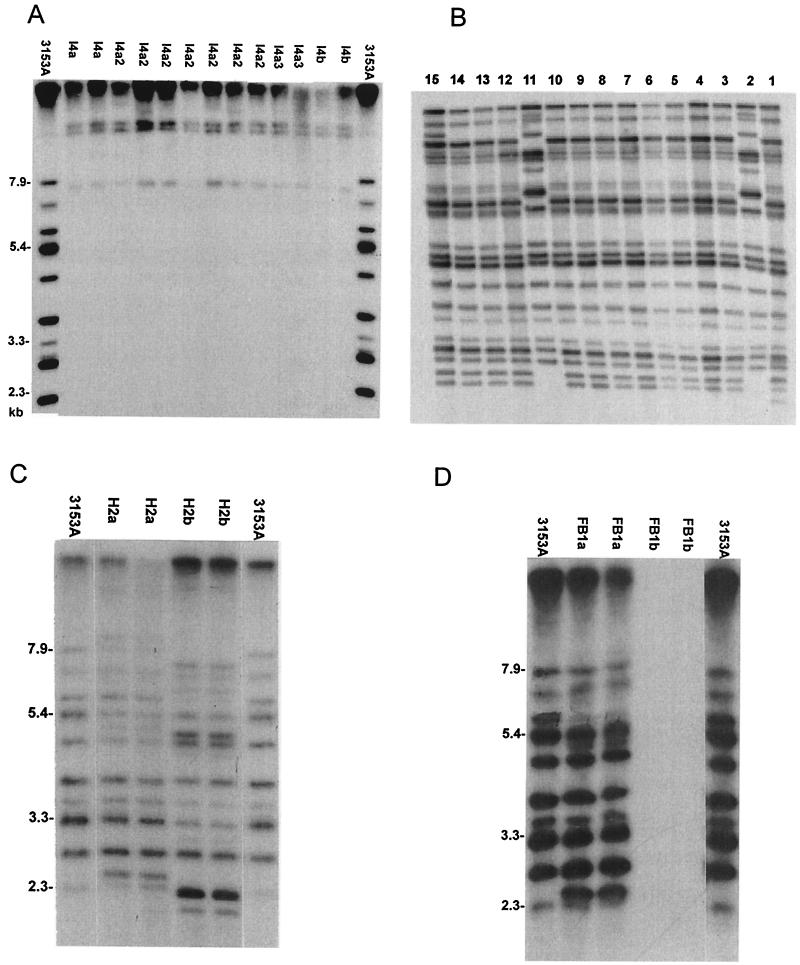
When originally probed with Ca3, the isolates from patient I4 did not give typical C. albicans patterns (Fig. 4A), indicating that they were non-C. albicans species. Typing with standard sugar assimilation techniques (ID32C kit; bioMérieux, Marcy l’Etoile, France) did not lead to any conclusive evidence as to the identity of this species. However, additional tests did reveal that the species was chlamydospore positive and formed hyphae. As the isolates from this patient could not be identified with certainty, it was thought that they were most likely C. dubliniensis. To verify this notion, DNA was reextracted from the isolates, and the resultant Southern blot was probed with mid-repeat sequence probe Cd25, specific for C. dubliniensis (12). Figure 4B shows the results for isolates obtained from the same patient and probed with the C. dubliniensis species-specific probe. Beginning from the right side of Fig. 4B, lanes 1 to 8 represent isolates obtained from the first swab upon entry into the study, lanes 9 and 10 represent isolates obtained from a second swab before any thrush was evident, and lanes 11 to 15 represent isolates obtained during an episode of thrush. Two different strains were evident by hybridization with this species-specific probe. A minor genotype can be seen in lanes 2 and 11 of Fig. 4B, and a more frequent genotype can be seen in the remainder of the lanes. A dendrogram based on Fig. 4B was generated (data not shown) which showed a major cluster consisting of the most frequent genotype and a minor cluster consisting of the minor genotype. The SAB for the two genotypes was 0.7, demonstrating their unrelatedness.
FIG. 4.
Southern blot hybridization patterns generated with probe Ca3 (A) and probe Cd25 (B) for colonies isolated from patient I4 and with probe Ca3 for colonies isolated from patients H2 (C) and patient FB1 (D). See the text for an explanation of the lanes in panel B. Molecular sizes in kilobases are denoted to the left of the Southern blots.
