Abstract
Obesity, diabetes, hypertension, and heart disease are highly heritable conditions that in aggregate are the major causes of morbidity and mortality in the developed world and are growing problems in developing countries. To map the causal genes, we conducted a population screen for these conditions on the Pacific Island of Kosrae. Family history and genetic data were used to construct a pedigree for the island. Analysis of the pedigree showed highly significant heritability for the metabolic traits under study. DNA samples from 2,188 participants were genotyped with 405 microsatellite markers with an average intermarker distance of 11 cM. A protocol using loki, a Markov chain Monte Carlo sampling method, was developed to analyze the Kosraen pedigree for height, a model quantitative trait. Robust quantitative trait loci for height were found on 10q21 and 1p31. This protocol was used to map a set of metabolic traits, including plasma leptin to chromosome region 5q35; systolic blood pressure to 20p12; total cholesterol to 19p13, 12q24, and 16qter; hip circumference to 10q25 and 4q23; body mass index to 18p11 and 20q13; apolipoprotein B to 2p24–25; weight to 18q21; and fasting blood sugar to 1q31–1q43. Several of these same chromosomal regions have been identified in previous studies validating the use of loki. These studies add information about the genetics of the metabolic syndrome and establish an analytical approach for linkage analysis of complex pedigrees. These results also lay the foundation for whole genome scans with dense sets of SNPs aimed to identifying causal genes.
Keywords: loki, quantitative trait locus, Syndrome X
The metabolic syndrome (also known as Syndrome X) includes central obesity, insulin resistance, hypertension, and dyslipidemia, all of which are risk factors for diabetes and vascular heart disease (1). In aggregate, these disorders represent major causes of morbidity and mortality in industrialized countries and are growing problems in the developing world (2). Familial aggregation, adoption, twin, and segregation analysis studies all indicate that the component disorders of Syndrome X are highly heritable with substantial genetic effects, interacting with environmental factors, leading to the development of disease. Although the genes for uncommon monogenic forms of obesity, insulin resistance/diabetes, hypertension, and dyslipidemia have been identified, these gene variants do not account for most of the heritability of these conditions in the general population. Although linkages to various chromosomal regions have been reported for these disorders in the general population, further studies will be necessary to identify the causal genes (3–5). Genetic heterogeneity and/or allelic variation may contribute to the difficulty in identifying these genes, a possibility that can be mitigated by studying the genetically isolated population of the Pacific Island of Kosrae, Federated States of Micronesia.
In 1994, working with the Department of Health on Kosrae, we initiated a genetic epidemiologic study of the metabolic syndrome with the hypothesis that the reduced genetic complexity of this population isolate would facilitate disease gene mapping. Kosrae was settled by a small number of founders ≈2,000 years ago, and there have been numerous bottlenecks since the time. These bottlenecks include typhoons and the introduction of communicable diseases by whalers in the 19th century. After World War II, the United States supplied aid to Micronesia in the form of high-fat and high-salt Western foods (6–9).
Before this time the population subsisted by fishing, foraging, and some farming, and they were not noted to be especially obese. Coincident with this change in lifestyle and environment on Kosrae, the prevalence of obesity and other metabolic diseases increased in a manner similar to other native populations, such as the Pima Indians of Arizona and the Nauruans of Oceania (10, 11). However, only a subset of the Kosraen population suffers from the component disorders of the metabolic syndrome, thus providing an opportunity to study the basis for gene–environment interactions leading to metabolic disease.
In this population-based screen, a series of quantitative and qualitative traits relevant to the metabolic syndrome were collected from 2,188 individuals, and DNA from these participants was genotyped for 405 microsatellite markers. Linkage analysis of the Kosraen population was performed by using loki (http://loki.homeunix.net), an analysis program that uses Markov chain Monte Carlo sampling. loki performs linkage analysis by using oligogenic quantitative trait locus (QTL) models and produces estimates of the best trait model, most likely location of linked QTLs, and the percentage of variance in the trait explained by each QTL (12). loki was used because linkage analysis of quantitative traits with covariate corrections in a large pedigree is computationally difficult, and most other analysis programs require that the pedigree be split into multiple subpedigrees, reducing the power to detect linkage. Here, we report the map locations for a number of loci segregating with stature and a series of metabolic traits in the Kosraen population.
Results
The 1994 Kosrae Screen.
After signing an informed consent, 2,188 adults participated in the population-based screen. Each participant completed medical and family questionnaires and had a physical examination and phlebotomy. The questionnaire noted the participant’s gender, family data, smoking status, village, age, and health status. Physical exams included measures of height (HT) and weight (WT), also used for BMI calculation, hip circumference (HIP), waist circumference (WST), fasting blood sugar (FBS), diastolic blood pressure (DBP), and systolic blood pressure (SBP), both also used for calculating mean arterial pressure. DNA was extracted from whole blood, and plasma concentrations for leptin (LEP), total cholesterol (TC), triglyceride (TG), apoA-I, and apoB were measured. To develop trait models for genetic analyses, environmental covariate effects (gender, parity, smoking status, village of residence, and age) were calculated for each trait by regression analysis. The clinical data were corrected for these covariates in the subsequent analyses.
A pedigree was constructed from the family data, genealogical records, and meetings with village elders. The pedigree includes 2,286 individuals, including 1,564 study participants, and traced back five generations. The average sibship size was four, with a range of 1–12 children. The pedigree included 110 loops, mostly marriage loops. The inbreeding coefficient for all individuals was zero except for one individual (0.008). Relationship errors were identified by using initial marker genotypes, and 55 modifications were made to the constructed pedigree before trait analysis.
Genome Scan.
A set of 405 microsatellite markers, assembled in 39 panels, was developed based on the Marshfield 9 screening set (www.marshfieldclinic.org/research/genetics). The 1,564 study individuals in the pedigree were genotyped by using the modified versions of the Marshfield panels. (Marker information and panel configurations are available from the authors upon request.) Marker statistics were as follows: average number of genotypes per marker was 1,453 (range 951–1,564), average genotyping (Mendelian) error 0.49% (range 0–2.2%), and average heterozygosity 0.67 (range 0.12–0.9). This error rate was similar to other studies, and heterozygosity was slightly less than that reported for Caucasian populations.
We used 360 of the 380 autosomal markers in the initial analysis for an average autosomal map density of 10 cM, with three gaps of >12 cM (due to removal of markers with high error rates). The other 20 markers were added subsequently to reduce the size of the gaps.
Genetic Analysis of Stature Using loki.
