Abstract
Pyrazinamide (PZA), an analog of nicotinamide, is a prodrug for tuberculosis which requires conversion to the bactericidal compound pyrazinoic acid by bacterial pyrazinamidase activity. Mutations leading to a loss of pyrazinamidase activity cause PZA resistance in Mycobacterium tuberculosis. Thus, the detection of pyrazinamidase activity makes the discrimination of PZA-resistant tuberculosis possible. However, the detection of the pyrazinamidase activity of M. tuberculosis isolates needs a large amount of bacilli and is therefore time consuming. In this paper, we describe a new method for the detection of pyrazinamidase activity with a PCR-based system. The genes encoding pyrazinamidase (pncA genes) in 30 resistant clinical isolates were amplified by PCR by using forward primers containing bacteriophage T7 promoter sequences at their 5" ends. Then the PCR products were directly subjected to an in vitro transcription-translation coupled system. All of the PZA-resistant isolates tested showed reduced pyrazinamidase activity compared to susceptible M. tuberculosis type strain H37Rv. In contrast, all of the 15 susceptible clinical isolates exhibited pyrazinamidase activities similar to that of H37Rv. This fact suggested the possibility of the usefulness of this system for the rapid detection of PZA-resistant M. tuberculosis.
Tuberculosis (TB) caused by members of Mycobacterium tuberculosis complex is one of the common human diseases, causing 3,000,000 deaths per year worldwide (23). While the disease is associated with economic impoverishment, TB is on the rise in many industrialized nations. The spread of TB is due to immigration, the emergence of drug-resistant strains (24), and the AIDS epidemics. The increasing number of drug-resistant M. tuberculosis strains has made the rapid identification of susceptibility clinically important because those patients suffering from these strains require specialized antibiotic treatment.
Recently, a number of genetic changes leading to rifampin, isoniazide, pyrazinamide (PZA), streptomycin, and kanamycin resistances have been characterized. Nearly 95% of rifampin-resistant M. tuberculosis strains carry mutations in a certain region of the beta subunit of the RNA polymerase encoded by the rpoB gene (20, 21). The mutations correlated to isoniazide resistance were found on the katG (25), inhA (1), and ahpC/oxyR (26) genes. Ethambutol-resistant M. tuberculosis strains carried mutations on a certain part of the gene coding for alabinosyl transferase (2, 17). Mutations on the rrs and rpsL genes coding for the 16S ribosomal RNA and ribosome protein S12, respectively, confer streptomycin resistance (8, 11). A part of the kanamycin-resistant phenotype can be detected by identifying the mutations on the rrs gene (19). Missense mutations in the DNA gyrase gene (gyrA) have been reported to be associated with increased levels of resistance to fluoroquinolones (5).
PZA is one of the most important drugs for anti-TB short-course chemotherapy. PZA is converted to pyrazinoic acid by mediation of pyrazinamidase in M. tuberculosis, and it inhibits fatty acid synthesis. Reduced pyrazinamidase activity correlates well with resistance to PZA. The mutations that confer PZA resistance have been investigated on the pyrazinamidase gene (pncA) by Scorpio and Zhang (15). Thus, finding mutations on the pncA gene makes the rapid detection of PZA-resistant M. tuberculosis possible.
A number of rapid and simple methods based on the PCR technique have been developed for the detection of a mutation on the gene responsible for the drug resistance. PCR single-strand conformational polymorphism (21) and molecular beacon sequence analysis (14) are suitable for the detection of a limited number of mutations on specific genes. PCR product direct sequencing and DNA microarray (9) can detect many mutations at the same time. The latter two were suitable for the detection of PZA-resistant M. tuberculosis because the mutations responsible for PZA resistance were dispersed on the pncA gene. However, as these methods require costly apparatus, such as a DNA sequencer or laser scanner, they can not be done in the small clinical laboratory.
In this report, we explain a new technique using PCR and an in vitro transcription-translation coupled system that enabled the rapid pyrazinamidase assay and PZA susceptibility test to be done in the small clinical laboratory. By this method, PZA-resistant M. tuberculosis can be detected within 8 h without costly apparatus.
MATERIALS AND METHODS
Bacterial strains.