(iii) Strain replacement and species replacement.
Figure 4C represents isolates from patient H2 representing two different strains of C. albicans. The first strain was obtained before an episode of thrush (Fig. 4C, lanes 1and 2), and the second strain was obtained during an episode of thrush (lanes 3 and 4). Two clusters were seen in the dendrogram generated from this fingerprint (data not shown). Cluster A consisted of isolates obtained during an episode of thrush (H2b), and cluster B consisted of isolates obtained before the onset of clinical thrush (H2a). The SAB for these two clusters was 0.61, again indicative of unrelatedness. This patient was the only one of the 11 HIV-positive patients who developed thrush and in whom the original commensal strain was replaced by a different strain upon the development of oral candidiasis, although many others had minor genetic variations of the same strain.
Two of the patients had either the original infecting species replaced by another species when thrush developed (Fig. 4D) or had multiple species present throughout the study period (data not shown for patient I2). The Ca3 Southern blot hybridization pattern of the isolates obtained from patient FB1 are shown in Fig. 4D; lanes 1 and 2 represent isolates obtained prior to an episode of thrush, and lanes 3 and 4 represent isolates obtained during an episode of thrush. No hybridization pattern is apparent in lanes 3 and 4 of Fig. 4D, indicating that these isolates are non-C. albicans species. In an attempt to identify the second species, standard sugar assimilation assays were conducted with an API 32C kit (BioMerieux, Inc.); the species was identified as C. parapsilosis.
Although the data are not shown, patient I2 had multiple species throughout the study period. These included C. guillermondii, C. tropicalis, C. albicans, and S. cerevisiae. Prior to the development of thrush, the majority of species isolated were C. guillermondii and C. tropicalis; however, when the patient developed thrush, C. tropicalis remained during the infection but was joined by C. albicans, which had not been present before. DNA fingerprints of the C. albicans isolates showed that all were identical.
DISCUSSION
Asymptomatic oral carriage of C. albicans is a common finding in HIV-infected patients (49). In 1989, Korting conducted a study in which the microbiological recoveries of oral C. albicans from 62 HIV-infected patients were 57.5% for CDC stage I patients, 76.5% for stage II patients, and 87.5% for stage III patients (14). Similarly, Fong et al. found yeast carriage rates in HIV-positive individuals to be 67% for those with CD4+-cell counts above 500 cells/μl, 86% for patients with counts of 200 to 500 cells/μl, and 82% for patients with CD4+-cell counts below 200 cells/μl (6). Although a substantial body of epidemiological data exists regarding the prevalence of C. albicans in this group and its increase with disease progression, no studies to date have monitored HIV-positive patients longitudinally to evaluate changes in yeast carriage, intensity of carriage, or frequency of carriage. Since oral candidiasis is a common opportunistic disease in HIV-infected patients (10, 11) and it has been suggested to result from yeast overgrowth in subjects with a history of colonization (5), it is important to understand all aspects of the disease.
To directly assess the relationship of yeast carriage to the development of oral thrush, the oral cavities of HIV-positive individuals with CD4+-cell counts above 200 cells/μl and no prior history of oral thrush were sampled at intervals varying from 1 to 6 months over a 2-year time period by using a swabbing technique. Although this technique may be less sensitive than the imprint or saliva collection technique, the carriage rates obtained do not vary significantly from those in previous reports (6, 9, 31). We found that the overall frequencies of carriage were 77% for the HIV-positive individuals and 55% for the control group. What was most interesting was that patients with CD4+-cell counts of <400/μl had a significantly higher level of yeast carriage.
We also showed that the intensity of carriage and the development of thrush were related to CD4+-cell counts. This finding correlates well with previously reported results showing an increase in spontaneous phenotypic switching frequencies during an episode of oral thrush (50).
It is known that the immune response against C. albicans and related species is primarily achieved by macrophages and neutrophils. Although HIV-positive patients maintain neutrophil function (which would explain why disseminated disease is rarely seen in this population), their macrophage function declines as the number of T-helper cells is compromised (11, 22, 30). Therefore, it follows that persons with low CD4+-cell counts would have increased numbers of colonizing yeasts in their oral cavities.
