Abstract
The PCR primers for O, H, and Vi antigen genes, tyv (rfbE), prt (rfbS), fliC-d, fliC-a, and viaB, were designed and used for the rapid identification of Salmonella enterica serovars Typhi and Paratyphi A with multiplex PCR. The results showed that all the clinical isolates examined of Salmonella serovars Typhi and Paratyphi A were accurately identified by this assay.
Enteric fever remains an important public health problem in many countries of the world. Typhoid fever and paratyphoid fever are still serious public health problems in many geographic areas and are endemic in most countries, especially those of Southeast Asia and Africa. Recently, multiple-drug- or fluoroquinolone-resistant strains of Salmonella enterica serovars Typhi and Paratyphi A have been emerging on the Indian subcontinent, spreading to, and becoming major problems, throughout the world (1, 4, 6). Typhoid fever and paratyphoid fever are sometimes-fatal infections of adults and children that cause bacteremia and inflammatory destruction of the intestine and other organs and that require urgent treatment by the administration of appropriate antibiotics. The diagnosis of typhoid fever or paratyphoid fever is made by ordinary culture methods and biochemical tests. The classic diagnosis method for typhoid fever or paratyphoid fever requires at least 4 or 5 days for positive results. A rapid, alternative method is needed for the diagnosis of typhoid fever or paratyphoid fever. Some researchers have already reported serovar Typhi detection methods with PCR that use the fliC-d gene (7), the Vi capsular antigen gene (3), and the 16S rRNA gene (9). As only one gene was targeted for the identification of serovar Typhi in these methods, strains of Salmonella serovars other than serovar Typhi were detected in some cases. In this study, we developed a more specific diagnosis method for both typhoid fever and paratyphoid fever based on a multiplex PCR technique that detected the Vi antigen gene (viaB), H antigen genes (fliC-d and fliC-a), and O antigen synthesis genes (tyv and prt). This system enabled us to identify and differentiate serovars Typhi and Paratyphi A, which are clinically important human pathogens, by only a single PCR, when we isolated the bacteria from blood or stool cultures from clinical patients.
The bacterial strains used in this study were collected from the regional public health office in Japan, and all isolates were identified by biochemical and serological tests. A suspension of bacteria was heated at 100°C for 10 min. The samples were then used for the PCRs. We designed the primers tyv-s and tyv-as for detection of the tyvelose epimerase gene (tyv, previously called rfbE) and the primers fliCcom-s and fliCd-as for detection of the fliC-d gene (phase-1 flagellin gene for d antigen [H:d]) of Salmonella serovar Typhi. The primers parat-s and parat-as were designed for detection of a paratose synthase gene (prt, previously called rfbS), and the primers fliCcom-s and fliCa-as were designed for detection of a fliC-a gene (phase-1 flagellin; H:a). The primer sequences used in this study are listed in Table 1. The gene prt encodes CDP-paratose synthase, which converts CDP-4-keto-3,6-dideoxyglucose to CDP-paratose. The gene prt is present in both serovars Typhi and Paratyphi A. The gene tyv encodes CDP-tyvelose epimerase, which converts CDP-paratose to CDP-tyvelose. The tyv gene is present in both serovars Typhi and Paratyphi A, but the tyv gene of serovar Paratyphi A does not produce active CDP-tyvelose epimerase due to the 1-bp deletion which causes the frameshift mutation and converts codon 4 of Tyv to a stop codon (8). We used this deleted region for the design of primer tyv-s to specifically detect the tyv gene of serovar Typhi but not of serovar Paratyphi A. The primers for the viaB gene were previously reported by Hashimoto et al. (3). The researchers reported the two kinds of primers. One specifically detects the Salmonella Vi antigen gene, and the other detects both the Salmonella and the Citrobacter freundii Vi antigen genes. We used the primers which detected only the Salmonella Vi antigen gene in this assay. The fliC-d and viaB genes are present in serovar Typhi, and the fliC-a gene is present in serovar Paratyphi A.
TABLE 1.
