Abstract
A new approach for the M-typing of Streptococcus pyogenes is reported. Oligonucleotide primers were used in a PCR to amplify the N-terminal region of the emm gene. The presence of the PCR amplification product is associated with the corresponding M serotype. This technique offers potential advantages over other molecular typing methods.
Since the first description of the streptococcal M protein (8, 9), many studies have elucidated the function, immunochemistry, genetic structure, and antigenic variations of this bacterial molecule (3). This protein antigen, in fact, is one of the most important epidemiological markers used to identify and characterize group A streptococci (GAS). At present, more than 81 serologically different types of this protein are known. However, M-typing antisera are difficult to prepare and are not commercially available. Hence, after the publication of the N-terminal nucleotide sequence of the M protein-coding genes (emm genes) from different types, several laboratories have developed different molecular typing methods (1, 4, 5, 6, 7, 14).
In view of the fact that severe infections caused by GAS, including their suppurative and nonsuppurative sequelae, are increasing and that typing methods are essential tools for outbreak investigation and surveillance, we describe here a new approach based on application of the simple PCR technology. The procedure is inexpensive and easy to perform, even on a large number of samples. Experiments have been run to assess specificity, sensitivity, and reproducibility. To our knowledge, this technique, as it is here presented, has never been reported before.
The specificity of a given M serotype resides in the N-terminal portion of the mature GAS M protein. The corresponding 5" end of the emm gene is also type specific (3). Therefore, we used oligonucleotides annealing to this region as reverse primers in standard PCRs (Table 1). In all the reactions, the forward primer was represented by an oligonucleotide pairing to a highly conserved sequence internal to the isp locus (10). In the present study, we tested 33 clinical isolates of GAS strains belonging to serotypes M1 (two strains), M2 (four strains), M3 (three strains), M4 (eight strains), M5 (one strain), M6 (five strains), M8 (two strains), M12 (six strains), M18 (one strain), and M24 (one strain).
TABLE 1.
Oligonucleotide probes used in this study
| Probe | Nucleotide sequencesa | Refer-ence |
|---|---|---|
| emm1 | 5" TTC TAT AAC TTC CCT AGG ATT ACC ATC ACC 3" | 5 |
| emm2 | 5" TGC TTC TTT TTT GAC AGG GAC AGG GTT CTT 3" | 5 |
| emm3 | 5" CAT GTC TAG GAA ATC CTC CAT TAA CAC TCC 3" | 5 |
| emm4 | 5" CCA CGC TGA ATC AGC CTC AGG CTT TTT AAT 3" | 6 |
| emm5 | 5" CGG GTC ATT TAT TGT ACC CCT AGT CAC GGC 3" | 5 |
| emm6 | 5" TGC TTT GTC CGG GTT TTC TAC CGT CCC CCT 3" | 5 |
| emm8 | 5" TCG TTA TTA GAA ATA CTA TGA GAT TTT GGG 3" | 6 |
| emm12 | 5" ACG TTG TTT TTC TGC GAC TAA ATC ACT ATG 3" | 5 |
| emm18 | 5" CGT CTT TAT TGT CTG CTG TAG CTC GAG TAA 3" | 5 |
| emm24 | 5" TTC TTG TAC TTT TTC CAG AGT ATC TGT CTG 3" | 5 |
| isp | 5"CCG TAT CTT TAG CGC ACT CGA GGA CAA TTT GCG AGA TTA G 3" | 3 |
For emm oligonucleodide probes, the sequences are complementary to the coding strand of the N-terminal region of the corresponding emm gene sequences.
Grid experiments covering all the possible crosses between emm-specific oligonucleotides and available streptococcal strains of known M serotype were performed. In brief, 2 μl of chromosomal DNA embedded in agarose plugs, prepared as described by Ripa et al. (12), was added to a PCR mix containing 67 mM Tris (pH 8.8), 16 mM (NH4)2SO4, 0.15 mg of bovine serum albumin per ml, 0.01% Tween 20, 2 mM MgCl2, 0.2 mM deoxynucleotide triphosphates, emm-specific reverse primer and isp forward primer (0.2 μM each), and a 25:1 mixture of Taq and Pfu DNA polymerases (0.5:0.02 U), in a total volume of 30 μl. It is not strictly required to prepare template DNA in agarose. We obtained comparable results using DNA released by cultured cells after freezing and thawing (2).
The amplification reaction included one cycle at 95°C for 1 min, followed by 25 cycles at 95°C for 15 s, 60°C for 2 min, and 68°C for 6 min. The reaction product was analyzed by agarose gel electrophoresis.
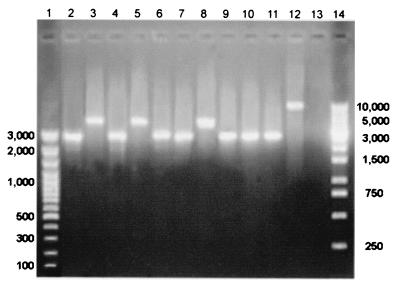
Experiments were performed in which every emm-specific oligonucleotide was used in an amplification reaction against the corresponding M type strain. In Fig. 1, the results obtained with a representative subset of strains are shown. All reactions gave a positive amplification signal. Reactions containing M1, M3, M5, M6, M12, M18, and M24 strains DNA produced a ≅3-kb amplicon, while for M2, M4, and M8, a ≅5-kb fragment was obtained. This difference can be explained by the well-documented hypervariability of this portion of the Streptococcus pyogenes chromosome.
