Abstract
Class I hydrophobins are a unique family of fungal proteins that form a polymeric, water-repellent monolayer on the surface of structures such as spores and fruiting bodies. Similar monolayers are being discovered on an increasing range of important microorganisms. Hydrophobin monolayers are amphipathic and particularly robust, and they reverse the wettability of the surface on which they are formed. There are also significant similarities between these polymers and amyloid-like fibrils. However, structural information on these proteins and the rodlets they form has been elusive. Here, we describe the three-dimensional structure of the monomeric form of the class I hydrophobin EAS. EAS forms a β-barrel structure punctuated by several disordered regions and displays a complete segregation of charged and hydrophobic residues on its surface. This structure is consistent with its ability to form an amphipathic polymer. By using this structure, together with data from mutagenesis and previous biophysical studies, we have been able to propose a model for the polymeric rodlet structure adopted by these proteins. X-ray fiber diffraction data from EAS rodlets are consistent with our model. Our data provide molecular insight into the nature of hydrophobin rodlet films and extend our understanding of the fibrillar β-structures that continue to be discovered in the protein world.
Keywords: amyloid, NMR, polymer
Hydrophobins are a large family of secreted, low-molecular-mass (7–9 kDa) proteins unique to filamentous fungi. There is little amino acid sequence similarity between hydrophobins, except for a characteristic pattern of eight cysteine residues that form four intramolecular disulfide bonds (1, 2). These proteins have remarkable biophysical properties and function by self-assembling into amphipathic polymeric films at the interface between hydrophobic and hydrophilic surfaces (3). The surfactive and amphipathic properties of hydrophobins facilitate the formation of essential aerial structures such as hyphae, spores, and fruiting bodies (4).
Two classes of hydrophobins have been identified based on their hydrophobicity plots and physical properties (5). For class I hydrophobins, the polymer film comprises cylindrical rodlets with dimensions of ≈10 × 100–250 nm and their outward-facing hydrophobic surface has extremely low wettability (6, 7). These films are very robust; they are resistant to boiling in detergents or strong alkalis (8, 9). The morphology of isolated rodlets is reminiscent of amyloid fibrils isolated from diseased tissue and formed in vitro. Reconstituted rodlets stained with Congo red give the green-gold birefringence characteristic of similarly stained amyloid fibers and circular dichroism (CD) data indicate that the rodlets contain extensive β-structure, suggesting that rodlets and amyloid fibrils have structural features in common (10, 11, ** ). Class II hydrophobin films are significantly less robust and lack the rodlet morphology of class I hydrophobins (12, 13).
Class I hydrophobins, once solubilized, will spontaneously reform native-like rodlet films at any hydrophilic/hydrophobic interface. Such films can reverse the wettability of a surface such Teflon and are considered to have great potential as biomaterials (14, 15). However, despite considerable efforts, our knowledge of the structure of hydrophobins and the mechanism through which they form rodlets is far from complete.
We have determined the solution structure of the class I hydrophobin EAS (the protein encoded by the easilywettable gene) from Neurospora crassa. The structure comprises a four-stranded β-barrel core as well as an additional two-stranded sheet and two sizeable disordered regions. Notably, all of the charged residues are localized to a single surface of the protein. To probe the role of the disordered regions in the structure and function of EAS, we have used site-directed mutagenesis to delete up to half of the largest loop, and we demonstrate that such mutants are competent to fold and form native-like rodlets. By using our structural and mutagenic data, we have constructed a model for the polymeric EAS rodlets that incorporates all previously existing biophysical data on hydrophobins, as well as information from newly recorded x-ray fiber diffraction experiments. These results represent detailed structural information on the nature of these remarkable rodlet layers and expand our understanding of the growing class of polymeric protein fibrils that are built on a β-sheet scaffold.
Results
Solution Structure of EAS.