Primers for multiplex PCR amplification of Salmonella enterica serovars Typhi and Paratyphi A
| Gene and primer (oligonucleotide sequence) | Length (bp) | Amplified fragment size (bp) | Sourceb |
|---|---|---|---|
| tyv (rfbE) | |||
| tyv-s (5"-GAG GAA GGG AAA TGA AGC TTT T-3") | 22 | 615 | M29682 |
| tyv-as (5"-TAG CAA ACT GTC TCC CAC CAT AC-3") | 23 | M29682 | |
| prt (rfbS) | |||
| parat-s (5"-CTT GCT ATG GAA GAC ATA ACG AAC C-3") | 25 | 258 | M29682 |
| parat-as, (5"-CGT CTC CAT CAA AAG CTC CAT AGA-3") | 24 | M29682 | |
| viaB | |||
| vi-s (5"-GTT ATT TCA GCA TAA GGA G-3") | 19 | 439 | D14156 |
| vi-as (5"-CTT CCA TAC CAC TTT CCG-3") | 18 | D14156 | |
| fliC | |||
| fliCcom-s (5"-AAT CAA CAA CAA CCT GCA GCG-3") | 21 | L21912 | |
| fliCd-as (5"-GCA TAG CCA CCA TCA ATA ACC-3") | 21 | L21912 | |
| fliCa-as (5"-TAG TGC TTA ATG TAG CCG AAG G-3") | 22 | X03393 | |
| fliCcom-fliCd-as | 750 (489)a | ||
| fliCcom-fliCa-as | 329 |
Number in parentheses represents size of PCR product of H:j gene.
Primers were designed using sequences corresponding to indicated GenBank-EMBL-DDBL nucleotide sequence database accession numbers.
The PCR was carried out with a 50 μl mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 1 U of Taq DNA polymerase (Promega, Madison, Wis.), 0.2 mM deoxynucleoside triphosphate, a 0.1 μM concentration (each) of primers (tyv-s, tyv-as, parat-s, fliCcom-s, fliCd-as, and fliCa-as, a 0.2 μM concentration (each) of primers parat-as, vi-s, and vi-as, and 5 μl of the DNA sample. The PCR was carried out under the following conditions: 25 cycles with heat denaturation at 95°C for 30 s, primer annealing at 55°C for 60 s, and DNA extension at 72°C for 90 by a DNA thermal cycler (model 9600; Applied Biosystems, Foster City, Calif.). The amplified DNA was separated by 2% agarose gel electrophoresis, stained with ethidium bromide, and visualized by UV transillumination.
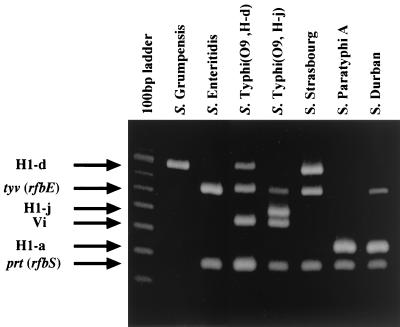
The multiplex PCR using five sets of primer pairs, which were targeted for the viaB, prt, tyv, fliC-d, and fliC-a genes, correctly identified Salmonella serovars Typhi and Paratyphi A and differentiated the two serovars by the combinations of the different-size bands produced: four positive bands, which consist of viaB, prt, tyv and fliC-d PCR products, in serovar Typhi and two positive bands, which consist of prt and fliC-a PCR products, in serovar Paratyphi A (Fig. 1). As expected, the prt primers in this study reacted with both serovars Typhi and Paratyphi A, yielding PCR products of the same size. The presence in both serovars Typhi and Paratyphi A of the prt gene was consistent with the findings of a previous report (8). The primers for tyv specifically detected the tyv gene of serovar Typhi. The prt primers also detected strains belonging to the O2 and O9 groups of Salmonella, and the tyv primers detected isolates of the Salmonella O9 group (Table 2). The primer pairs for fliC-d and fliC-a specifically detected the fliC-d and fliC-a genes, respectively, for the Salmonella serovars, and were able to distinguish fliC-d and fliC-a genes from other Salmonella serovar fliC genes. The primers for fliC-d also detected the fliC-j gene, which is an alternate phase of serovar Typhi H-1 antigen genes (5). Since fliC-j is a 261-bp deletion derivative of the fliC-d gene (2), the PCR product was smaller (Fig. 1).