M. tuberculosis strain H37Rv and Mycobacterium bovis BCG strain Tokyo have been maintained in our laboratory. The 30 PZA-resistant clinical strains used in this study were isolated in Osaka Prefectural Habikino Hospital and National Sanatorium Minami-Okayama Hospital and maintained in our laboratory. The susceptibilities to PZA of the clinical isolates were determined on 7H10 agar (Difco) slants (pH 5.5) containing 100 μg of PZA/ml. The TB bacilli grown on the Ogawa medium were suspended in phosphate-buffered saline, spread onto the slants, and incubated for 4 weeks at 37°C to determine their susceptibilities. The pyrazinamidase tests were performed according to the method of McClatchy et al. (13).
DNA preparation.
M. tuberculosis strains were grown on Ogawa medium. Mycobacterial DNA was extracted from a single colony by a combination of chloroform extraction and mechanical disruption (12).
Amplification of the pncA gene carrying the T7 promoter.
The pncA gene fragments carrying the T7 promoter sequence were amplified by PCR with the primers shown in Fig. 1. Amplification was performed in a 0.2-ml PCR tube with a total reaction volume of 50 μl by using a personal thermal cycler (Takara Shuzo Co. Ltd., Kyoto, Japan). Each reaction tube contained 2.5 U of Taq DNA polymerase; 200 μM (each) dATP, dCTP, dGTP, and dTTP; 20 mM Tris-HCl; 50 mM KCl; 2.5 mM MgCl2; sample DNA; and the primers presented in Fig. 1 and Table 1. A PCR round was conducted with an initial 1-min denaturation step at 98°C coupled to a repeating cycle of 5 s at 98°C, 10 s at 55°C, and 1 min at 72°C for 35 cycles, followed by a 7-min final extension step at 72°C.
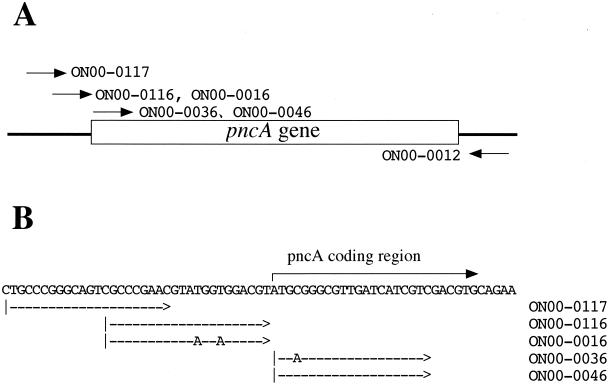
FIG. 1.
Primers used for amplification of the pncA gene of M. tuberculosis. (A) Schematic presentation of the positions of the primers used. (B) Exact positions of the primers used. The sequence TGGT in primer ON00-0016 was converted to AGGA to give an optimum ribosome binding sequence (Shine-Dalgarno sequence) in primer ON00-0116. The sequence ATGC in primer ON00-0046 was converted to ATGA to give a Kozak consensus sequence in primer ON00-0036.
TABLE 1.
Primers used for amplification of the pncA gene
| Primer | Sequencea | Remark |
|---|---|---|
| ON00-0117 | GGATCCTAATACGACTCACTATAGGGAGCCCTGCCCGGGCAGTCGCCCGAAC | |
| ON00-0016 | GGATCCTAATACGACTCACTATAGGGAGCCCGCCCGAACGTATGGTGGACGT | |
| ON00-0116 | GGATCCTAATACGACTCACTATAGGGAGCCCGCCCGAACGTAAGCAGGACGT | Good SDb |
| ON00-0046 | GGATCCTAATACGACTCACTATAGGGAGCCACCATGCGGGCGTTGATCATCGTC | |
| ON00-0036 | GGATCCTAATACGACTCACTATAGGGAGCCACCATGAGGGCGTTGATCATCGTC | Good Kozakc |
| ON00-0012 | GGGGTCGACCACCGCCGCCAACAGTTCATCCC |
The sequence encoding the T7 phage promoter is shown in boldface type.
The sequence TGGT in ON00-0016 was converted to AGGA to give an optimum ribosome binding sequence (Shine-Dalgarno [SD] sequence [16]).
The sequence ATGC in ON00-0046 was converted to ATGA to give a Kozak consensus sequence.
In vitro synthesis of pyrazinamidase from PCR product.
In vitro transcription-translation coupled systems (TNT T7 quick for PCR DNA, TNT-coupled Wheat Germ Extract systems, and E. coli T7 S30 Extract system; Promega Corporation, Madison, Wis.) were used for the synthesis of pyrazinamidase from the PCR-amplified pncA gene carrying the bacteriophage T7 promoter sequence. These systems contain the cell lysates of rabbit reticulocyte, wheat germ, or Escherichia coli with bacteriophage T7 RNA polymerase. mRNAs are transcribed from the T7 promoter existing upstream of the pncA gene PCR product. Then pyrazinamidases are translated from the mRNAs by the machinery in the lysates.