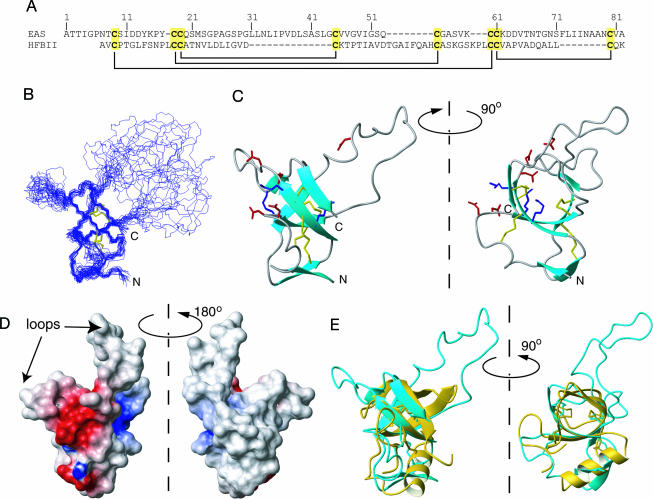
The triple-resonance data sets collected on monomeric EAS isoform Ib (for definition of isoform Ib, see Supporting Text, which is published as supporting information on the PNAS web site) were of good quality; we were able to assign 97% of backbone and 96% of side-chain nuclei. The disulfide bond linkages (Fig. 1A) were determined chemically (see Supporting Text, Fig. 5, and Table 1, which are published as supporting information on the PNAS web site) by using the method of Wu and Watson (16), and this information was combined with distance and angle restraints determined from the NMR data to calculate the structure of EAS. The 20 lowest-energy structures from the final aria (www.pasteur.fr/recherche/unites/Binfs/aria) calculations were chosen to represent the solution structure of EAS (Fig. 1B). Restraint densities and structural statistics for the ensemble are given in Fig. 6 and Table 2, which are published as supporting information on the PNAS web site. The structure of EAS is centered on an irregular four-stranded β-barrel, comprising K15–C19, V46–V47, S57–K62, and N79–V81, with an additional two-stranded antiparallel β-sheet (T2–T3 and Q53–C54) appended to one face. All of the β-sheet secondary structure is antiparallel. An overlay over the backbone atoms of residues for which the φ and ψ angle order parameters are >0.9 has an rms deviation of 0.67 Å. Two of the four disulfide bonds (19–45 and 61–80) lie in the center of the barrel, whereas the other two (9–60 and 18–54) connect the outside surface of the barrel to the additional sheet and a nearby loop (see Fig. 6C).
Fig. 1.
Solution structure of EAS. (A) Sequences of EAS and the class II hydrophobin HFBII, indicating the conserved disulfide bonding pattern. (B) Overlay of the 20 lowest-energy conformers of EAS. (C) Ribbon diagram of EAS. Cys side chains are shown as orange sticks. Positively and negatively charged residues are shown as blue and red sticks, respectively. (D) Electrostatic surface of EAS. The clustering of charged residues is apparent. (E) Overlay of EAS with the x-ray crystal structure of HFBII. The minimized average structure of EAS is shown in cyan, and HFBII (Protein Data Bank ID code 1R2M) is shown in yellow. Cys residues are shown in neon.
In addition to the well ordered regions, two disordered loops exist: M22–S42 and V65–F72. Backbone rms deviations, the number of nuclear Overhauser effects (NOEs) observed for these residues (see Fig. 6) and 1H-15N heteronuclear NOE data (12) attest to the poor definition of these sequences. The inside of the β-barrel is a tightly packed hydrophobic core comprising residues from all four strands, whereas a “secondary” hydrophobic core connects the outside surface of the barrel to the additional short β-sheet. It is also notable that the large unstructured loop between M22 and S42 is extremely hydrophobic in nature, whereas the shorter loop is predominantly made up of uncharged polar residues.
One of the features of the EAS sequence is the unusually low number of charged residues (≈10% of the sequence): five aspartates and three lysines. Of these eight residues, six (four Asp and two Lys) are clustered on a single surface of the protein (Fig. 1 C and D). The remainder of the surface of the well ordered core, as well as the M22–S42 loop, is essentially uncharged.
Comparison with the Class II Hydrophobin HFBII.
None of the β-barrel domains listed in the SCOP database (17) display the same topology as EAS. The only other known structure that shares the same barrel topology is the class II hydrophobin from Trichoderma reesei, HFBII (18). The sequence of HFBII is shown in Fig. 1A, and Fig. 1E shows an overlay of the two structures over the heavy atoms of the eight Cys residues. Although the barrel part of the EAS fold is very similar to HFBII, the remainder of the structures differs significantly. HFBII does not possess the very long disordered loop found in EAS, and the additional two-stranded β-sheet in EAS is replaced with an α-helix in HFBII. This helix occupies basically the same region of space as the small sheet in EAS and comprises residues that, in EAS, form one of the strands of the sheet (including the fifth Cys residue). HFBII also lacks six residues at the N terminus, which form the second strand of the small sheet in EAS.