FIG. 1.
Identification of Salmonella serovars Typhi, Paratyphi A, Enteritidis, Grumpensis, Strasbourg, and Durban by multiplex PCR. After the PCR, the PCR products were separated by 2% agarose gel electrophoresis.
TABLE 2.
Bacterial strains used to evaluate the specificities of multiplex PCRs and the multiplex PCR results
| Bacterium | Strain no. | Antigen structure
|
PCR resulta
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phage type | O antigen | H-1 | H-2b | tyv | fliC-d | viaB | fliC-a | prt | ||
| Salmonella enterica | ||||||||||
| Serovar Typhi | 990116 | D1 | 9,12,[Vi] | d | − | + | + | + | − | + |
| Serovar Typhi | 990120 | E1 | 9,12,[Vi] | d | − | + | + | + | − | + |
| Serovar Typhi | 990005 | UVS1 | 9,12,[Vi] | d | − | + | + | + | − | + |
| Serovar Typhi | 990006 | A | 9,12,[Vi] | d | − | + | + | + | − | + |
| Serovar Typhi | 990007 | E1 | 9,12,[Vi] | d | − | + | + | + | − | + |
| Serovar Typhi | 990008 | E1 | 9,12,[Vi] | d | − | + | + | + | − | + |
| Serovar Typhi | 990009 | E1 | 9,12,[Vi] | d | − | + | + | + | − | + |
| Serovar Typhi | 990012 | E1 | 9,12,[Vi] | d | − | + | + | + | − | + |
| Serovar Typhi | 990014 | E1 | 9,12,[Vi] | d | − | + | + | + | − | + |
| Serovar Typhi | 990037 | D1 | 9,12,[Vi] | d | − | + | + | + | − | + |
| Serovar Typhi | 980096 | 46 | 9,12,[Vi] | d | − | + | + | + | − | + |
| Serovar Typhi | 980111 | DVS | 9,12,[Vi] | d | − | + | + | + | − | + |
| Serovar Typhi | 980077 | UVS1 | 9,12,[Vi] | d | − | + | + | + | − | + |
| Serovar Typhi | 980014 | UVS1 | 9,12,[Vi] | j | − | + | +c | + | − | + |
| Serovar Typhi | GIFU9954 | Rough | d | − | + | + | + | − | + | |
| Serovar Paratyphi A | 000055 | 1 | 1,2,12 | a | [1,5] | − | − | − | + | + |
| Serovar Paratyphi A | 000056 | 1 | 1,2,12 | a | [1,5] | − | − | − | + | + |
| Serovar Paratyphi A | 990110 | 2 | 1,2,12 | a | [1,5] | − | − | − | + | + |
| Serovar Paratyphi A | 970083 | 2 | 1,2,12 | a | [1,5] | − | − | − | + | + |
| Serovar Paratyphi A | 960007 | 3 | 1,2,12 | a | [1,5] | − | − | − | + | + |
| Serovar Paratyphi A | 000001 | 4 | 1,2,12 | a | [1,5] | − | − | − | + | + |
| Serovar Paratyphi A | 000041 | 4 | 1,2,12 | a | [1,5] | − | − | − | + | + |
| Serovar Paratyphi A | 990081 | 5 | 1,2,12 | a | [1,5] | − | − | − | + | + |
| Serovar Paratyphi A | 970032 | 5 | 1,2,12 | a | [1,5] | − | − | − | + | + |
| Serovar Paratyphi A | 990046 | 6 | 1,2,12 | a | [1,5] | − | − | − | + | + |
| Serovar Paratyphi A | 990103 | 6 | 1,2,12 | a | [1,5] | − | − | − | + | + |
| Serovar Chester | 99023 | 1,4,[5],12 | e,h | e,n,x | − | − | − | − | − | |
| Serovar Agona | 99076 | 1,4,[5],12 | f,g,s | [1,2] | − | − | − | − | − | |
| Serovar Oranienburg | 99026 | 6,7,14 | m,t | [z57] | − | − | − | − | − | |