FIG. 1.
PCR M typing sensitivity. M serotypes versus emm-specific oligonucleotides. Lanes 1 and 14, molecular weight standards (in base pairs); lane 2, M1-2 versus emm1; lane 3, M2 versus emm2; lane 4, M3 versus emm3; lane 5, M4 versus emm4; lane 6, M5 versus emm5; lane 7, M6 versus emm6; lane 8, M8 versus emm8; lane 9, M12 versus emm12; lane 10, M18 versus emm18; lane 11, M24 versus emm24; lane 12, internal control for forward primer, amplification of the vir regulon according to Gardiner et al. (3); lane 13, no-DNA control.
In particular, the template chromosomal DNA amplified by our PCRs may contain the fcrA gene, which encodes an immunoglobulin G (IgG)-Fc binding protein (11). The fact that the region included between the vir and emm genes has an estimated length of about 1.7 to 1.9 kb (11) and that the isp locus maps upstream to the vir gene can fully explain the difference of our PCR product length (5 kb versus 3 kb). Moreover, the fcrA gene has been previously reported to be present in M2, M4, and M8 strains (11) and not in the other M type strains used in the present work.
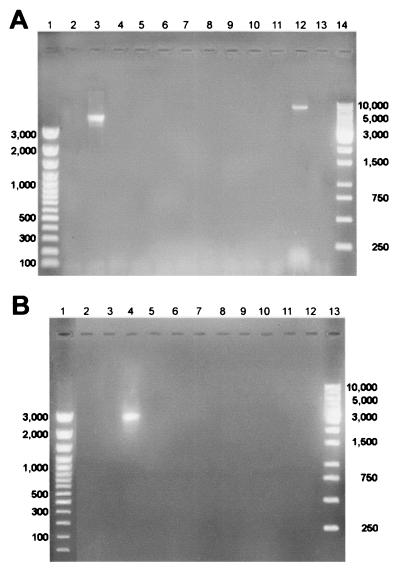
In parallel experiments, the whole set of oligonucleotides were used in PCRs with template DNA isolated from a selected M serotype strain. In Fig. 2A, results of a typical experiment in which M2 strain DNA was added are shown; only the PCR containing emm2 oligoprimer gave an amplification product. Subsequently, a countertest was performed in which a selected emm-specific oligonucleotide was used as the reverse primer in reactions where chromosomal DNA isolated from different M serotype strains was added as the template. Results for experiments conducted using the emm3 primer are presented in Fig. 2B, where the only PCR positive for amplification was that containing M3 strain chromosomal DNA. Identical analyses were performed on the other M type strains (M1, M4, M5, M6, M8, M12, and M18), with comparable results (not shown). Taken together, these data support the high grade of specificity of the technique.
FIG. 2.
PCR M typing specificity. (A) emm-specific oligonucleotides in reaction containing M2 serotype template chromosomal DNA. Lanes 1 and 14, molecular size standards (in base pairs); lane 2, M2 versus emm1; lane 3, M2 versus emm2; lane 4, M2 versus emm3; lane 5, M2 versus emm4; lane 6, M2 versus emm5; lane 7, M2 versus emm6; lane 8, M8 versus emm8; lane 9, M2 versus emm12; lane 10, M2 versus emm18; lane 11, M2 versus emm24; lane 12, internal control for forward primer, amplification of the vir regulon according to Gardiner et al.; lane 13, no-DNA control. (3). (B) emm3 oligonucleotide in amplification reactions with different M serotype chromosomal DNAs. Lanes 1 and 14, molecular size standards (in base pairs); lane 2, emm3 versus M1; lane 3, emm3 versus M2; lane 4, emm3 versus M3; lane 5, emm3 versus M4; lane 6, emm3 versus M5; lane 7, emm3 versus M6; lane 8, emm3 versus M8; lane 9, emm3 versus M12; lane 10, emm3 versus M18; lane 11, emm3 versus M24; lane 12, no-DNA control.