Role of the Disordered Regions.
The presence of the two disordered loops in EAS, in particular the large M22–S42 loop, is striking given that the protein functions by forming an ordered polymeric fibril. A sequence alignment of type I hydrophobins (see Fig. 7, which is published as supporting information on the PNAS web site) reveals that this sequence, between Cys-3 and -4, is the least conserved portion of the protein in terms of both size and make-up. To explore the function of this loop, we used site-directed mutagenesis. Because hydrophobins have only ever been isolated from natural sources, we first had to set up an expression protocol in bacteria; this process necessitated the oxidative refolding of EAS from inclusion bodies. The integrity of refolded recombinant EAS was established by RP-HPLC, and 1H NMR (see Fig. 8, which is published as supporting information on the PNAS web site), and it was shown to form native-like rodlets (data not shown). Deletion of either 7 or 11 residues (residues 29–35 or 27–37, inclusive) yielded EAS mutants that still formed stable, well ordered structures (Fig. 8) and retained the ability to form rodlets. This finding is consistent with the sequence data and indicates that the rodlet topology must be able to accommodate significant sequence diversity in this region.
Constraints on Rodlet Assembly.
We next sought to understand how EAS monomers assemble into the polymeric, amyloid-like structures typical of class I hydrophobins. Our data, together with a number of published biophysical studies (reviewed in ref. 19), provide many restrictions on how the monomers can stack to form rodlets.
Restraints imposed by the EAS core structure.
The global conformation of the EAS core is cylindrical, with a flexible loop at each end of the cylinder. Given the tight packing of the core and the disulfide bonds, it is unlikely that the core will unfold substantially during rodlet assembly; the lack of change in both 1H NMR and CD spectra of the monomer under a range of harsh conditions (11, 20) supports this conclusion. Any structural model for the rodlets also must take into account the sequence variability of the loop regions, given the observed morphological similarity of rodlets formed from different hydrophobins.
Restraints imposed by the charge distribution in the monomer.
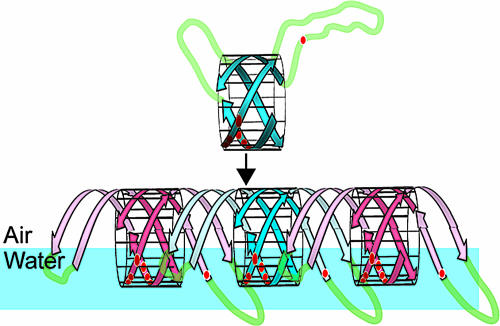
Class I hydrophobin rodlets form a monolayer at air:water interfaces with a hydrophobic side that faces air and a hydrophilic side that contacts water (21). Given that there is a single continuous charged patch on the surface of the EAS core and that the diametrically opposite face is completely hydrophobic, the simplest way of arranging monomers in the monolayer is for the charged side to face the water (Fig. 2). This arrangement is consistent with the way other small surfactive molecules align at air:water interfaces (22).
Fig. 2.
Possible model for EAS rodlet formation. Schematic representation of EAS monomers stacking at an air:water interface. Red circles indicate charged residues. The two disordered loops might “add on” to the barrel, forming additional β-structure that H-bonds to the next monomer. Adjacent monomers are colored blue and red for clarity.
Hydrogen-bonding (H-bonding) orientation.
In the orientation of the monomer noted above, the H-bonds in the β-barrel are parallel to the air–water interface. This idea is supported by appearance of the positive amide I band in the polarization modulation infrared reflection-absorption spectroscopy spectra recorded during polymerization of SC3 (23).
Changes in β-sheet content upon polymerization.
CD, attenuated-total-reflectance–Fourier transform infrared (ATR-FTIR), and fiber diffraction data all point toward increases in β-structure upon polymerization. This result is most easily explained by either regularization of the existing β-structure or the adoption of a β conformation by the disordered loops. It is notable that the first disordered loop (residues 20–42) of EAS is predicted from sequence analysis to be in a β-sheet conformation and also that a mass spectrometry study of SC3 revealed an increase in protection for residues in the corresponding region after polymerization (24). Indeed, residues in the corresponding loop are predicted to form some β-structure in all class I hydrophobins.