| Serovar Infantis | 99063 | 6,7,14 | r | 1,5 | − | − | − | − | − | |
| Serovar Litchfield | 99087 | 6,8 | l,v | 1,2 | − | − | − | − | − | |
| Serovar Hadar | 99114 | 6,8 | z10 | e,n,x | − | − | − | − | − | |
| Serovar Enteritidis | 99109 | 1,9,12 | [f],g,m,[p] | [1,7] | + | − | − | − | + | |
| Serovar Javiana | 99112 | 1,9,12 | l,z28 | 1,5 | + | − | − | − | + | |
| Serovar Senftenberg | 99017 | 1,3,19 | g,[s],t | − | − | − | − | − | − | |
| Serovar Grumpensis | 99089 | 13,23 | d | 1,7 | − | + | − | − | − | |
| Serovar Poona | 99108 | 1,13,22 | z | 1,6 | − | − | − | − | − | |
| Serovar Typhimurium | 1363 | 1,4,[5],12 | i | 1,2 | − | − | − | − | − | |
| Serovar Enteritidis | 1364 | 1,9,12 | [f],g,m,[p] | [1,7] | + | − | − | − | + | |
| Serovar Weltereden | 1365 | 3,10[15] | r | z6 | − | − | − | − | − | |
| Serovar Durban | S-222 | 9,12 | a | e,n,Z15 | + | − | − | + | + | |
| Serovar Strasbourg | S-214 | 9,46 | d | 1,7 | + | + | − | − | + | |
| Serovar Ndolo | S-154 | 1,9,12 | d | 1,5 | + | + | − | − | + | |
| Serovar Paratyphi C | GIFU12823 | 6,7,[Vi] | c | 1,5 | − | − | + | − | − | |
| Serovar Dublin | GIFU13011 | 1,9,12[Vi] | g,p | − | + | − | + | − | + | |
| Citrobacter freundii | Vi+ | − | −d | − | − | − | ||||
| Yersinia pseudotuberculosis | 1b | − | − | − | − | − | ||||
| Yersinia pseudotuberculosis | 2a | − | − | − | − | − | ||||
| Yersinia pseudotuberculosis | 2b | − | − | − | − | − | ||||
| Yersinia pseudotuberculosis | 4a | − | − | − | − | − | ||||
| Yersinia pseudotuberculosis | 4b | − | − | − | − | − | ||||
| Yersinia pseudotuberculosis | 5b | − | − | − | − | − | ||||
| Yersinia enterocolitica | O3 | − | − | − | − | − | ||||
| Yersinia enterocolitica | O5 | − | − | − | − | − | ||||
| Yersinia enterocolitica | O8 | − | − | − | − | − | ||||
| Yersinia enterocolitica | O9 | − | − | − | − | − | ||||
| Vibrio cholerae eltor Ogawa | O1 | − | − | − | − | − | ||||
| Vibrio cholerae eltor Inaba | O1 | − | − | − | − | − | ||||
| Vibrio cholerae | O139 | − | − | − | − | − | ||||
| Vibrio cholerae | non-O1, non-O139 | − | − | − | − | − | ||||
| Vibrio mimicus | − | − | − | − | − | |||||
| Vibrio parahaemolyticus | − | − | − | − | − | |||||
| Vibrio fluvialis | − | − | − | − | − | |||||
| Aeromonas hydrophila | − | − | − | − | − | |||||
| Aeromonas sobria | − | − | − | − | − | |||||
| Aeromonas caviae | − | − | − | − | − | |||||
| Escherichia coli | − | − | − | − | − | |||||
| Shigella dysenteriae | − | − | − | − | − | |||||
| Shigella flexneri | − | − | − | − | − | |||||
| Shigella boidyii | − | − | − | − | − | |||||
| Shigella sonnei | − | − | − | − | − | |||||
+, PCR positives −, PCR negative.
−, no H-2 phase.
H1-j antigen.
Our primers for the viaB gene did not react with Vi antigen genes of C. freundii.