The average intensity of carriage for the 48 subjects recruited from Iowa tended to increase over time, culminating with the development of oral thrush in five of the patients. This increase in intensity of carriage suggests that the innate immune response is negatively affected by the progression of HIV and that yeast numbers can be predictors of the development of candidiasis if the person is monitored over time. Torssander et al. found that the rate of carriage of C. albicans in HIV-seropositive men was significantly higher than that in HIV-seronegative men; however, they noted no correlation with CD4+-cell counts (48). Felix and Wray also noted a higher rate of oral C. albicans carriage in HIV-positive subjects (93.1%) than in control subjects (57.4%) but did not examine the effect of CD4+ lymphocytopenia (4). Similarly, DeBernardis et al. noted that increased oral colonization correlated with clinical disease and low CD4+-cell counts but found that the rates of colonization were similar for asymptomatic HIV-positive patients and control subjects (about 30%) (3). In a more recent study, Fong et al. also found a strong correlation between asymptomatic yeast carriage, development of thrush, and CD4+-cell counts (6). The results from their study most closely resemble the results found for the patients who developed thrush in this study. The one main difference was that they did not find a temporal correlation between the quantity of yeasts isolated and the development of clinical thrush. We showed that there was a definite tendency toward increased yeast colonization over time, leading to the development of thrush. The difference in results, however, is most likely due to the fact that they examined Candida carriage at only one point in time; whereas we monitored subjects over a 2-year period.
Although it would be useful to use yeast carriage intensity as a predictor for the onset of clinical thrush, we saw much individual variation in the level of yeast carriage throughout the study period, and unless a patient is monitored closely for yeast carriage over time, a one-time swab will not be a good indicator of the likelihood for the development of oral thrush.
The majority of the isolates obtained from the HIV-positive subjects who developed thrush were C. albicans (84%); however, several other species were identified. These included C. parapsilosis, C. humicola, C. dubliniensis, C. guillermondii, C. tropicalis, C. glabrata, and S. cerevisiae. Altogether, non-C. albicans strains accounted for 16.4% of the strains isolated from the 11 HIV-positive patients who developed thrush. In comparison, the control population of healthy subjects had 75% C. albicans and 25% other species (C. parapsilosis, C. zelanoides, T. beigelii, and C. famata). These findings are in agreement with those of Barchiesi et al. (2), who found that approximately 16% of isolates from HIV-positive patients were non-C. albicans. Although carriage rates were similar for both study subjects and control subjects, mixed populations of yeasts were seen in three of the HIV-positive subjects, whereas none of the control subjects were populated with more than one yeast species. As it is known that the efficacy of colonization for a microorganism depends not only on host immune factors but also on existent microflora, colonization with multiple yeast species in HIV-infected individuals might be due not only to their immune status but also to the changing oral environment present in these individuals. This change in the oral environment would lead to shifts in existing bacterial populations which may not be able to compete as effectively for mucosal colonization, allowing transient yeast species to attach themselves more readily to tissues in the oral cavity.
We also found that of the three oral areas swabbed, the floor of the mouth had the fewest yeast colonies. This finding was most likely due to a combination of factors: the constant exposure of the yeast cells to saliva, with its antimicrobial peptides, and the increased likelihood of cells being swept away with the normal cleansing actions of saliva (8, 24).
Although a control group was used to compare carriage rates and intensities, the healthy subjects were not monitored over time as the test subjects were. It would be interesting to conduct a future study where control subjects are monitored longitudinally to determine if such drastic changes in yeast populations and carriage rates are noted for them as well.
In spite of the importance of oral candidiasis as a complication of HIV disease, its molecular epidemiology still has not been completely elucidated. The combined results of a number of studies have shown four basic scenarios for the genetic relatedness of strains obtained from patients with sequential episodes of recurrent oropharyngeal candidiasis (16, 26, 34, 36). In the first scenario, the same strain is responsible for each recurrent episode and remains genetically invariant. In the second scenario, the same strain is responsible for recurrent episodes, but small variations occur in the genotype. In the third scenario, the infecting strain is replaced by an unrelated strain in a subsequent episode. In the fourth scenario, the infecting species is replaced by an unrelated species in a subsequent episode. All of these studies have taken sequential isolates from recurrent infections; therefore, the question that still remains unanswered as to what happens to yeast strains isolated from HIV-positive individuals in the transition from a healthy state to a disease state. In an attempt to answer this question, we isolated yeast samples from HIV-positive individuals who had never experienced an episode of oral thrush and monitored these patients over time until the development of oral candidiasis.