Microscopy data.
Atomic force micrographs of class I hydrophobins have shown the rodlets to be composed of two straight tracks of subrodlets, each track with dimensions of ≈10 × 100–250 nm. For SC3, it has been reported that two or three protofilaments, each with a diameter of 2.5 nm, make up each track (20, 25), and ellipsometry measurements have shown that the SC3 film is ≈3 nm thick (26). The diameter of the β-barrel of EAS is ≈25 Å, supporting the idea that the rodlets are a molecular monolayer. The rodlets also have a very high length-to-width ratio and are straight, indicating that the stacking of hydrophobin monomers within the rodlet does not incur much twisting, because at least 30 monomers must able to stack together without a change in the orientation of the charged face.
Modeling of Rodlet Assembly.
These data allow inferences to be drawn about the topology of the rodlet superstructure. First, extension along the rodlet long axis is most likely to arise from monomers in a head-to-tail arrangement, so that “infinite” extension can occur. Second, the individual monomer barrels will be aligned such that their charged face will contact water. Third, the most effective stacking has β-barrels stacking end-to-end along the rodlet long axis (Fig. 2). In this orientation, the protofilament will have a diameter of ≈25 Å, which agrees well with the thickness of the SC3 films. In this orientation, the “leading” edge of one β-barrel is in a position that allows H-bonding to the “trailing” edge of the next barrel. This assembly mode provides additional H-bonds that are parallel to the air:water interface. It is additionally possible that the loop regions extend the barrel at each end by forming two additional β-hairpins that H-bond to the barrel core (pale hairpins in Fig. 2). These hairpins would be positioned to contact the next monomer.
To test the geometric plausibility of this model, structure calculations were carried out for an EAS dimer in aria. For each monomer, the NOE and angle constraints from the original EAS structure calculation were used, together with additional intramolecular and intermolecular H-bond restraints designed to join two monomers to make a dimer. In some cases, the loop regions were restrained to form additional β-structure (see Supporting Text). Many sets of such restraints were trialed (representing different relative geometries of the monomers), but only a few resulted in structures that satisfied the following criteria: (i) low total energies (less than three times that of the monomer); (ii) no distance restraint violations >0.5 Å and angle violations >5°; (iii) presentation of the charged face on one side of the dimer and the hydrophobic face on the opposite side; and (iv) minimal twisting and bending between the two monomers.
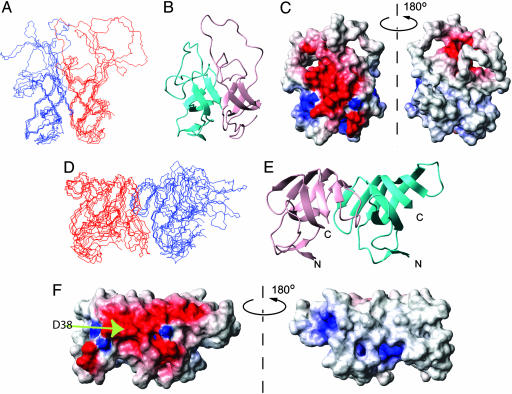
Models from the calculations without and with incorporation of the flexible loops in the repeating β-structure are shown in Fig. 3. Both models are compatible with the EAS core structure and display highly amphipathic surfaces, consistent with the properties of the rodlets (Fig. 3 C and E). It is notable that incorporation of the loops into the barrel structure maintains the amphipathic nature of the cylinder. In particular, residue D38, the only charged residue in the two flexible loops, becomes positioned along the same face as other charged residues that make up the charged face of the core β-barrel (Fig. 3F). The rodlet structure can be readily built up from these dimer models: individual protofilaments are created from end-to-end stacking of monomers, and the bundling of these filaments creates the observed monolayer film.
Fig. 3.
Molecular structures of EAS rodlet models. Structures of EAS dimers were calculated by using the monomer NMR restraint list supplemented with putative H-bonds. (A) Five lowest-energy structures and ribbon diagram of the dimeric model without incorporation of the flexible loops in the repeating β-structure. (D) As in A, but with incorporation of the flexible loops in the repeating β-structure in the dimeric model. (B and E) Ribbon diagram of the models in the same orientation as A and D, respectively. (C and F) Electrostatic potential diagram of the dimeric models, in the same orientation as A and D, respectively.