To examine possible cross-reactions of the selected viaB, prt, tyv, and fliC primers among major enteric pathogens, including the several genera of the family Enterobacteriaceae, some strains were tested by the multiplex PCR assay; none showed positive results (Table 2). To further evaluate the primer specificities for Salmonella species, we tested several kinds of salmonella serovars. Detection of both prt and fliC-a correctly identified serovar Paratyphi A. Detection of the combination of viaB, tyv, and fliC-d correctly identified serovar Typhi. Hashimoto et al. (3) reported that the PCR primers specific for the Vi antigen gene also reacted with the chromosomal DNAs of serovars Paratyphi C and Dublin which possessed Vi antigen genes. However, our system, which targeted not only the Vi antigen gene but also serogroup O9 and H:d genes for identification, discriminated serovar Typhi from serovars Paratyphi C and S. Dublin. Furthermore, our system differentiated serovar Typhi from serovar Strasbourg (9,46:d:1,7), and serovar Ndolo (1,9,12:d:1,5) possessed O9 and H:d genes but not the Vi antigen gene (Table 2). Similarly, other O9 and H:d group strains without the Vi antigen, such as serovars Tarshyne (9,12:d:1,6), Eschberg (9,12:d:1,7), Bangui (9,12:d:e,n,z15), Zega(9,12:d:z6), Jaffna (1,9,12:d:z35), Ontario (9,46:d:1,5), Quentin (9,46:d:1,6), Olten (9,46:d:e,n,z15), and Plymouth (9,46:d:z6), may be also discriminated from serovar Typhi by this assay. Here we used purified colonies of each strain for our PCR. We plan to examine whether our system is usable for direct detection from clinical samples.
Taken together, the methods described here may make possible the detection and/or identification of clinically important strains of Salmonella serovars Typhi and Paratyphi A strains within a few working days of the arrival of specimens in the diagnostic microbiology laboratory.
In this study, we used the primers for viaB, tyv, prt, fliC-a, and fliC-d genes only. However, if specific primers are designed for the amplification of other flagellin genes and/or other O-antigen synthase genes, it might be possible to identify other human-pathogenic Salmonella serovars by the combination of O- and H-antigen-specific gene-targeted PCR primers with multiplex PCR.
Acknowledgments
This work was partially supported by a grant for International Health Cooperation Research (12A-1) from the Ministry of Health, Labor, and Welfare (to K.H.).
REFERENCES
- 1.Anand, A. C., V. K. Kataria, W. Singh, and S. K. Chatterjee. 1990. Epidemic multiresistant enteric fever in eastern India. Lancet 335:352.. [DOI] [PubMed] [Google Scholar]
- 2.Frankel, G., S. M. Newton, G. K. Schoolnik, and B. A. Stocker. 1989. Intragenic recombination in a flagellin gene: characterization of the H1-j gene of Salmonella typhi. EMBO J. 8:3149-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hashimoto, Y., Y. Itho, Y. Fujinaga, A. Q. Khan, F. Sultana, M. Miyake, K. Hirose, H. Yamamoto, and T. Ezaki. 1995. Development of nested PCR based on the ViaB sequence to detect Salmonella typhi. J. Clin. Microbiol. 33:775-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirose, K., K. Tamura, H. Sagara, and H. Watanabe. 2001. Antibiotic susceptibilities of Salmonella enterica serovar Typhi and S. enterica serovar Paratyphi A isolated from patients in Japan. Antimicrob. Agents Chemother. 45:956-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kauffman, F. 1936. Über die diphasische Natur der Typhusbacillen. Z. Hyg. 119:103-118. [Google Scholar]
- 6.Rowe, B., L. R. Ward, and E. J. Threlfall. 1995. Ciprofloxacin-resistant Salmonella typhi in the UK. Lancet 346:1302.. [DOI] [PubMed] [Google Scholar]
- 7.Song, J.-H., H. Cho, M. Y. Park, D. S. Na, H. B. Moon, and C. H. Pai. 1993. Detection of Salmonella typhi in the blood of patients with typhoid fever by polymerase chain reaction. J. Clin. Microbiol. 31:1439-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verma, N., and P. Reeves. 1989. Identification and sequence of rfbS and rfbE, which determine antigenic specificity of group A and group D salmonellae. J. Bacteriol. 171:5694-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu, Q., C. K. Lim, and Y. N. Chan. 1996. Detection of Salmonella typhi by polymerase chain reaction. J. Appl. Bacteriol. 80:244-251. [DOI] [PubMed] [Google Scholar]