Protein synthesis was performed mainly according to the manufacturer's manual. Briefly, the amplified pncA gene fragment was first qualified by analysis by agarose gel electrophoresis with a DNA standard of known concentration, then precipitated by adding 5 μl of 3 M sodium acetate and 1.00 μl of ethanol, and centrifuged at 9,000 × g for 5 min. After being washed with 70% ethanol, the resulting precipitate was dissolved in an appropriate volume of distilled water to bring the solution to a concentration of 100 ng/μl. Then, 2 μl of DNA solution was added to 20 μl of either rabbit reticulocyte lysate, wheat germ lysate, or E. coli S30 lysate containing T7 RNA polymerase. After the addition of 1 μl of 0.5 mM methionine, the in vitro RNA and protein synthesis was performed at 30°C for 0, 60, 90, 120, and 240 min. The resulting lysates were directly used for the micropyrazinamidase assay.
Micropyrazinamidase assay.
Measurement of pyrazinamidase activities was performed with some modification of the method described by McClatchy et al. (13). Twenty microliters of the lysate, after the in vitro synthesis of pyrazinamidase, was added to 100 μl of 10 mg of PZA solution/ml in distilled water and incubated for 1 h at 37°C. After incubation, 10 μl of 10% ammonium iron(II) sulfate was added to the reaction mixture, followed by further incubation for 2 h at 37°C. In the case of the experiments with the wheat germ and E. coli lysates, direct measuring of the optical density at 450 nm gave the pyrazinamidase activity because the maximum absorbance of brown product from the pyrazinamidase test was observed at 450 nm. In the case of the experiment with the rabbit reticulocyte lysate, 130 μl of 25% polyethylene glycol 6000 solution was added to the resulting reaction mixture and the red colored hemoglobin interfering with the accurate measurement of pyrazinamidase activity was removed by passing the mixture through a Micropure-EZ centrifuge filter (Millipore Corporation, Bedford, Mass.). Pyrazinamidase activity was then measured by looking at the optical density at 450 nm.
RESULTS
Amplification of T7 promoter-tailed pncA gene.
Five kinds of T7 promoters 5" attached to pncA gene fragments were amplified by PCR with the primers shown in Fig. 1 and Table 1. All of the primers used in this study amplified the necessary amount of DNA fragment when the DNA extracted from M. tuberculosis strain H37Rv was used (data not shown). The primer pairs ON00-0036 plus ON00-0012 and ON00-0046 plus ON00-0012 were tested for the amplification of the pncA gene of M. bovis BCG and PZA-resistant M. tuberculosis clinical isolates to get similar amounts of PCR products (data not shown).
Monitoring of in vitro pyrazinamidase synthesis.
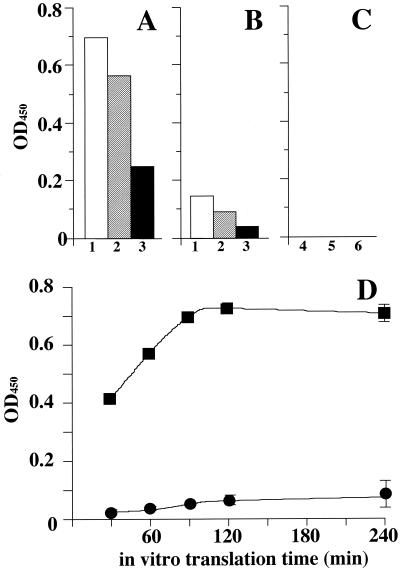
Pyrazinamidase synthesized by an in vitro transcription-translation coupled system was analyzed by a micropyrazinamidase assay. Figure 2 shows a comparison of the pyrazinamidase activities in the three systems, rabbit reticulocyte, wheat germ, and E. coli S30, with five kinds of PCR products. The highest level of pyrazinamidase activity was obtained by the rabbit reticulocyte lysate system with the DNA amplified by using ON00-0036 and ON00-0012 as primers. The next highest activity was obtained by the rabbit reticulocyte lysate with the DNA amplified by using ON00-0046 and ON00-0012 as primers. Reduced activity was observed with the wheat germ lysate system, and no pyrazinamidase activity was observed with the E. coli S30 system (Fig. 2A to C). Thus, in vitro pyrazinamidase synthesis was optimized by using PCR products amplified with ON00-0036 and ON00-0012 as primers. A time course study of the in vitro pyrazinamidase synthesis was performed with the pncA gene fragment carrying the T7 promoter amplified from M. tuberculosis H37Rv and M. bovis BCG. The 0.2 μg of PCR products was mixed with 20 μl of rabbit reticulocyte lysate and incubated for 30, 60, 90, 120, and 240 min. The pyrazinamidase activities in the reaction mixtures were measured by a micropyrazinamidase assay. The pyrazinamidase activity of the reaction mixture containing M. tuberculosis pncA PCR product increased from 30 to 90 min in a time-dependent manner. In contrast, faint pyrazinamidase activity was observed in the reaction mixture containing the M. bovis BCG pncA PCR product (Fig. 2B). This observation exhibited a good correlation with the pyrazinamidase activity of these bacilli (Table 2). Because the difference between the pyrazinamidase activities reached its maximum at the incubation time of 90 min, we used 90 min for in vitro pyrazinamidase synthesis in the experiments with the clinical isolates.