Although loki has been used on simulated data from the Genetic Analysis Workshop 12 (13) and with real data sets (14), it has not been used to analyze a complex pedigree such as that on Kosrae. To develop a strategy to use loki for quantitative trait analysis of the Kosrae pedigree, we focused on HT as a model complex trait. HT is a highly heritable oligogenic trait with clear covariate effects (e.g., childhood nutrition) (15, 16). HT was chosen because it is an extremely robust measure that can be determined precisely and reproducibly and does not change significantly after adulthood. In Kosrae, the heritability of HT was 0.64 after covariate correction (calculated using morgan). This result is similar to that reported in other populations and suggests significant genetic loading (17). Segregation analysis showed four to six major genes, each explaining 5–25% of the trait variance, and a polygenic effect (calculated by using loki). Thus, a number of genes for HT, with large effect sizes, appeared to be segregating in the Kosraen population.
loki results are reported as L-scores, which, as estimates of the Bayes factor, show the ratio between the probability that a QTL signal is “real” and that it is due to chance alone. An L-score of ≥ 20 indicates strong evidence for linkage, as suggested by Bayes factor calibration tables (18). However, L-scores can vary in repeated analyses because of differences in sampling, mixing, or convergence, and the magnitude of the L-score at a real locus can differ with model changes. Previously, loki was used to analyze a simulated data set by employing a hierarchical scheme to identify the most likely trait loci (13). This approach was adapted for the analysis of the Kosrae data set as follows: (i) Initial genome scans were performed on a sample subset (1,102 individuals) with two trait models, 1 or 2 chromosomes at a time, with and without a polygenic effect being factored into the analysis; (ii) reanalysis of regions with L-scores ≥10 on the complete sample set (1,564 individuals), with additional markers; (iii) joint analysis of chromosomes with strong linkage regions on the complete sample (to identify possible interactions among loci); (iv) inspection of L-graphs for peak localization and shape; and (v) estimation of QTL parameters.
HT was analyzed in this fashion by using two trait models: Model.HT_A, with corrections for gender, parity, smoking, age, and LEP (which mapped to the same region on chromosome 5; see below), and Model.HT_B, with corrections for those covariates and additional quantitative traits (WT, HIP, WST, FBS, DBP, SBP, TC, TG, apoA, and apoB). We included Model.HT_B because HT correlated with most of the metabolic traits and clustered with an obesity/diabetes factor identified by using principal component analysis (19). This new factor had the following significant loadings: HT (0.3942), LEP (0.6708), WT (0.8086), WST (0.8247), HIP (0.8782), and TG (0.2600). Using models with corrections for correlated metabolic traits (such as Model.HT_B) can have two outcomes. In one scenario, these corrections reduce the trait variance, enabling identification of QTLs not identified with the simpler model (or that were identified but with lower L-scores). Alternatively, if a QTL influences the trait through its interaction with other related traits, when these traits are “corrected out” and this QTL might no longer be evident. Thus, we might expect different models to result in both overlapping and nonoverlapping loci.
HT Results.
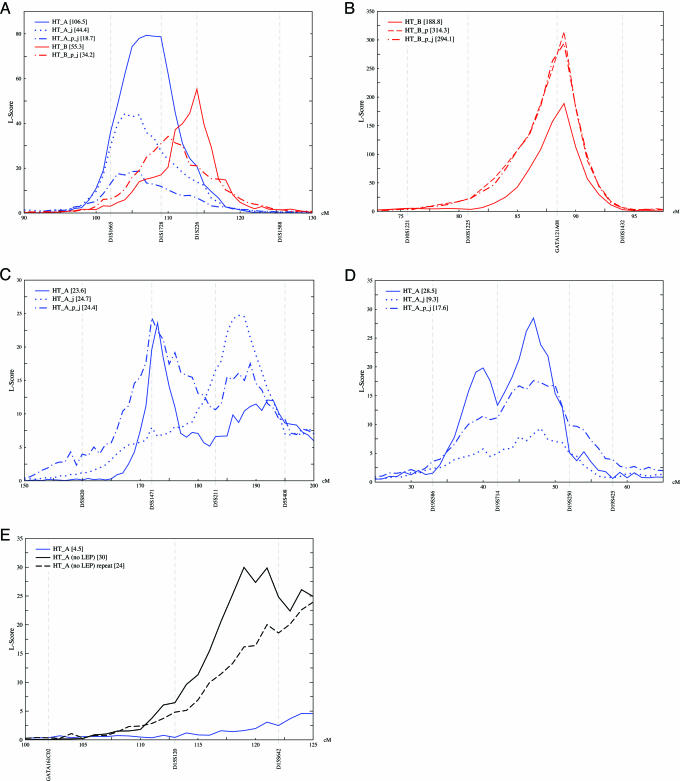
The results for both models are shown in Table 1. Model HT_A identified three loci with strong evidence for linkage (L-scores ≥ 20): chromosomes 1 (L-score of 35.7 at 106.5 cM), 5 (33.9 at 171.5 cM), and 15 (43.7 at 115.5 cM), and one suggestive L-score (≥ 10) on chromosome 19 (10.8 at 40.5 cM). When polygenes were included in the model (polygenic effect), the scores decreased but remained high on chromosomes 1 (20 at 104.5 cM) and 5 (21.1 at 164.5 cM). Model.HT_B identified two loci with strong linkage evidence, on chromosomes 1 (37.9 at 107.5 cM) and 10 (124.6 at 88.5 cM), and a suggestive signal on chromosome 18 (10.1 at 29.5). As noted above, the observation that different models yield different results is not unexpected. These chromosomes then were reanalyzed on the entire pedigree (Table 2). The scores increased on chromosome 1 for both models, although the precise map position differed slightly: Model.HT_A (L-score 79.4 at 106.5 cM) and Model.HT_B (55.3 at 113.5 cM). The locus on chromosome 5 (detected for Model.HT_A) decreased (23.6) but remained strong. The locus on chromosome 10 (for Model.HT_B) increased (188.8), and the polygenic effect resulted in a further increase (314.3). On chromosome 15 (Model.HT_A), the L-score decreased to 0.5 but increased when the LEP correction was excluded (30.0 at 118.5 cM). The suggestive signal on chromosome 18 (Model.HT_B) decreased, whereas the suggestive signal on chromosome 19 (Model.HT_A) increased (28.5 at 46.5 cM).
Table 1.
Highest L-scores and cM (Marshfield map) on each chromosome, for two height models
| Chr. | MODEL.HT_A |
MODEL.HT_B |
||
|---|---|---|---|---|
| L-score | cM | L-score | cM | |
| 1 | 35.7 | 106.5 | 37.9 | 107.5 |
| 2 | 0.7 | 174.5 | 0.9 | 52.5 |
| 3 | 1.4 | 104.5 | 0.9 | 100.5 |
| 4 | 2.7 | 159.5 | 8.1 | 55.5 |
| 5 | 33.9 | 171.5 | 5.4 | 171.5 |
| 6 | 2.5 | 176.5 | 7.7 | 93.5 |
| 7 | 4.9 | 124.5 | 5.5 | 183.5 |
| 8 | 3.2 | 67.5 | 4.5 | 84.5 |
| 9 | 9.7 | 169.5 | 6.6 | 29.5 |
| 10 | 0.6 | 72.5 | 124.6 | 88.5 |
| 11 | 1.0 | 57.5 | 2.3 | 42.5 |
| 12 | 3.0 | 96.5 | 1.3 | 18.5 |
| 13 | 0.9 | 95.5 | 0.8 | 23.5 |
| 14 | 1.2 | pter | 0.9 | pter |
| 15 | 43.7 | 115.5 | 0.9 | 44.5 |
| 16 | 0.8 | pter | 3.5 | 71.5 |
| 17 | 3.6 | 31.5 | 3.0 | 84.5 |
| 18 | 2.9 | 114.5 | 10.1 | 29.5 |
| 19 | 10.8 | 40.5 | 7.4 | 86.5 |
| 20 | 0.6 | pter | 6.5 | 49.5 |
| 21 | 0.5 | 58.5 | 1.3 | 1.5 |
| 22 | 4.8 | 12.5 | 0.9 | pter |
Model.HT_A has corrections for age, gender, parity, smoking, and leptin levels, and Model.HT_B has the same corrections as A and for WT, HIP, WST, FBS, blood pressure, TC, TG, apoA1, and apoB. L-scores ≥ 20 are in bold, and L-scores ≥ 10 are in italics. Chr., Chromosome.