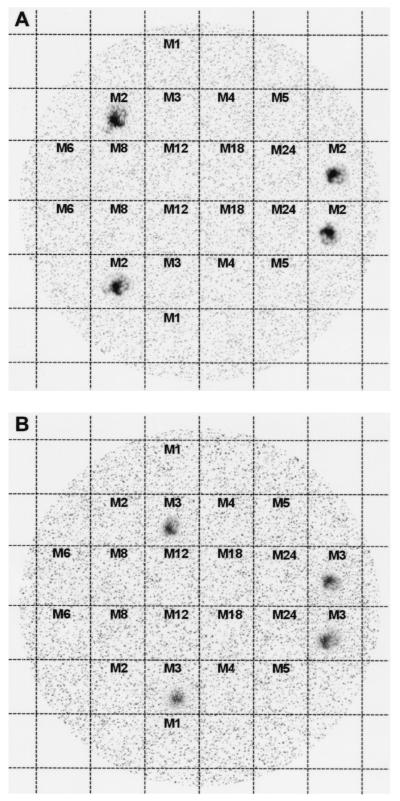
To definitely rule out nonspecificity of the PCR, we conducted colony blot hybridization experiments. Following described protocols (13), DNA immobilized on a nylon filter by UV light was hybridized to the same set of 32P-labeled emm gene-specific probes used in the PCR experiments (Table 1). The results of this set of experiments are shown in Fig. 3, where it can be seen that the emm2 oligonucleotide hybridizes only with M2 strain DNA (Fig. 3A). The same relative result was obtained when the emm3 oligonucleotide was used (Fig. 3B).
FIG. 3.
Colony hybridization. The numbers in the grids correspond to the M serotypes. (A) Hybridization experiment with the 32P-labeled emm2-specific probe (corresponding to the emm2 oligonucleotide reported in Table 1). (B) Hybridization experiment with the 32P-labeled emm3 oligonucleotide (corresponding to the emm3 oligonucleotide reported in Table 1).
With this simple protocol, typing of the M protein could be attempted by any laboratory in which PCR is routinely in use. Results are not as informative as those obtainable with the sequencing approach; nevertheless, the technique offers high specificity, since it is based on sequencing data. We think that with a representative set of emm-specific oligonucleotides and a good experimental design, it is possible to carry out good molecular epidemiology studies without the need to adopt any expensive and/or time-consuming techniques. For example, one could start by ranking M types in potential order of frequency and testing common types first. All typed isolates could then be excluded from subsequent analyses.
Acknowledgments
We thank G. Orefici (Istituto Superiore di Sanità, Rome) for supplying clinical isolates of S. pyogenes with known M serotype. We are also grateful to C. L. Pon for helpful review of the manuscript.
REFERENCES
- 1.Beall, B., R. Facklam, and T. Thompson. 1996. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J. Clin. Microbiol. 34:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandi, L., M. Falconi, and S. Ripa. 2000. Plasmid curing effect of trovafloxacin. FEMS Microbiol. Lett. 184:297-302. [DOI] [PubMed] [Google Scholar]
- 3.Fischetti, V. A. 1989. Streptococcal M protein: molecular design and biological behavior. Clin. Microbiol. Rev. 2:285-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardiner, D. L., J. Hartas, B. Currie, J. D. Mathews, D. J. Kempt, and K. S. Sriprakash. 1995. Vir typing: a long-PCR typing method for group A streptococci. PCR Methods Appl. 4:288-293. [DOI] [PubMed] [Google Scholar]
- 5.Gardiner, D. L., and K. S. Sriprakash. 1996. Molecular epidemiology of impetiginous group A streptococcal infections in aboriginal communities of northern Australia. J. Clin. Microbiol. 34:1448-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaufhold, A., A. Podbielski., D. R. Johnson, E. L. Kaplan, and R. Lutticken. 1992. M protein gene typing of Streptococcus pyogenes by nonradioactively labeled oligonucleotide probes. J. Clin. Microbiol. 30:2391-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaufhold, A., A. Podbielski., G. Baumgarten, M. Blokpoel, J. Top, and L. Schouls. 1994. Rapid typing of group A streptococci by use of DNA amplification and nonradioactive allele specific oligonucleotide probes. FEMS Microbiol. Lett. 119:19-26. [DOI] [PubMed] [Google Scholar]
- 8.Lancefield, R. C. 1928. The antigenic complex of Streptococcus haemolyticus. I. Demonstration of the type-specific substance in extracts of Streptococcus haemolyticus. J. Exp. Med. 47:91-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lancefield, R. C. 1962. Current knowledge of the type specific M antigens of group A streptococci. J. Immunol. 89:307-313. [PubMed] [Google Scholar]
- 10.McIver, K. S., S. Subbarao, E. M. Kellner, A. S. Heath, and J. R. Scott. 1996. Identification of isp, a locus encoding an immunogenic secreted protein conserved among group A streptococci. Infect. Immun. 64:2548-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Podbielski, A. 1993. Three different types of organization of the vir regulon in group A streptococci. Mol. Gen. Genet. 237:287-300. [DOI] [PubMed] [Google Scholar]
- 12.Ripa, S., C. Zampaloni, L. A. Vitali, E. Giovanetti, M. P. Montanari, M. Prenna, and P. E. Varaldo. 2001. SmaI macrorestriction analysis of Italian isolates of erythromycin-resistant Streptococcus pyogenes and correlations with macrolide-resistance phenotypes. Microb. Drug Resist. 7:55-61. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Saunders, N. A., G. Hallas, E. T. Gaworzewska, L. Metherell, A. Efstratiou, J. V. Hookey, and R. C. George. 1997. PCR-enzyme-linked immunosorbent assay and sequencing as an alternative to serology for M-antigen typing of Streptococcus pyogenes. J. Clin. Microbiol. 35:2689-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]