FIG. 2.
Activities of in vitro-synthesized pyrazinamidase. The pyrazinamidase activities synthesized in rabbit reticulocyte lysate (A), wheat germ lysate (B), and E. coli S30 lysate (C) were examined as described in Materials and Methods. The white, hatched, and filled bars in panels A and B represented the pyrazinamidase activities obtained from the experiments using ON00-0036, ON00-0046, and ON00-0016 as forward primers, respectively. As shown in panel C, although ON00-0116 (bar 4), ON00-0016 (bar 5), and ON00-0017 (bar 6) were used as forward primers for the time course study, no pyrazinamidase activity was observed. (D) The time course study of the pyrazinamidase activities synthesized in rabbit reticulocyte lysate with ON00-0036 as the forward primer was performed. Templates used for the PCR whose results are shown in panel D were M. tuberculosis H37Rv (PZA susceptible; filled square) and M. bovis BCG (PZA resistant; filled circle). OD450, optical density at 450 nm.
TABLE 2.
Characteristics of M. tuberculosis strains examined in this study
| Strain | PZA susceptibilitya | PZase activityb | Mutation(s) |
|---|---|---|---|
| 98-076 | R | − | 14-T→A |
| 98-075 | R | − | 100-T→G |
| 98-073 | R | − | Base 486-496 deleted |
| 98-069 | R | − | Deletion of the entire gene |
| 98-064 | R | − | 403-A→C |
| 98-062 | R | − | 169-C→G |
| 98-060 | R | − | 260/261 ACc |
| 98-053 | R | − | Base 449 deleted |
| 98-052 | R | − | 181-A→C, 416-T→C |
| 98-048 | R | − | 385/386 CGc |
| 98-047 | R | − | 184/185 Tc 290-G→T |
| 98-009 | R | − | 412-T→C |
| 97-001s | R | − | Bases 182-183 deleted |
| 97-020 | R | − | 24-C→G |
| 97-019 | R | − | 104-T→C |
| 97-015 | R | − | 415-G→T |
| 97-012 | R | − | 152-A→C |
| 97-010 | R | − | 350/351 Tc |
| 97-009 | R | − | 146-A→C |
| 97-008 | R | − | 467-T→A |
| 97-006 | R | − | 212-A→C |
| 97-004 | R | − | 297-C→A |
| 97-001 | R | − | 383-T→G |
| 96-004 | R | − | Base 412 deleted |
| 94-067 | R | − | 230/231 IS 6110c |
| 94-054 | R | − | 8-C→A |
| 94-053 | R | − | 391/392 GGc |
| 94-050 | R | − | 407/408 GCc |
| 94-048 | R | − | 170-A→C |
| 94-018 | R | − | 40-T→C |
| 98-120 | S | + | None |
| 98-119 | S | + | None |
| 98-113 | S | + | None |
| 98-110 | S | + | None |
| 98-103 | S | + | None |
| 98-108 | S | + | None |
| 98-107 | S | + | None |
| 98-105 | S | + | None |
| 98-104 | S | + | None |
| 98-099 | S | + | None |
| 98-095 | S | + | None |
| 98-071 | S | + | None |
| 98-063 | S | + | None |
| 98-058 | S | + | None |
| 98-057 | S | + | None |
| 98-056 | S | + | None |
| H37Rv | S | + | None |
| BCG | R | − | 169-C→G |
R, resistant; S, susceptible.
PZase, pyrazinamidase; −, absent; +, present.