Table 2.
L-scores and position (in cM) for strong loci for two height models (as in Table 1)
| Chr. | Model | Initial set |
Whole pedigree |
||
|---|---|---|---|---|---|
| L-score | cM | L-score | cM | ||
| 1 | A | 35.7 | 106.5 | 79.4 | 106.5 |
| B | 37.9 | 107.5 | 55.3 | 113.5 | |
| 5 | A | 33.9 | 171.5 | 23.6 | 172.5 |
| 10 | B | 124.6 | 88.5 | 188.8 | 88.5 |
| 15 | A | 43.7 | 115.5 | 0.5 | 118.5 |
| 18 | B | 10.1 | 29.5 | 4.3 | 18.5 |
| 19 | A | 10.8 | 40.5 | 28.5 | 46.5 |
Table is comparing analysis of the initial sample set (1,102 samples, as reported in Table 1) with analysis using the whole pedigree (1,564 samples). L-scores ≥ 20 are shown in bold. Chr., chromosome.
The results of the joint analyses (j appended to model name) for chromosomes 1, 5, and 19 (using Model.HT_A) and chromosomes 1 and 10 (using Model.HT_B), without or with a polygenic correction (_p appended to model name) are shown in Table 3. On chromosome 1, all scores were strong, with nearly equivalent map positions for HT_A (103.5–105.5 cM) and B (≈110 cM). The L-scores for chromosome 5 did not change appreciably but showed slightly different map positions without (Model.HT_A, 186.5 cM) and with the polygenic effect (Model.HT_A_p, 171.5 cM). The chromosome 19 scores decreased, with suggestive linkage (17.6) for Model.HT_A_p_j only. The L-scores for the locus on chromosome 10 remained extremely high, for Model.HT_B (117.0) and Model.HT_B_p (294.1).
Table 3.
Joint chromosome analyses for two height models, using the whole pedigree
| Chr. | MODEL.HT_A_j |
MODEL.HT_A_p_j |
MODEL.HT_B_j |
MODEL.HT_B_p_j |
||||
|---|---|---|---|---|---|---|---|---|
| L-score | cM | L-score | cM | L-score | cM | L-score | cM | |
| 1 | 44.4 | 103.5 | 18.7 | 105.5 | 60.0 | 110.5 | 34.2 | 109.5 |
| 5 | 24.7 | 186.5 | 24.4 | 171.5 | — | — | — | — |
| 19 | 9.3 | 47.5 | 17.6 | 46.5 | — | — | — | — |
| 10 | — | — | — | — | 117.0 | 88.5 | 294.1 | 88.5 |
Joint chromosome analyses (“j” denotes joint analysis; “p” includes polygenic effect) for two height models (see Table 1), using the whole pedigree. L-scores ≥ 20 are shown in bold. Chr., chromosome.
Strong evidence for linkage for two (or more) loci in a joint analysis suggests that those QTLs interact to influence trait variance. For Model.HT_A, chromosomes 1 and 5 appeared together in 55% of the iterative analyses, chromosomes 1 and 19 appeared together in 24% of the iterations, and chromosomes 5 and 19 appeared together in 19% of the iterations. For Model.HT_B, chromosomes 1 and 10 appeared together in 68% of the iterations. These results strongly suggest that the QTLs on 1 and 5 (in Model.HT_A) and 1 and 10 (Model.HT_B) jointly influence HT trait variance.
The linkage regions for HT and the percent variance attributable to these loci are summarized in Table 4. The chromosome 1 locus was consistently found with both models, in joint analysis and with the polygenic correction, with very similar chromosomal locations (Fig. 1A). This finding strongly suggests that there is a HT locus on Kosrae on chromosome 1. The chromosome 10 locus was identified by using Model.HT_B (Fig. 1B), which suggests a QTL for HT, without metabolic trait interactions. This locus had extremely high L-scores, which increased with joint analysis and correction for a polygenic effect. This result further demonstrates that reduction of nonmajor gene variance by accounting for (i) major genes on other chromosomes and (ii) residual genetic effect (polygenes) enhances the ability to map genes.
Table 4.
Final loci for height
| Chr. | Model | Range, cM | % Variance |
Other studies | |
|---|---|---|---|---|---|
| Genetic | Total | ||||
| 1 | A and B | 100–120 | 10–30 | 5–20 | Refs. 17 and 20 |
| 10 | B | 80–95 | 10–30 | 5–20 | Ref. 21 |
| 5 | A | 160-qter | 10–20 | 10–15 | Refs. 22 and 23 |
| 19 | A | 30–55 | 10–40 | 20 | Ref. 21 |
| 15 | A without LEP | 110-qter | 15–30 | 15–20 | Ref. 17 |
For each locus, the linkage region (“range”), percent genetic and total trait variance explained by that QTL, and other studies that identified the same region are shown. The percent variance is shown as a range, because in each loki iteration the result may be slightly different.
Fig. 1.
HT loci. L-scores are on the y axis; cM (Marshfield) and markers are on the x axis. Model.HT_A (in blue), HT corrected for environmental covariates (gender, parity, smoking, and age) and LEP; Model.HT_A (no LEP) (in black), same corrections as Model.HT_A but not LEP; Model.HT_B (in red), HT corrected for environmental covariates and WT, HIP, WST, LEP, FBS, SBP, DBP, TC, TG, and apoA-I and apoB levels. _p denotes polygenic correction and _j denotes joint chromosome analysis. (A) Chromosome 1. (B) Chromosome 10. (C) Chromosome 5. (D) Chromosome 19. (E) Chromosome 15.
The other loci are provisional. Chromosome 5 was consistently found for Model.HT_A, with similar scores in joint analysis and including the polygenic effect. However, the scores were of borderline strength and showed a split peak (Fig. 1C), potentially revealing a problem with the model. Chromosome 19 also was identified by using Model.HT_A, but the statistical support was strongest only when the entire cohort was analyzed. The L-scores decreased with joint analysis and polygenic effect, and the signal was poorly localized and also split on a marker (Fig. 1D). Chromosome 15 was only found by using Model.HT_A without LEP; the L-scores were low and decreased with the polygenic effect, and mapped off the end of the chromosome (Fig. 1E). Nonetheless, all five HT loci were also found in other studies (see Table 4 and Discussion), validating the analytical approach. Further studies, with additional markers and alternative analytical approaches, are needed to establish whether these loci are reproducible. If true, these results would suggest that loki is quite sensitive.
Familial Aggregation of the Metabolic Syndrome Traits.