X-Ray Fiber Diffraction of the EAS Rodlets.
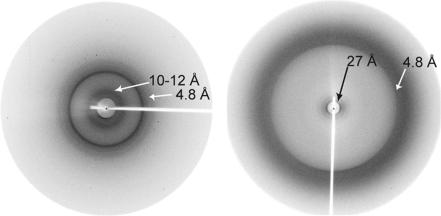
We carried out x-ray diffraction studies to assess the validity of our model. Pelleted rodlet preparations lacking in alignment displayed two reflections, a relatively intense one at 4.8 Å and a much weaker, diffuse reflection at 10–12 Å (Fig. 4). Partially aligned rodlet samples, which were prepared in a capillary situated in a magnetic field, displayed birefringence between cross-polarizers and gave rise to a diffraction pattern with different features. The maximum intensity of the 4.8-Å reflection was centered on the meridian of the pattern, and no reflection was observed at 10–12 Å. An additional low-angle reflection was detected at ≈27 Å.
Fig. 4.
X-ray fiber diffraction of EAS rodlets. (Left) Pattern obtained from pelleted, preformed rodlets. (Right) Pattern obtained from rodlets prepared in a capillary in a magnetic field. Both patterns have a strong reflection at 4.7 Å, from the spacing between adjacent β-strands in a β-sheet. A very weak and diffuse inner reflection is seen at ≈10 Å (left), probably arising from an intersheet spacing. The 27-Å reflection is consistent with the dimensions of hydrophobin monomers arranged along the long axis of the fibril.
Discussion
Hydrophobin Monomer Structure.
A previous structural study of cerato-ulmin (a class II hydrophobin) concluded that the cysteines were linked consecutively (i.e., Cys-1–Cys-2 etc) (27). In contrast, the recent crystal structure of HFBII revealed the pattern Cys-1–Cys-6, Cys-2–Cys-5, Cys-3–Cys-4, Cys-7–Cys-8 (18). We have shown that the pattern in the class I protein EAS matches that of HFBII; it is likely that this arrangement is common to all hydrophobins.
Despite the low level of sequence similarity between class I and II hydrophobins, our data reveal that they share essentially the same core fold. It is also clear from our structure of EAS how other class I family members could readily adopt the same fold: both flexible loops in EAS have ends that are close in space and should therefore tolerate the substantial variation that exists in these regions (Fig. 6C and Supporting Text); for example, the Cys-3–Cys-4 loop can vary between 4 and 29 residues. Our mutagenesis data show unambiguously that the identity of the Cys-3–Cys-4 loop is not important for the folding or structure of the EAS monomer.
Formation of Rodlets.
In the models presented here, the EAS monomer structure is essentially maintained in the protofilaments, and the “sticky” leading and trailing edges of the β-barrel core provide a means of linking monomers through backbone H-bonding. This finding is analogous to the situation in amyloid and amyloid-like fibrils, where intermolecular H-bonding between edge strands links adjacent monomers into a β-scaffold that can extend for many hundreds of nanometers along the fibril length. In the classical “cross-β” structure, β-strands run perpendicular to the fiber axis and form β-sheets that associate face-to-face, and H-bond formation is likely to provide a substantial driving force for polymerization. It has been proposed that this finding explains why such a diverse range of sequences can form amyloid-like fibrils. The same reasoning applies to hydrophobins: the homology between class I members is low, but the same polymeric structure is in principle accessible because it is driven by backbone interactions. The other factor that assists polymerization is the localization of monomers to the air:water interface. This event restricts diffusion of monomers to two dimensions and also orients them relative to each other, making for more frequent and effective collisions and thereby allowing rapid polymerization, potentially with concomitant ordering of the flexible loops.
It is notable that, although the EAS and HFBII monomers share very similar overall folds, class II hydrophobins such as HFBII do not appear to form ordered rodlets. Two possible reasons for this result are (i) the less hydrophobic nature of class II proteins (e.g., ≈400 Å2 for HFBII out of a total surface area of 1,600 Å2), which would reduce the driving force for alignment at the air:water interface, and (ii) the lack of flexible loops in class II proteins (as shown for HFBII), which might make it harder to fit the monomers together.
X-Ray Fiber Diffraction.