Insertion of base(s).
Activities of in vitro-synthesized pyrazinamidase in PZA-resistant M. tuberculosis clinical isolates.
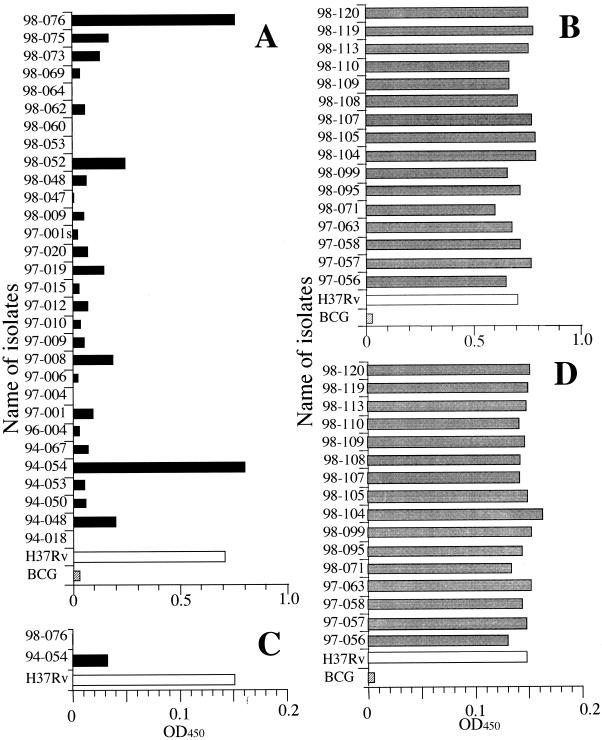
The pyrazinamidase activities of in vitro-synthesized pyrazinamidase from 30 M. tuberculosis clinical isolates were examined by the method developed as described above. Of 30 samples tested, 28 exhibited pyrazinamidase activity lower than that of PZA-susceptible M. tuberculosis H37Rv (Fig. 3A). The samples from strains TB94-054 and TB98-076 exhibited high levels of pyrazinamidase activity. The reason for the high levels of pyrazinamidase activity in these samples was that the positions of the mutations were contained in the 5" primer used. Thus, we used the primer ON00-0016 instead of ON00-0036 (Fig. 1 and Table 1) for further studies. Though the pyrazinamidase activity of M. tuberculosis H37Rv exhibited in this system was low, the activities of TB94-054 and TB98-076 were much lower and more distinguishable (Fig. 3B).
FIG. 3.
In vitro pyrazinamidase activities of PZA-resistant clinical isolates. (A) The pyrazinamidase activities synthesized with rabbit reticulocyte lysate obtained from the experiments with ON00-0036 as a forward primer were examined. Templates used for the PCR were DNAs from M. tuberculosis H37Rv (PZA susceptible; white bar), M. bovis BCG (PZA resistant; hatched bar), and PZA-resistant clinical isolates (black bars). (B) The same experiments were performed with PZA-susceptible clinical isolates (gray bars), and the results were compared with the results obtained with M. tuberculosis H37Rv (white bar) and M. bovis BCG (hatched bar). (C) The pyrazinamidase activities synthesized with rabbit reticulocyte lysate obtained from the experiments with ON00-0016 as a forward primer were examined. Templates used for the PCR were DNAs from M. tuberculosis H37Rv (white bar) and PZA-resistant clinical isolates (black bar). (D) The same experiments were performed with PZA-susceptible clinical isolates (gray bars), and the results were compared with the results obtained with M. tuberculosis H37Rv (white bar) and M. bovis BCG (hatched bar). OD450, optical density at 450 nm.
DISCUSSION
We have been interested in studying the mutations on a certain gene responsible for drug resistance of M. tuberculosis. Numerous efforts have been made in this field. A lot of mutations related to the resistance to anti-TB drugs were investigated for rifampin (18, 20, 21), isoniazide (1, 25, 26), ethambutol (2, 17), streptomycin (8, 11), kanamycin (19), and fluoroquinolones (5). The same kind of work on PZA is especially important because the susceptibility test for M. tuberculosis with media is sometimes unsettled due to the low level of pH dependence of this method (3). Lots of experiments have been performed with special emphasis on the gene encoding pyrazinamidase (the pncA gene) as a responsible gene (15). Nearly 90% of PZA-resistant M. tuberculosis isolates carry mutations in the coding region of pncA, and about 5% carried mutations in the ribosome binding site of this gene (6, 7, 10). Theoretically, sequencing the pncA gene by using a DNA sequencer or a DNA microarray makes the rapid detection of PZA-resistant M. tuberculosis possible. However, these methods require costly apparatus. The newly reported methods for the detection of single-point mutations, such as PCR single-strand conformational polymorphism (21) and molecular beacon sequence analysis (14), were applied for the rapid detection of rifampin- or other drug-resistant TB. However, they are not suitable for the detection of PZA-resistant TB because of the dispersed existence of the mutations on the pncA gene. Therefore, a rapid method that did not involve finding the mutations seemed to be important.