Significant heritability (h2) was seen for all metabolic traits after covariate correction, as calculated by using the morgan package (Table 5). This finding suggests strong genetic effects on these traits in the Kosraen population, with similar heritabilities to other family studies (24–26). Segregation analyses (using loki) of particular trait models suggested that a significant percent of trait variance was attributable to major genes, ranging from 78% (WT) to 16% (SBP), with at least one major gene for each trait, and ≈14–32% total trait variance explained by the largest QTL (Table 5). With the suggestion of major genes, we applied the analytical approach used to map HT loci to map QTLs segregating the Kosrae population for a series of metabolic traits. As above, in each case we analyzed a number of models. When different models resulted in signals at the same locus, we used the model with the strongest support for further analysis; in the other instances, all trait models were used. The most likely loci are described below and reported in Table 6.
Table 5.
Heritability of stature and metabolic traits
| Trait | h2 | Segregation analysis |
||
|---|---|---|---|---|
| Number of QTLs | Mean % variance attributable to: |
|||
| Total genetic effects | Largest QTL | |||
| HT | 0.64 | 5.5 | 81 | 28 |
| WT | 0.53 | 3 | 78 | 32 |
| BMI | 0.45 | 3 | 63 | 27 |
| WST | 0.40 | 3.5 | 74 | 31 |
| HIP | 0.45 | 3.5 | 52 | 26 |
| LEP* | 0.20 | 1.5 | 23 | 20 |
| FBS* | 0.24 | 2.5 | 54 | 31 |
| DBP | 0.29 | 3 | 39 | 20 |
| SBP* | 0.25 | 1 | 16 | 14 |
| MAP | 0.26 | 3 | 49 | 24 |
| TC | 0.41 | 2 | 36 | 26 |
| TG* | 0.21 | 2 | 40 | 24 |
| APOB | 0.31 | 3 | 38 | 18 |
| APOA-I | 0.36 | 2.5 | 51 | 29 |
For each trait, heritability (h2), or percent trait variance due to genetic effects, is shown in column 2. Columns 3–5 show the results of the segregation analysis, averaged over all iterations: number of QTLs, percent variance due to all QTLs, and percent variance due to the QTL with the largest effect.
*Log transformed.
Table 6.
Significant QLTs for metabolic traits, in order of priority
| Trait | Chr. | Max L-score | Peak, range, cM | % Variance |
Other studies | |
|---|---|---|---|---|---|---|
| Genetic | Total | |||||
| LEP | 5 | 104.2 | 183.5, 170–200 | 20–100 | 10–25 | Refs. 27–30 |
| SBP | 20 | 142.5 | 32.5, 25–40 | 30–50 | 15–20 | Refs. 31–34 |
| TC | 19 | 69.2 | 46.5, 35–50 | 40–50 | 20–30 | Refs. 14, 35–38 |
| HIP | 10 | 50.0 | 135.5, 125–140 | 50–70 | 10–20 | Refs. 28 and 39 |
| BMI | 18 | 49.0 | 16.5, pter-40 | 30–70 | 15–30 | Refs. 29 and 40 |
| TC | 12 | 32.0 | 152.5, 140–170 | 40–80 | 20–25 | Ref. 37 |
| APOB | 2 | 37.0 | 32.5, 25–40 | 40–60 | 20 | Refs. 36, 41, and 42 |
| HIP | 4 | 20.1 | 105.5, 90–115 | 30–50 | 5–15 | Ref. 33 |
| TC | 16 | 24.4 | 139.5, 125-qter | 40–60 | 25 | |
| BMI | 20 | 20.1 | 96.5, 85-qter | 40–70 | 15–25 | Refs. 3, 23, and 43–45 |
| WT | 18 | 20.0 | 83.5, 75–105 | 20–40 | 5–15 | Refs. 29 and 46–48 |
| FBS | 1 | 20.0 | 261.5, 210–275 | 100 | 15–25 | Refs. 49–56 |
The chromosome, signal location (peak location and extended linkage range in cM), percent of genetic and total trait variance, and other genome scan studies that found this region for the same or related traits are shown.
Strong Evidence for Linkage.
After correction for environmental and other metabolic covariates, QTLs on chromosomes 18 and 20 were found for BMI (h2 = 0.45). The chromosome 18 locus was found consistently and increased in the joint analysis (L-score 49.0). A diffuse peak was split over marker GATA88A12 (Fig. 2A), peaked at 16.5 cM/D18S967, and explained 30–70% of the genetic and 15–30% of the total variance. The chromosome 20 L-score was lower and decreased in repeat and joint analyses.
Fig. 2.
Selected QTLs for metabolic traits. L-scores are on the y axis; cM (Marshfield) and markers are on the x axis. _p denotes polygenic correction, and _j denotes joint chromosome analysis. Repeat refers to the same analysis repeated. (A) BMI on chromosome 18, using BMI_B, corrected for gender, parity, smoking, village, and age, and the other quantitative traits. (B) LEP on chromosome 5. Three models for LEP, all corrected for gender, smoking, village, and age, were used. Additional corrections for MAP and WT (blue), HT and MAP (red), and MAP (black), were made. (C) SBP on chromosome 20. SBP is corrected for gender, parity, smoking, village, and age. MAP is corrected for gender, parity, smoking, village, age, WST, and LE. (D) TC on chromosome 19. TC is corrected for gender, parity, village, age, and the other quantitative traits.
QTLs on chromosomes 10 and 4 were found for HIP (h2 = 0.45), corrected for all covariates. The chromosome 10 signal was found consistently in repeat and joint analyses, with L-scores of 20–50, and overlapped with a low signal (L-score 9) in the same region for WT. The HIP peak mapped directly on D10S1817 at 135.5 cM and explained 50–70% of the genetic and 10–20% of the total variance. The chromosome 4 locus was less robust for the original and joint analyses. One borderline signal was found for WT (h2 = 0.53) corrected for all covariates, on chromosome 18. Analysis of plasma LEP (h2 = 0.20) resulted in one robust signal on chromosome 5, which was consistently found by using several models and in joint analysis. Several of these models are shown in Fig. 2B and show the differences in signal strength and peak shape for different trait models. This linkage peak was at 183.5 cM, close to D5S211, and explained 20–100% of the genetic and 10–25% of the total variance. For diabetes traits, a borderline signal (L-score 20) was found for FBS (h7 = 0.24) on chromosome 1 but was poorly localized.
In analyses of hypertension, a signal for SBP (h2 = 0.25), corrected for environmental covariates, was identified on chromosome 20 (L-score 142.5). This locus was consistently found, although the score fell slightly in repeat analyses, possibly because of sampling variation. The signal was tightly localized on GATA81E09 at 32.5 cM (Fig. 2C) and explained 30–50% of the genetic and 15–20% of the total variance. Initially, this locus mapped off the end of the chromosome, near a signal for MAP. When GATA81E09 was added and the whole pedigree was analyzed, the linkage peak was localized to its current position.
The only significant scores for MAP (h2 = 0.26) were on the end of chromosome 2 (as above) where it was correlated with SBP (Fig. 2C), and at the end of chromosome 9. DBP (h2 = 0.29) similarly mapped to the ends of chromosome 9, suggesting a blood pressure locus that was diffusely localized in this region.