Unlike many other reported studies, this study showed that although 54% (n = 6) of the study subjects maintained genetically similar strains throughout the study period, the complex banding pattern given with the Ca3 probe demonstrated minor genetic variations (microevolution) in all cases except one. This result could have been due to the fact that techniques such as restriction fragment length polymorphism, randomly amplified polymorphic DNA, and karyotyping analysis may not be sensitive enough to detect single band variations. Using Ca3, Lockhart et al. (17, 20) demonstrated microevolution and substrain shuffling of genetically similar C. albicans strains.
The remaining 46% of the study subjects had either multiple strains throughout the study period (n = 2), strain replacement (n = 1), or species replacement (n = 2). Of the subjects with multiple strains, one (I4) was infected with two different strains of C. dubliniensis distinguished by a recently developed species-specific probe (12). In both patients, one of the strains was a minor inhabitant, and the other was the predominant strain found. In the remaining three patients (H2, FB1, and I2), either the original strain was replaced with a different strain when candidiasis developed or the original species was replaced with a new species. Redding et al. evaluated the DNA subtype and fluconazole susceptibility of isolates from AIDS patients and found that for one patient in whom fluconazole resistance was detected, the original infecting C. albicans strain was replaced by fluconazole-resistant C. guillermondii and C. albicans strains (36). Using randomly amplified polymorphic DNA analysis, Metzgar et al. (26) evaluated the genotypes of yeast isolates obtained from 12 HIV-positive individuals before and after treatment with fluconazole and found that in one patient, the original infecting C. albicans strain was replaced by a strain of C. dubliniensis. The scenarios of both strain replacement and species replacement may mean that multiple strains and/or species are present throughout the process of infection and are just being overlooked by current sampling techniques, that environmental changes in HIV-positive individuals are selecting for strains or species that are more capable of surviving in hostile environments, or that the loss of host defenses against fungi is allowing usually more vulnerable species to escape the immune system.
It has been reported that genetic diversity among C. albicans strains from patients with AIDS is significantly reduced compared to that of commensal strains from healthy individuals (41). Although the data were not presented in this study, isolates from all 11 HIV-positive individuals who developed thrush were combined in a dendrogram to evaluate similarities among the various patients. Unlike Schmid et al. (41), we did not find a reduction in genetic diversity. However, this result may have been due to the fact that the patients in our study came from three different geographical regions. Kleinegger et al., who assessed genotypic relatedness among commensal isolates in Iowa City, found a reduction in diversity; however, when these isolates were compared to those from another geographical region, no common clusters between the two different locales were seen (13).
In conclusion, a longitudinal study of HIV-positive patients over a 2-year period allowed us to monitor the dynamics of Candida oral colonization and its evolution toward thrush and to evaluate the prognostic value of Candida carriage in the development of AIDS. Combined with a molecular approach, our results demonstrated that some drastic changes associated with disease and immune status occurred in the oral cavities of HIV-positive patients and that these changes involved not one but several different scenarios. An understanding of the different scenarios involved in the development of oral candidiasis in HIV-positive patients may play an important role in the management of this disease.
Acknowledgments
This research was funded by Public Health Service grant DE00364 from the NIH/NIDCR.