As mentioned above, PZA is a drug that is only active at a low pH of around 5, and the identification of PZA resistance on media is not easy. So, the PZA susceptibility test now depends on the pyrazinamidase test because of the strong correlation of reduced pyrazinamidase activity with the PZA resistance phenotype (4, 13, 22). However, the pyrazinamidase assay method has shortcomings; e.g., it takes at least 3 weeks to prepare a sufficient amount of bacilli and it also takes 4 days for the pyrazinamidase assay. Thus, the in vitro pyrazinamidase synthesis and assay method was designed and examined in this study.
The DNA fragments of the pncA gene of M. tuberculosis carrying the bacteriophage T7 promoter were amplified by PCR, and the pyrazinamidase was synthesized by in vitro transcription-translation coupled systems. Five kinds of PCR products were examined by this system for the ability to produce pyrazinamidase. The DNA sequences attached to the pncA gene were as follows. (i) The T7 promoter sequence and an additional ACC were attached just upstream of the initiation codon, and the sequence around the initiation codon was converted to a Kozak consensus sequence (25) (from ATGC to ATGA, primer ON00-0036). (ii) The T7 promoter sequence and an additional ACC were attached just upstream of the pncA gene initiation codon (primer ON00-0046). (iii) The T7 promoter sequence was attached to the −22 position of the pncA gene (primer ON00-0016). (iv) The T7 promoter sequence was attached to the −22 position of the pncA gene, and the sequence of the putative ribosome binding region was converted to a Shine-Dalgarno consensus sequence (26) (−10 to −7, from TGGT to AGGA, primer ON00-0116). (v) The T7 promoter sequence was attached to the −35 position of the pncA gene (primer ON00-0117). The amplification efficiencies of the pncA gene with each of these primers were almost the same. Thus, the pyrazinamidase activities expressed with the three kinds of lysates, rabbit reticulocyte, wheat germ, and E. coli S30, were examined. From the results, the pncA PCR product carrying the T7 promoter, followed by the initiation codon with a Kozak consensus sequence worked most efficiently with the rabbit reticulocyte lysate. The reason why no pyrazinamidase activity was observed with the E. coli S30 system is not clear. There might exist some incompatibility, such as codon usage and/or molecular chaperon of the amplified pncA gene, with the E. coli S30 system. By using this system, 30 resistant isolates were tested with the in vitro system. Of 30 resistant strains, 28 exhibited reduced pyrazinamidase activity compared to the PZA-susceptible strain H37Rv (Fig. 3A), whereas activities similar to that of H37Rv were obtained with PZA-susceptible clinical isolates (Fig. 3B). Two isolates, TB94-054 and TB98-076, carrying the mutations 8-C→A and 14-T→A, respectively, exhibited pyrazinamidase activities similar to that of strain H37Rv (Table 2) with this method. These mutated bases were included in primer ON00-0036, and the PCR products from these strains had the wild-type sequence. Therefore, the pyrazinamidase activity was exhibited at a level similar to that of the PZA-susceptible strain H37Rv. Although the pyrazinamidase activity expressed was lower with primer ON00-0016 (Fig. 2A), it was necessary to use this primer for looking at the pyrazinamidase activity of TB94-054 and TB98-076. Expectedly, reduced pyrazinamidase activities compared to that of H37Rv were observed with this system (Fig. 3C), whereas activities similar to that of H37Rv were obtained with PZA-susceptible clinical isolates (Fig. 3D). These results indicated the possibility of detecting more than 90% of PZA-resistant M. tuberculosis with the in vitro pyrazinamidase assay method established in this study. In addition, some of the resistant isolates demonstrated weak pyrazinamidase activities by the in vitro method. This might correlate with the MIC, and PZA might still be effective, particularly if it is administered as part of combination therapy. This should be further investigated in future studies.