Analysis of apoB (h2 = 0.31) corrected for all covariates identified one QTL (L-score 37.0) on chromosome 2. This locus showed a narrow peak at 32.5 cM, between D2S1400 and D2S1360, and explained 40–60% of the genetic and 20% of the total variance. This region also had suggestive signals for MAP, SBP, and the BP factor and is close to the apoB gene. Three loci were found for TC (h2 = 0.41), by using two models. TC_A (corrected for environmental covariates) revealed one locus on chromosome 12 (32.0). This linkage peak was found consistently over many analyses, with higher scores for joint chromosome analysis. It overlapped with a signal for a dyslipidemia factor, previously identified by using principal component analysis, suggesting important quantitative trait correlations (19). This locus was not found by using TC_B, possibly because it is “corrected out” by the related traits that are included in this model. However, QTLs were found on chromosomes 19 and 16 for TC, by using TC_B (corrected for all covariates). The chromosome 19 locus (68.2) was consistently seen with the polygenic correction and in joint analysis (Fig. 2D). It mapped at 46.5 cM, between D19S714 and D19S250, and explained 40–50% of the genetic and 20–30% of the total variance. This linkage region is close to apoE, a gene known to affect TC levels in many populations and in Kosrae (57–59). We went on to account for a possible apoE effect by calculating the trait means for each genotype (e2, e3, e4) in the trait logistic regression and reanalyzed this model multiple times. L-scores were more stable with this correction (data not shown), suggesting that the causal gene is distinct from apoE and that reducing genetic variance from apoE makes the mapping of this distinct QTL more robust. ApoB had a suggestive signal at this locus. Although the linkage peak on chromosome 16 (24.4) was consistently found, with similar scores for multiple and polygenic analyses, and overlap with signal for the dyslipidemia factor, it mapped to the extreme end of the chromosome.
Discussion
Here, we report the results of an initial genetic analysis of stature and components of the metabolic syndrome on the Pacific Island of Kosrae and show evidence for linkage to a number of relevant traits. Most of the adult population participated (2,188 individuals) in a 1994 genetic study, revealing a high prevalence of obesity, diabetes, hypertension, and dyslipidemia (19). The heritability of traits related to these conditions was determined from a comprehensive pedigree of the population, and the impact of environmental factors has been assessed. In addition, possible confounding effects of uncorrected environment covariates are mitigated by the similar lifestyle that most people on Kosrae share. These studies add useful information about the genetics of stature and the metabolic syndrome and further describe an analytical approach using loki for identifying QTLs in large, complex pedigrees.
Linkage Analysis Using loki.
We used loki to identify loci contributing to variance in stature and metabolic quantitative traits in the Kosrae Study. The Genetic Analysis Workshop 10 recommendations, which suggested maximizing power to identify complex trait genes by multipoint analysis of quantitative traits, with corrections for covariates, in large, extended pedigrees, informed our analysis of this data set (60). However, because of computational difficulties, linkage methods cannot be used on a pedigree of the size and complexity of the Kosrae pedigree. In contrast, sampling-based approaches are able, in theory, to perform linkage analysis without simplifying either the model or the pedigree.
loki is a Markov chain Monte Carlo linkage analysis method that samples from all possible complete data states, with the most probable states more likely to be sampled. This program can handle missing data and allows for multilocus inheritance models (61). loki has been previously validated in analyses of simulated data sets in Genetic Analysis Workshop 12 when it correctly identified QTLs as well as or better than other programs (13). For comparative analyses with other programs, see Cheng et al. (62) and Pankratz et al. (63). loki also performed as well as solar in identifying linkage regions for apoliprotein B (apoB) levels (14). However, although loki has been able to identify loci in simulated and real data sets, computational limitations associated with this sampling method limit the ability to directly correlate L-scores to P values or logarithm of odds scores. Thus, the loci identified in this work represent candidate regions that will need to be further evaluated. One way of addressing this challenge would be to generate LOP scores, which are the ratio of the logarithm of two L-scores (posterior probability of linkage), one for the chromosome being tested and a second for an unlinked pseudochromosome (64). Thus, LOP scores relate the L-score to a negative locus as compared with loki (which relates the signal to randomness) and thus provide an output more easily related to a P value. A more definitive way of pursuing the loki peaks would be to use dense marker sets and other analytical approaches such as transmission disequilibrium test or sib pair analyses. This approach will allow us to better understand the robustness of a particular L-score in predicting the location of a true rather than false-positive locus.
We refined the analytic protocol used in Genetic Analysis Workshop 12 by analyzing height (HT) (a model complex trait, influenced by multiple interacting genes and early environmental effects, with little measurement error) and then used this protocol to analyze metabolic syndrome traits. This procedure enhances the power of loki to detect real linkage and prioritizes QTLs for follow-up studies. Our results replicate linkages from other studies, further validating this approach.
Replication of Linkage Regions.
Two robust QTLs for HT were found on 10q21 and 1p31, and three less robust signals were found on 5q34, 19p13, and 15q26. These loci were identified previously in Caucasian populations, and these studies now provide an opportunity to assess whether Caucasian variants increase HT, because Micronesians are generally shorter than Caucasians (see Table 4) (17, 20–23).
The following loci were identified for traits associated with the metabolic syndrome (listed in order of priority): serum leptin (LEP) on 5q35, systolic blood pressure (SBP) on 20p12, total cholesterol (TC) on 19p13, hip circumference (HIP) on 10q25, body mass index (BMI) on 18p11, TC on 12q24, apoB on 2p24–25, HIP on 4q23, TC on 16qter, BMI on 20q13, weight (WT) on 18q21, and fasting blood sugar (FBS) on 1q31–1q43. All of the results confirm linkage regions from previous studies, except TC on 16. In Caucasian populations, obesity traits mapped to the same regions on chromosome 5 (27–30), on chromosome 10 (28, 29), on chromosome 20 (3, 23, 43–45), on chromosome 18 [both the BMI region (29, 40) and the WT region (29, 46–48)], and on chromosome 4 (65). Diabetes traits produced signals on chromosome 1q in a region that has been implicated by many studies, including Caucasian, American Indian, and Asian populations (49–56). Hypertension traits mapped to chromosome 20 in Caucasian and mixed Caucasian/American Indian populations (31–34). Dyslipidemia traits localized to chromosomes 19 (14, 35–38) and 2 (36, 41, 42) in studies of Caucasian and American Indian populations, whereas chromosome 12 was found in a Caucasian population (37).
Gene Mapping in Kosrae.
Most of the loci identified in this work were previously found in Caucasian cohorts, and it is possible that the same alleles are relevant in both Micronesians and Caucasians. Alternatively, it may be that we identified loci with susceptibility alleles of Caucasian origin in the Kosrae population (resulting from Caucasian admixture). However, recent data analyses of Y chromosome and mitochondrial haplotypes and a structure analysis of select microsatellite markers indicate that the rate of admixture is relatively low (J.M.F., M.S., J.L.B., P. Bonnen, I. Peer, and D. Altshuler, unpublished data). This finding suggests that the same susceptibility loci are segregating on Kosrae as in other populations. In some cases the segregation of Caucasian alleles or chromosomal segments also may contribute to phenotypic differences. For example, because of the typically short stature of native Kosraens, we might expect that the inheritance of tallness would be of Caucasian origin, whereas those with the opposite effect might be of Micronesian origin.