REFERENCES
- 1.Anderson, J., T. Srikantha, B. Morrow, S. H. Miyasaki, T. C. White, N. Agabian, J. Schmid, and D. R. Soll. 1993. Characterization and partial nucleotide sequence of the DNA fingerprinting probe Ca3 of Candida albicans. J. Clin. Microbiol. 31:1472-1480. [DOI] [PMC free article] [PubMed]
- 2.Barchiesi, F., M. Del Poeta, V. Morbiducci, F. Ancarani, and G. Scalise. 1993. Turbidimetric and visual criteria for determining the in vitro activity of six antifungal agents against Candida spp. and Cryptococcus neoformans. Mycopathologia 124:19-25. [DOI] [PubMed] [Google Scholar]
- 3.DeBernardis, F., M. Boccanera, L. Rainaldi, C. E. Guerra, I. Quinti, and A. Cassone. 1992. The secretion of aspartyl proteinase, a virulence enzyme, by isolates of Candida albicans from the oral cavity of HIV-infected subjects. Eur. J. Epidemiol. 8:362-367. [DOI] [PubMed] [Google Scholar]
- 4.Felix, D. H., and D. Wray. 1993. The prevalence of oral candidiasis in HIV-infected individuals and dental attenders in Edinburgh. J. Oral Pathol. Med. 22:418-420. [DOI] [PubMed] [Google Scholar]
- 5.Fetter, A., M. Partisani, H. Koenig, M. Kremer, and J. M. Lang. 1993. Asymptomatic oral Candida albicans carriage in HIV-infection: frequency and predisposing factors. J. Oral Pathol. Med. 22:57-59. [DOI] [PubMed] [Google Scholar]
- 6.Fong, I. W., M. Laurel, and A. Burford-Mason. 1997. Asymptomatic oral carriage of Candida albicans in patients with HIV infection. Clin. Investig. Med. 20:85-93. [PubMed] [Google Scholar]
- 7.Gallagher, P. J., D. E. Bennett, M. C. Henman, R. J. Russell, S. R. Flint, Shanley, D. B., and D. C. Coleman. 1992. Reduced azole susceptibility of oral isolates of Candida albicans from HIV-positive patients and a derivative exhibiting colony morphology variation. J. Gen. Microbiol. 138:1901-1911. [DOI] [PubMed] [Google Scholar]
- 8.Guthmiller, J. M., K. G. Vargas, R. Srikantha, L. L. Schomberg, P. L. Weistroffer, P. B. McCray, Jr., and B. F. Tack. 2001. Susceptibility of oral bacteria and yeast to mammalian cathelicidins. Antimicrob. Agents Chemother. 45:3216-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hester, C., J. Hauman, B. Medsci, M. Medsci, I. Thompson, F. Theunissen, and P. Wolfardt. 1993. Oral carriage of candida in healthy and HIV-seropositive persons. Oral Surg. Oral Med. Oral Pathol. 76:570-572. [DOI] [PubMed] [Google Scholar]
- 10.Holmberg, K., and R. D. Meyer. 1986. Fungal infections in patients with AIDS and AIDS-related complex. Scand. J. Infect. Dis. 18:179-185. [DOI] [PubMed] [Google Scholar]
- 11.Imam, N., C. C. Carpenter, K. H. Mayer, A. Fisher, M. Stein, and S. B. Danforth. 1990. Hierarchical pattern of mucosal candida infections in HIV-seropositive women. Am. J. Med. 89:142-146. [DOI] [PubMed] [Google Scholar]
- 12.Joly, S., C. Pujol, M. Rysz, K. Vargas, and D. R. Soll. 1999. Development and characterization of complex DNA fingerprinting probes for the infectious yeast Candida dubliniensis. J. Clin. Microbiol. 37:1035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleinegger, C. L., S. R. Lockhart, K. Vargas, and D. R. Soll. 1996. Frequency, intensity, species, and strains of oral Candida vary as a function of host age. J. Clin. Microbiol. 34:2246-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korting, H. C. 1989. Clinical spectrum of oral candidosis and its role in HIV-infected patients. Mycoses 32(Suppl. 2):23-29. [DOI] [PubMed] [Google Scholar]
- 15.Korting, H. C., M. Ollert, A. Georgii, and M. Froschl. 1988. In vitro susceptibilities and biotypes of Candida albicans isolates from the oral cavities of patients infected with human immunodeficiency virus. J. Clin. Microbiol. 26:2626-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Lee, K. L., H. R. Buckley, and C. Campbell. 1975. An amino acid liquid synthetic medium for development of mycelial and yeast forms of Candida albicans. Sabouradia 13:148-153. [DOI] [PubMed] [Google Scholar]
- 16.Lischewski, A., D. Harmsen, K. Wilms, G. Baier, U. Gunzer, H. Klinker, M. Wilhelm, A. Schwinn, and J. Hacker. 1999. Molecular epidemiology of Candida albicans isolates from AIDS and cancer patients using a novel standardized CARE-2 DNA fingerprinting technique. Mycoses 42:371-383. [DOI] [PubMed] [Google Scholar]