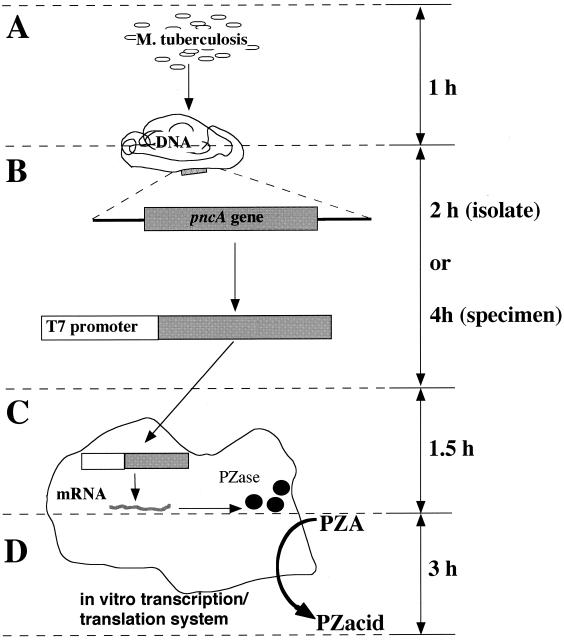
The steps of the in vitro pyrazinamidase assay method established in this study are presented schematically in Fig. 4. The amounts of time required for DNA extraction, pncA gene amplification, pyrazinamidase in vitro synthesis, and the pyrazinamidase assay were 1, 2 to 4, 1.5, and 3 h, respectively. By this method, the pyrazinamidase activity of M. tuberculosis can be detected within 8 h from clinical isolates and within 10 h from clinical specimens (using nested PCR for amplification of the pncA gene), whereas more than 3 weeks are necessary (3 weeks for the culture and 4 days for the pyrazinamidase test) for the conventional method. The method developed in this study can dramatically reduce the time for the detection of PZA-resistant M. tuberculosis. As the procedure of this method is very simple, safe, and cost saving, the in vitro pyrazinamidase assay method can serve as the standard method for the detection of PZA-resistant M. tuberculosis.
FIG. 4.
Schematic representation of the in vitro pyrazinamidase assay. The in vitro pyrazinamidase assay consisted of four steps. The total time required to complete the procedure is less than 8 h. (A) Extraction of DNAs from samples takes 1 h. (B) PCR amplification of the pncA gene carrying the T7 promoter takes 2 or 4 h. (C) In vitro synthesis of pyrazinamidase takes 1.5 h. (D) The micropyrazinamidase assay to look at the conversion of PZA to pyrazinoic acid (PZacid) takes 3 h. PZase, pyrazinamidase.
Acknowledgments
We thank Chiyoji Abe for kind advice.
This work was supported in part by the Health Science Grant “Research on Emerging and Reemerging Diseases” from the Ministry of Health and Welfare, Tokyo, Japan.
REFERENCES
- 1.Banerjee, A., E. Dubnau, A. Quemard, V. Balasubramanian, K. S. Um, T. Wilson, D. Collins, G. de Lisle, and W. R. Jacobs, Jr. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227-230. [DOI] [PubMed] [Google Scholar]
- 2.Belanger, A. E., G. S. Besra, M. E. Ford, K. Mikusova, J. T. Belisle, P. J. Brennan, and J. M. Inamine. 1996. The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc. Natl. Acad. Sci. USA 93:11919-11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler, W. R., and J. O. Kilburn. 1982. Improved method for testing susceptibility of Mycobacterium tuberculosis to pyrazinamide. J. Clin. Microbiol. 16:1106-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler, W. R., and J. O. Kilburn. 1983. Susceptibility of Mycobacterium tuberculosis to pyrazinamide and its relationship to pyrazinamidase activity. Antimicrob. Agents Chemother. 24:600-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cambau, E., W. Sougakoff, M. Besson, C. Truffot-Pernot, J. Grosset, and V. Jarlier. 1994. Selection of a gyrA mutant of Mycobacterium tuberculosis resistant to fluoroquinolones during treatment with ofloxacin. J. Infect. Dis. 170:479-483. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, S. J., L. Thibert, T. Sanchez, L. Heifets, and Y. Zhang. 2000. pncA mutations as a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis: spread of a monoresistant strain in Quebec, Canada. Antimicrob. Agents Chemother. 44:528-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies, A. P., O. J. Billington, T. D. McHugh, D. A. Mitchison, and S. H. Gillespie. 2000. Comparison of phenotypic and genotypic methods for pyrazinamide susceptibility testing with Mycobacterium tuberculosis. J. Clin. Microbiol. 38:3686-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finken, M., P. Kirschner, A. Meier, A. Wrede, and E. C. Bottger. 1993. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol. Microbiol. 9:1239-1246. [DOI] [PubMed] [Google Scholar]