The ultimate goal of these studies is to identify the causal genes for a series of metabolic traits. Although we might consider compiling dense maps around each of the peaks reported here individually, it is more efficient to employ a whole-genome approach. Recent data have indicated that the SNPs on the Affymetrix 100 K arrays are highly informative on Kosrae and that extended linkage disequilibrium and reduced haplotype diversity significantly reduce the number of SNP markers needed to sample a significant fraction of the genome. Thus, with existing technology we should be able to sample a significant fraction of the genome in the Kosrae cohort and test the possible contribution of SNPs in regions of the genome reported here, or other regions, by using sib pair analysis, transmission disequilibrium test or other complementary analytical approaches. Analysis of the Kosrae pedigree has identified 4,584 sibpairs and 885 two-parent trios, suggesting that there should be sufficient power to fine map metabolic disease on Kosrae.
In conclusion, we developed a protocol for using loki to analyze data from a complex pedigree and used it to identify quantitative loci in the Kosraen population. Similar regions of linkage have been reported in studies of stature and metabolic syndrome disorders in other populations. These results suggest that further genetic analysis of the Kosrae cohort is likely to yield important results. Future studies should seek to establish the population origins of the contributing haplotypes and to facilitate ultimate identification of causal genes and alleles. The Kosrae study provides a potentially powerful data set to identify causative genes for complex traits and disorders with clinical implications.
Methods
Data Collection.
The data collection and phenotype definitions for the Kosrae study of the metabolic syndrome are described in Shmulewitz et al. (19) and are summarized below. The Rockefeller University Institutional Review Board and the Kosrae Department of Health approved this study. All 2,188 participants signed the informed consent before filling out family and medical questionnaires. The pedigree was constructed from the questionnaires, family visits, and genealogical records.
HT, WT, waist (WST), and HIP circumference, FBS, diastolic blood pressure (DBP), and SBP were measured. BMI was calculated as WT (kg) divided by HT squared (m2). The average of three DBP and SBP measures was used in the analysis. Mean arterial pressure (MAP) was calculated as DBP + ([SBP − DBP]/3). Blood was drawn for DNA isolation and for determination of LEP, TC, triglycerides (TG), and apoA-I and apoB. LEP levels were determined by RIA (Linco Research, St. Louis, MO), TC and TG by enzymatic colorimetric measurements (Boehringer Mannheim and Sigma), and apoAI and apoB by a double-antibody immunoassay technique. LEP, FBS, and TG were natural log transformed before analysis, and diabetics were excluded from FBS analysis.
Genotyping.
Genomic DNA was isolated from blood as described in ref. 66. A total of 1,564 samples were genotyped for the entire genome scan marker set at the Genotyping Core Facility at The Rockefeller University. We modified the Marshfield marker set 9 by replacing ≈100 markers, to develop a marker set more suitable for a non-Caucasian population. Markers with low heterozygosity, poor amplification, or high error rates in the Kosrae population were replaced. We added markers in chromosomal regions with gaps of >12 cM. Forty percent of the markers had alleles not found in the Centre d’Etude Polymorphisme Humain reference population. Therefore, we set up our own panels to allow for multiplex gel loading. Details of the screening set, panels, and genotyping protocol are available from the authors upon request.
We used the Applied Biosystems genotyper program to score the genotypes. The templates were user-defined with the identity (size) of each marker’s alleles, and the fluorescent dye; alleles were labeled automatically on the basis of these parameters. Two investigators independently checked the allele calls. Each plate had four Centre d’Etude Polymorphisme Humain controls; if any of these controls had incorrect genotypes, the genotyping was repeated. Each genotype data set was imported into a filemaker (FileMaker, Santa Clara, CA) database for comparison. The consensus data were screened for Mendelian inconsistencies in nuclear families. These results were used to identify pedigree errors (individuals in the wrong place), and then genotyping errors were checked manually. For each marker, loki identified likely Mendelian inconsistencies by using the whole pedigree, and the respective genotypes were deleted for that marker. Markers with observed Mendelian error rates of ≥2% were rescored or regenotyped to rule out a systematic allele-calling error. This method reduced the observed error rate for all but four of these markers, which were not used in analysis. To avoid the analyses taking an excessive amount of time, we broke pedigree loops by duplicating selected individuals.
Preliminary and Linkage Analyses.
For details on preliminary and linkage analyses, see Supporting Methods, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
Foremost, we thank the people of Kosrae (especially Tholman Alik, Matchugo Talley, Vita Skilling, and Hiroshi Ismael), who made this study possible. We also thank members of the Genotyping Core Facility (Lynn Petukhova, Sandra Oplanich, and Merav Yifrach), Jeff Winick, Stephanie Boger, Elizabeth Stephenson, Chavie Wolfish, Jason Kim, Rebecca Breslow, Zoltan Takacs, Brandi Galke, and Maria Karayiorgou for help with genotyping and DNA preparation; Peter Verlander for his filemaker pro database; Thomas Lehner, Derek Gordon, Mark Levenstein, Dale Nyholt, and Jurg Ott for analysis advice; Peter Herndon for computer help; and Susan Korres for all her assistance. This work was supported by National Research Service Award Training Grant GM07982 and National Institute of Mental Health Training Program Grant T32MH65213 (to D.S.).