- 17.Lockhart, S. R., J. J. Fritch, A. S. Meier, K. Schroppel, T. Srikantha, R. Galask, and D. R. Soll. 1995. Colonizing populations of Candida albicans are clonal in origin but undergo microevolution through C1 fragment reorganization as demonstrated by DNA fingerprinting and C1 sequencing. J. Clin. Microbiol. 33:1501-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lockhart, S. R., S. Joly, C. Pujol, J. D. Sobel, M. Pfaller, and D. R. Soll. 1997. Development and verification of fingerprinting probes for Candida glabrata. Microbiology 143:3746.. [DOI] [PubMed] [Google Scholar]
- 19.Lockhart, S. R., S. Joly, K. Vargas, J. Swails-Wenger, L. Enger, and D. R. Soll. 1999. Defenses against oral Candida carriage break down in the elderly. J. Dent. Res. 78:857-868. [DOI] [PubMed] [Google Scholar]
- 20.Lockhart, S. R., B. D. Reed, C. L. Pierson, and D. R. Soll. 1996. Most frequent scenario for recurrent Candida vaginitis is strain maintenance with “substrain shuffling”: demonstration by sequential DNA fingerprinting with probes Ca3, C1, and CARE2. J. Clin. Microbiol. 34:767-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marco, F., S. R. Lockhart, M. A. Pfaller, C. Pujol, M. S. Rangel-Frausto, T. Wiblin, H. M. Blumberg, J. E. Edwards, W. Jarvis, L. Saiman, J. E. Patterson, M. Rinaldi, R. P. Wenzel, and D. R. Soll. 1999. Elucidating the origins of nosocomial infections with Candida albicans by DNA fingerprinting with the complex probe Ca3. J. Clin. Microbiol. 37:2817-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marodi, L., H. M. Korchak, and R. B. Johnston. 1991. Mechanisms of host defence against Candida species. I. Phagocytosis by monocytes and monocyte derived macrophages. J. Immunol. 146:2783-2789. [PubMed] [Google Scholar]
- 23.Masia Canuto, M. M., F. Gutierrez Rodero, V. Ortiz de la Tabla Durcasse, C. Martin Gonzalez, C. M. Escolano Hortelano, A. Mora Rufete, and A. Martin Hidalgo. 1999. Epidemiology of yeast colonization and oropharyngeal infection other than Candida albicans in patients with HIV infection. Med. Clin. 112:211-214. [PubMed] [Google Scholar]
- 24.Mathews, M., H. P. Jia, J. M. Guthmiller, G. Losh, S. Graham, G. K. Johnson, B. F. Tack, and P. B. McCray. 1999. Production of beta-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect. Immun. 67:2740-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meiller, T., M. A. Jabra-Rizk, A.a. Baqui, J. I. Kelley, V. I. Meeks, W. G. Merz, and W. A. Falkler. 1999. Oral Candida dubliniensis as a clinically important species in HIV-seropositive patients in the United States. Oral Surg. Oral Med. Oral Pathol. 88:573-580. [DOI] [PubMed] [Google Scholar]
- 26.Metzgar, D., A. van Belkum, D. Field, R. Haubrich, and C. Wills. 1998. Random amplification of polymorphic DNA and microsatellite genotyping of pre- and posttreatment isolates of Candida spp. from human immunodeficiency virus-infected patients on different fluconazole regimens. J. Clin. Microbiol. 36:2308-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyasaki, S. H., J. B. Hicks, D. Greenspan, I. Polacheck, L. A. MacPhail, White, T. C., N. Agabian, and J. S. Greenspan. 1992. The identification and tracking of Candida albicans isolates from oral lesions in HIV-seropositive individuals. J. Acquir. Immune Defic. Syndr. 5:1039-1046. [PubMed] [Google Scholar]
- 28.Morace, G., E. Tamburrini, S. Manzara, A. Antinori, G. Maiuro, and G. Dettori. 1990. Epidemiological and clinical aspects of mycoses in patients with AIDS-related pathologies. Eur. J. Epidemiol. 6:398-403. [DOI] [PubMed] [Google Scholar]
- 29.Moss, A. R. 1988. Predicting who will progress to AIDS. Br. Med. J. 297:617-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen, H., K. D. Bentsen, L. Hojtved, E. H. Willemoes, F. Scheutz, M. Schiodt, K. Stoltze, and J. J. Pindborg. 1994. Oral candidiasis and immune status of HIV-infected patients. Oral Pathol. Med. 23:140-143. [DOI] [PubMed] [Google Scholar]
- 31.Odds, F. C. 1988. Candida and candidosis. The W. B. Saunders Co., Philadelphia, Pa.