- 9.Gingeras, T. R., G. Ghandour, E. Wang, A. Berno, P. M. Small, F. Drobniewski, D. Alland, E. Desmond, M. Holodniy, and J. Drenkow. 1998. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Res. 8:435-448. [DOI] [PubMed] [Google Scholar]
- 10.Hirano, K., M. Takahashi, Y. Kazumi, Y. Fukasawa, and C. Abe. 1998. Mutation in pncA is a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis. Tuber. Lung Dis. 78:117-122. [DOI] [PubMed] [Google Scholar]
- 11.Katsukawa. C., A. Tamaru, Y. Miyata, C. Abe, M. Makino, and Y. Suzuki. 1997. Characterization of the rpsL and rrs genes of streptomycin-resistant clinical isolates of Mycobacterium tuberculosis in Japan. J. Appl. Microbiol. 83:634-640. [DOI] [PubMed] [Google Scholar]
- 12.Kozak, M. 1987. An analysis of 5"-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15:8125-8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McClatchy, J. K., A. Y. Tsang, and M. S. Cernich. 1981. Use of pyrazinamidase activity in Mycobacterium tuberculosis as a rapid method for determination of pyrazinamide susceptibility. Antimicrob. Agents Chemother. 20:556-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piatek, A. S., S. Tyagi, A. C. Pol, A. Telenti, L. P. Miller, F. R. Kramer, and D. Alland. 1998. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol. 16:359-363. [DOI] [PubMed] [Google Scholar]
- 15.Scorpio, A., and Y. Zhang. 1996. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 2:662-667. [DOI] [PubMed] [Google Scholar]
- 16.Shine, J., and L. Dalgarno. 1974. The 3"-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 71:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sreevatsan, S., K. E. Stockbauer, X. Pan, B. N. Kreiswirth, S. L. Moghazeh, W. R. Jacobs, Jr., A. Telenti, and J. M. Musser. 1997. Ethambutol resistance in Mycobacterium tuberculosis: critical role of embB mutations. Antimicrob. Agents Chemother. 41:1677-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki, Y., C. Katsukawa, K. Inoue, Y. Yin, H. Tasaka, N. Ueba, and M. Makino. 1995. Mutations in rpoB gene of rifampicin resistant clinical isolates of Mycobacterium tuberculosis in Japan. J. Jpn. Assoc. Infect. Dis. 69:413-419. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki, Y., C. Katsukawa, A. Tamaru, C. Abe, M. Makino, Y. Mizuguchi, and H. Taniguchi. 1998. Detection of kanamycin-resistant Mycobacterium tuberculosis by identifying mutations in the 16S rRNA gene. J. Clin. Microbiol. 36:1220-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. Cole, M. J. Colston, L. Matter, K. Schopfer, and T. Bodmer. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647-650. [DOI] [PubMed] [Google Scholar]
- 21.Telenti, A., P. Imboden, F. Marchesi, T. Schmidheini, and T. Bodmer. 1993. Direct, automated detection of rifampin-resistant Mycobacterium tuberculosis by polymerase chain reaction and single-strand conformation polymorphism analysis. Antimicrob. Agents Chemother. 37:2054-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trivedi, S. S., and S. G. Desai. 1987. Pyrazinamidase activity of Mycobacterium tuberculosis—a test of sensitivity to pyrazinamide. Tubercle 68:221-224. [DOI] [PubMed] [Google Scholar]
- 23.World Health Organization. 1996. Report of the tuberculosis epidemic. World Health Organization, Geneva, Switzerland.
- 24.World Health Organization. 1997. Anti-tuberculosis drug resistance in the world. The WHO/IUATLD global project on anti-tuberculosis drug-resistance surveillance, 1994-1997. World Health Organization, Geneva, Switzerland.
- 25.Zhang, Y., B. Heym, B. Allen, D. Young, and S. T. Cole. 1992. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591-593. [DOI] [PubMed] [Google Scholar]
- 26.Zhang, Y., S. Dhandayuthapani, and V. Deretic. 1996. Molecular basis for the exquisite sensitivity of Mycobacterium tuberculosis to isoniazid. Proc. Natl. Acad. Sci. USA 93:13212-13216. [DOI] [PMC free article] [PubMed] [Google Scholar]