Abbreviations
- apoB
apoliprotein B
- BMI
body mass index
- DBP
diastolic blood pressure
- SBP
systolic blood pressure
- FBS
fasting blood sugar
- HT
height
- LEP
serum leptin
- MAP
mean arterial pressure
- QTL
quantitative trait locus
- TC
total cholesterol
- TG
triglycerides
- WST
waist
- WT
weight.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Reaven G. Diabetes. 1988;37:1595–1607. doi: 10.2337/diab.37.12.1595. [DOI] [PubMed] [Google Scholar]
- 2.American Heart Association. Heart Disease and Stroke Statistics: 2004 Update. Dallas: Am. Heart Assoc; 2003. [Google Scholar]
- 3.Comuzzie A. G., Allison D. B. Science. 1998;280:1374–1377. doi: 10.1126/science.280.5368.1374. [DOI] [PubMed] [Google Scholar]
- 4.Guo S.-W. Theor. Popul. Biol. 2000;57:1–11. doi: 10.1006/tpbi.2000.1449. [DOI] [PubMed] [Google Scholar]
- 5.Altmuller J., Palmer L. J., Fischer G., Scherb H., Wjst M. Am. J. Hum. Genet. 2001;69:936–950. doi: 10.1086/324069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hezel F. The First Taint of Civilization: A History of the Caroline and Marshall Islands in Pre-Colonial Days. Honolulu: Univ. of Hawaii Press; 1983. pp. 1521–1885. [Google Scholar]
- 7.Segal H. Kosrae: The Sleeping Lady Awakens. FSM: Kosrae State Government; 1989. [Google Scholar]
- 8.Cordy R. Asian and Pacific Archaeology Series. Vol. 10. Honolulu: Univ. of Hawaii Press; 1993. [Google Scholar]
- 9.Irwin G. The Prehistoric Exploration and Colonization of the Pacific. Cambridge, U.K.: Cambridge Univ. Press; 1994. [Google Scholar]
- 10.Zimmet P., Taft P., Guinea A., Guthrie W., Thoma K. Diabetologia. 1977;13:111–115. doi: 10.1007/BF00745137. [DOI] [PubMed] [Google Scholar]
- 11.Ravussin E., Valencia M. E., Esparza J., Bennett P. H., Schulz L. O. Diabetes Care. 1994;17:1067–1074. doi: 10.2337/diacare.17.9.1067. [DOI] [PubMed] [Google Scholar]
- 12.Heath S. Am. J. Hum. Genet. 1997;61:748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shmulewitz D., Heath S. C. Genet. Epidemiol. 2001;21(Suppl.):S686–S691. doi: 10.1002/gepi.2001.21.s1.s686. [DOI] [PubMed] [Google Scholar]
- 14.Yuan B., Neuman R., Duan S. H., Weber J. L., Kwok P. Y., Saccone N. L., Wu J. S., Liu K., Schonfeld G. Am. J. Hum. Genet. 2000;66:1699–1704. doi: 10.1086/302904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Preece M. Horm. Res. 1996;45:56–58. doi: 10.1159/000184849. [DOI] [PubMed] [Google Scholar]
- 16.Silventoinen K. J. Biosoc. Sci. 2003;35:263–285. doi: 10.1017/s0021932003002633. [DOI] [PubMed] [Google Scholar]
- 17.Hirschhorn J. N., Lindgren C. M., Daly M. J., Kirby A., Schaffner S. F., Burtt N. P., Altshuler D., Parker A., Rioux J. D., Platko J., et al. Am. J. Hum. Genet. 2001;69:106–116. doi: 10.1086/321287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raftery A. Biometrika. 1996;83:251–266. [Google Scholar]
- 19.Shmulewitz D., Auerbach S. B., Lehner T., Blundell M., Winick J. D., Youngman L. D., Skilling V., Heath S. C., Ott J., Stoffel M., et al. Hum. Hered. 2001;51:8–19. doi: 10.1159/000022953. [DOI] [PubMed] [Google Scholar]
- 20.Xu J., Bleecker E., Jongepier H., Howard T., Koppelman G., Postma D., Meyers D. Am. J. Hum. Genet. 2002;71:646–650. doi: 10.1086/342216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiltshire S., Frayling T. M., Hattersley A. T., Hitman G. A., Walker M., Levy J. C., O’Rahilly S., Groves C. J., Menzel S., Cardon L., et al. Am. J. Hum. Genet. 2002;70:543–546. doi: 10.1086/338760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perola M., Ohman M., Hiekkalinna T., Leppavuori J., Pajukanta P., Wessman M., Koskenvuo M., Palotie A., Lange K., Kaprio J., et al. Am. J. Hum. Genet. 2001;69:117–123. doi: 10.1086/321286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng H.-W., Xu F.-W., Liu Y.-Z., Shen H., Deng H., Huang Q.-Y., Liu Y.-J., Conway T., Li J.-L., Davies K., et al. Am. J. Med. Genet. 2002;113:29–39. doi: 10.1002/ajmg.10742. [DOI] [PubMed] [Google Scholar]
- 24.Kissebah A., Sonnenberg G., Myklebust J., Goldstein M., Broman K., James R., Marks J., Krakower G., Jacob H., Weber J., et al. Proc. Natl. Acad. Sci. USA. 2000;97:14478–14483. doi: 10.1073/pnas.97.26.14478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freeman M., Mansfield M., Barrett J., Grant P. Diabet. Med. 2002;19:994–999. doi: 10.1046/j.1464-5491.2002.00843.x. [DOI] [PubMed] [Google Scholar]
- 26.McQueen M., Bertram L., Rimm E., Blacker D., Santangelo S. BMC Genet. 2003;4:S96. doi: 10.1186/1471-2156-4-S1-S96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clement K., Philippi A., Jury C., Pividal R., Hager J., Demenais F., Basdevant A., Guy-Grand B., Froguel P. Int. J. Obes. Relat. Metab. Disord. 1996;20:507–512. [PubMed] [Google Scholar]
- 28.Hager J., Dina C., Francke S., Dubois S., Houari M., Vatin V., Vaillant E., Lorentz N. Nat. Genet. 1998;20:304–308. doi: 10.1038/3123. [DOI] [PubMed] [Google Scholar]
- 29.Perusse L., Rice T., Chagnon Y. C., Despres J. P., Lemieux S., Roy S., Lacaille M., Ho-Kim M., Chagnon M., Province M., et al. Diabetes. 2001;50:614–621. doi: 10.2337/diabetes.50.3.614. [DOI] [PubMed] [Google Scholar]
- 30.Feitosa M. F., Borecki I. B., Rich S. S., Arnett D. K., Sholinsky P., Myers R. H., Leppert M., Province M. Am. J. Hum. Genet. 2002;70:72–82. doi: 10.1086/338144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krushkal J., Ferrell R., Mockrin S. C., Turner S. T., Sing C. F., Boerwinkle E. Circulation. 1999;99:1407–1410. doi: 10.1161/01.cir.99.11.1407. [DOI] [PubMed] [Google Scholar]
- 32.Shearman A. M., Ordovas J. M., Cupples L. A., Schaefer E. J., Harmon M. D., Shao Y., Keen J. D., DeStefano A. L., Joost O., Wilson P., et al. Hum. Mol. Genet. 2000;9:1315–1320. doi: 10.1093/hmg/9.9.1315. [DOI] [PubMed] [Google Scholar]
- 33.Atwood L. D., Samollow P. B., Hixson J. E., Stern M. P., MacCluer J. W. Genet. Epidemiol. 2001;20:373–382. doi: 10.1002/gepi.7. [DOI] [PubMed] [Google Scholar]
- 34.Kristjansson K., Manolescu A., Kristinsson A., Hardarson T., Knudsen H., Ingason S., Thorleifsson G., Frigge M., Kong A., Gulcher J., et al. Hypertension. 2002;39:1044–1049. doi: 10.1161/01.hyp.0000018580.24644.18. [DOI] [PubMed] [Google Scholar]
- 35.Rainwater D. L., Almasy L., Blangero J., Cole S. A., VandeBerg J. L., MacCluer J. W., Hixson J. E. Arterioscler. Thromb. Vasc. Biol. 1999;19:777–783. doi: 10.1161/01.atv.19.3.777. [DOI] [PubMed] [Google Scholar]