- 32.Odds, F. C., and R. Bernaerts. 1994. CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J. Clin. Microbiol. 32:1923-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Odds, F. C., J. Schmid, and D. R. Soll. 1990. Epidemiology of Candida infections in AIDS, p. 67-74. In H. Vanden Bosshe (ed.), Mycoses in AIDS patients. Plenum Press, New York, N.Y.
- 34.Powderly, W. G., K. Robinson, and E. J. Keath. 1992. Molecular typing of candida albicans isolated from oral lesions of HIV-infected individuals. AIDS 6:81-84. [DOI] [PubMed] [Google Scholar]
- 35.Pujol, C., S. Joly, S. R. Lockhart, S. Noel, M. Tibayrenc, and D. R. Soll. 1997. Parity among the randomly amplified polymorphic DNA method, multilocus enzyme electrophoresis, and Southern blot hybridization with the moderately repetitive DNA probe Ca3 for fingerprinting Candida albicans. J. Clin. Microbiol. 35:2348-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Redding, S. W., M. A. Pfaller, S. A. Messer, J. A. Smith, J. Prows, L. L. Bradley, A. W. Fothergill, and M. G. Rinaldi. 1997. Variations in fluconazole susceptibility and DNA subtyping of multiple Candida albicans colonies from patients with AIDS and oral candidiasis suffering one or more episodes of infection. J. Clin. Microbiol. 35:1761-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sadhu, C., M. J. McEachern, B. E. Rustchenko, J. Schmid, D. R. Soll, and J. Hicks. 1991. Telomeric and dispersed repeat sequences in Candida yeasts and their use in strain identification. J. Bacteriol. 173:842-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samaranayake, L. P. 1992. Oral mycoses in HIV infection. Oral Surg. Oral Med. Oral Pathol. 73:171-180. [DOI] [PubMed] [Google Scholar]
- 39.Samaranayake, L. P., and P. Holmstrup. 1989. Oral candidiasis and human immunodeficiency virus infection. J. Oral Pathol. Med. 18:554-564. [DOI] [PubMed] [Google Scholar]
- 40.Scherer, S., and D. A. Stevens. 1987. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J. Clin. Microbiol. 25:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmid, J., F. C. Odds, M. J. Wiselka, K. G. Nicholson, and D. R. Soll. 1992. Genetic similarity and maintenance of Candida albicans strains from a group of AIDS patients, demonstrated by DNA fingerprinting. J. Clin. Microbiol. 30:935-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmid, J., M. Rotman, B. Reed, C. Pierson, and D. R. Soll. 1993. Genetic similarity of Candida albicans strains from vaginitis patients and their partners. J. Clin. Microbiol. 31:39-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmid, J., E. Voss, and D. R. Soll. 1990. Computer-assisted methods for assessing strain relatedness in Candida albicans by fingerprinting with the moderately repetitive sequence Ca3. J. Clin. Microbiol. 28:1236-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy. The principles and practice of numerical classification. W. H. Freeman & Co., San Francisco, Calif.
- 45.Soll, D. R. 1993. DNA fingerprinting of Candida albicans. J. Mycol. Med. 3:37-44. [Google Scholar]
- 46.Soll, D. R. 2000. The “ins and outs” of DNA fingerprinting the infectious fungi. Clin. Microbiol. Rev. 13:332-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan, D., K. Haynes, J. Bille, P. Boerlin, L. Rodero, S. Lloyd, M. Henman, and D. Coleman. 1997. Widespread geographic distribution of oral Candida dubliniensis strains in human immunodeficiency virus-infected individuals. J. Clin. Microbiol. 35:960-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torssander, J., L. Morfeldt-Manson, G. Biberfeld, A. Karlsson, P. O. Putkonen, and J. Wasserman. 1987. Oral candida albicans in HIV infection. Scand. J. Infect. Dis. 19:291-295. [DOI] [PubMed] [Google Scholar]
- 49.Tylenda, C. A., J. Larsen, C. K. Yeh, H. C. Lane, and P. C. Fox. 1989. High levels of oral yeasts in early HIV-1 infection. J. Oral Pathol. Med. 18:520-524. [DOI] [PubMed] [Google Scholar]
- 50.Vargas, K., S. A. Messer, M. Pfaller, S. R. Lockhart, J. T. Stapleton, J. Hellstein, and D. R. Soll. 2000. Elevated phenotypic switching and drug resistance of Candida albicans from human immunodeficiency virus-positive individuals prior to first thrush episode. J. Clin. Microbiol. 38:3595-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]