- 36.Imperatore G., Knowler W. C., Pettitt D. J., Kobes S., Fuller J. H., Bennett P. H., Hanson R. L. Arterioscler. Thromb. Vasc. Biol. 2000;20:2651–2656. doi: 10.1161/01.atv.20.12.2651. [DOI] [PubMed] [Google Scholar]
- 37.Klos K. L., Kardia S. L. R., Ferrell R. E., Turner S. T., Boerwinkle E., Sing C. F. Arterioscler. Thromb. Vasc. Biol. 2001;21:971–978. doi: 10.1161/01.atv.21.6.971. [DOI] [PubMed] [Google Scholar]
- 38.Soro A., Pajukanta P., Lilja H., Ylitalo K., Hiekkalinna T., Perola M., Cantor R., Viikari J., Taskinene M., Peltonen L. Am. J. Hum. Genet. 2002;70:1333–1340. doi: 10.1086/339988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmer L., Buxbaum S., Larkin E., Patel S., Elston R., Tishler P., Redline S. Am. J. Hum. Genet. 2003;72:340–350. doi: 10.1086/346064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chagnon Y. C., Chen W. J., Perusse L., Chagnon M., Nadeau A., Wilkison W. O., Bouchard C. Mol. Med. 1997;3:663–673. [PMC free article] [PubMed] [Google Scholar]
- 41.Aouizerat B. E., Allayee H., Cantor R. M., Davis R. C., Lanning C. D., Wen P., Dallinga-Thie G. M., de Bruin T. W. A., Rotter J., Lusis A. Am. J. Hum. Genet. 1999;42:397–412. doi: 10.1086/302490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pajukanta P., Cargill M., Viitanen L., Nuotio I., Kareinen A., Perola M., Terwilliger J. D., Kempas E., Daly M., Lilja H., et al. Am. J. Hum. Genet. 2000;67:1481–1493. doi: 10.1086/316902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lembertas A. V., Perusse L., Chagnon Y. C., Fisler J. S., Warden C. H., Purcell-Huynh D. A., Dionne F. T., Gagnon J., Nadeau A., Lusis A., et al. J. Clin. Invest. 1997;100:115–121. doi: 10.1172/JCI119637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee J. H., Reed D. R., Li W. D., Xu W., Joo E. J., Kilker R. L., Nanthakumar E., North M., Sakul H., Bell C., et al. Am. J. Hum. Genet. 1999;64:196–209. doi: 10.1086/302195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong C., Wang S., Li W.-D., Li D., Zhao H., Price R. Am. J. Hum. Genet. 2003;72:115–124. doi: 10.1086/345648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohman M., Oksanen L., Kaprio J., Koskenvuo M., Mustajoki P., Rissanen A., Salmi J., Kontula K., Peltonen L. J. Clin. Endocrinol. Metab. 2000;85:3183–3190. doi: 10.1210/jcem.85.9.6797. [DOI] [PubMed] [Google Scholar]
- 47.Gudmundsson G., Matthiasson S., Arason H., Johannsson H., Runarsson F., Bjarnason R., Helgadottir K., Thorisdottir S., Ingadottir G., Lindpaintner K., et al. Am. J. Hum. Genet. 2002;70:586–592. doi: 10.1086/339251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arya R., Duggirala R., Jenkinson C., Almasy L., Blangero J., O’Connell P., Stern M. Am. J. Hum. Genet. 2004;74:272–282. doi: 10.1086/381717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hanson R. L., Ehm M. G., Pettitt D. J., Prochazka M., Thompson D. B., Timberlake D., Foroud T., Kobes S., Baier L., Burns D., et al. Am. J. Hum. Genet. 1998;63:1130–1138. doi: 10.1086/302061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pratley R. E., Thompson D. B., Prochazka M., Baier L., Mott D., Ravussin E., Sakul H., Ehm M. G., Bruns D., Foroud T., et al. J. Clin. Invest. 1998;101:1757–1764. doi: 10.1172/JCI1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elbein S. C., Hoffman M. D., Teng K., Leppert M. F., Hasstedt S. J. Diabetes. 1999;48:1175–1182. doi: 10.2337/diabetes.48.5.1175. [DOI] [PubMed] [Google Scholar]
- 52.Vionnet N., Hani El H., Dupont S., Gallina S., Francke S., Dotte S., De Matos F., Durand E., Lepretre F., Lecoeur C., et al. Am. J. Hum. Genet. 2000;67:1470–1480. doi: 10.1086/316887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe R. M., Ghosh S., Langefeld C. D., Valle T. T., Hauser E. R., Magnuson V. L., Mohlke K. L., Silander K., Ally D., Chines P., et al. Am. J. Hum. Genet. 2000;67:1186–1200. [PMC free article] [PubMed] [Google Scholar]
- 54.Wiltshire S., Hattersley A. T., Hitman G. A., Walker M., Levy J. C., Sampson M., O’Rahilly S., Frayling T. M., Bell J., Lathrop G., et al. Am. J. Hum. Genet. 2001;69:553–569. doi: 10.1086/323249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lindgren C. M., Mahtani M. M., Widen E., McCarthy M. I., Daly M. J., Kirby A., Reeve M. P., Kruglyak L., Parker A., Meyer J., et al. Am. J. Hum. Genet. 2002;70:509–516. doi: 10.1086/338629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiang K., Wang Y., Zheng T., Jia W., Li J., Chen L., Shen K., Wu S., Lin X., Zhang G., et al. Diabetes. 2004;53:228–234. doi: 10.2337/diabetes.53.1.228. [DOI] [PubMed] [Google Scholar]
- 57.Frikke-Schmidt R. Scand. J. Clin. Lab. Invest. 2000;2000(Suppl.):3–25. [PubMed] [Google Scholar]
- 58.Eichner J., Dunn S., Perveen G., Thompson D., Stewart K., Stroehla B. Am. J. Epidemiol. 2002;155:487–495. doi: 10.1093/aje/155.6.487. [DOI] [PubMed] [Google Scholar]
- 59.Han Z., Heath S., Shmulewitz D., Li W., Auerbach S., Blundell M., Lehner T., Ott J., Stoffel M., Friedman J., et al. Am. J. Med. Genet. 2002;110:234–242. doi: 10.1002/ajmg.10445. [DOI] [PubMed] [Google Scholar]
- 60.Wijsman E., Amos C. Genet. Epidemiol. 1997;14:719–735. doi: 10.1002/(SICI)1098-2272(1997)14:6<719::AID-GEPI28>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 61.Heath S., Snow G., Thompson E., Tseng C., Wijsman E. Genet. Epidemiol. 1997;14:1011–1016. doi: 10.1002/(SICI)1098-2272(1997)14:6<1011::AID-GEPI75>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 62.Cheng R., Ma J., Elston R., Li M. Genet. Epidemiol. 2001;21(Suppl.):S748–S753. doi: 10.1002/gepi.2001.21.s1.s748. [DOI] [PubMed] [Google Scholar]
- 63.Pankratz N., Kirkwood S., Flury L., Koller D., Foroud T. Genet. Epidemiol. 2001;21(Suppl.):S732–S737. doi: 10.1002/gepi.2001.21.s1.s732. [DOI] [PubMed] [Google Scholar]
- 64.Daw E., Wijsman E., Thompson E. Genet. Epidemiol. 2003;24:181–190. doi: 10.1002/gepi.10230. [DOI] [PubMed] [Google Scholar]
- 65.Atwood L., Heard-Costa N., Cupples L., Jaquish C., Wilson P., D’Agostino R. Am. J. Hum. Genet. 2002;71:1044–1050. doi: 10.1086/343822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hopcroft D. W., Mason D. R., Scott R. S. Horm. Metab. Res. 1985;17:559–561. doi: 10.1055/s-2007-1013606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.