X-ray fiber diffraction data for EAS rodlets are consistent with both of the models proposed here. The 4.8-Å reflection (the distance between strands in a β-sheet) indicates that EAS rodlets contain repeating β-structure, whereas the ≈27-Å signal is consistent with the dimensions of the EAS monomer and with a rodlet structure in which the EAS monomers associate end-to-end. Amyloid samples typically display a reflection at 10–12 Å, which is interpreted as the spacing between β-sheets stacked perpendicular to the fibril long axis. The absence of this reflection for EAS is consistent with our model, which does not incorporate multiple parallel β-sheets, but could also be a consequence of the monolayer nature of the sample used for diffraction measurements. In other cases where diffraction patterns do not show the 10- to 12-Å spacing, it has been proposed that the fibrils are composed of β-helices (28, 29). The model of EAS hydrophobin rodlets proposed here has some similarities with β-helical structures. In the classical cross-β structure, the β-strands lie perpendicular (within 5°), to the fibril axis, whereas for EAS the β-strands that form the EAS barrel lie at ≈30° to the fibril long axis (30). However, the fact that rodlets have ordered underlying β-structure and bind Congo red to display yellow-green birefringence suggests that they share some structural elements with amyloid fibrils.
A Functional Fibril.
In many diseases, amyloid fibrils represent the endpoint of the conversion of soluble, active proteins into a form that is inactive and aggregates in an ordered, β-sheet-dependent fashion. The EAS hydrophobin rodlets are an example of another insoluble, β-sheet structure, but one in which a β-scaffold, with its ability to form extendable regular structures regardless of sequence, is used to present an active surface; i.e., an amphipathic monolayer with biological function. The fast kinetics of rodlet formation, compared with the rate of amyloid formation by many proteins, is also a functional feature of the system and might arise from a combination of the existence of preformed β-structure and the surface localization noted above.
A number of bacterial proteins appear to exhibit similar properties to hydrophobins. Chaplins are secreted on the surface of Streptomyces and also function in the formation of aerial structures by assembling into fibrils (19). Although these fibrils bear a strong visual resemblance to EAS rodlets, there is little similarity in the amino acid sequences, and there is no structural information available on the chaplin proteins. Curli and Tafi proteins from Escherichia coli and Salmonella, respectively, form a rather different loose network of amyloid-like fibrils (19) but again are poorly characterized.
In summary, our data show that EAS forms a β-barrel and have allowed the construction of a model for the rodlets that reproduces many of their observed physical properties, including overall dimensions, creation and maintenance of hydrophobic and hydrophilic faces along the rodlet, and minimal overall twisting and bending between monomers. The model is also consistent with experimental evidence from many biophysical studies of class I hydrophobins. Further data from techniques such as solid-state NMR will be required to fully validate the model.
Materials and Methods
Growth of N. crassa and Purification of EAS.
N. crassa STA4 was grown on solid Vogel’s N-medium as described in ref. 31. For the preparation of doubly labeled 13C/15N EAS, the medium was modified to remove other carbon sources and compensate for the loss of buffering activity. The modified medium contained K2HPO4 (50 mM, pH 7.0), MgSO4 (0.2%), 15NH4Cl (0.1%), CaCl2 (0.05%), N-trace elements [with FeSO4 substituted for Fe(NH4)2(SO4)2], biotin (5 μg/liter), and 13C6-d-glucose (1%). EAS was purified from mature conidia as described in ref. 11.
Production of Recombinant EAS and Mutagenesis.
A PCR-amplified DNA fragment encoding EAS cDNA was subcloned into the pHUE vector (32). This vector produced recombinant EAS (rEAS) fused to the C terminus of human ubiquitin, with an additional His-6 tag at the very N terminus. Mutants were constructed with deletions of residues 28–35 or 27–37 from EAS. All constructs were expressed as inclusion bodies in E. coli and purified under denaturing conditions by using Ni-NTA agarose (Qiagen, Hilden, Germany). rEAS and mutants were refolded in redox buffer, then dialyzed into cleavage buffer for the removal of the His-6-ubiquitin tag before subjected to RP-HPLC. The identity of the proteins was confirmed by MALDI-TOF/MS. See Supporting Text for more details.
Mapping the Disulfide Bonds of EAS.
The disulfide linkages of EAS were mapped by the method of Wu and Watson (16). Detailed conditions are provided in Supporting Text.
NMR Spectroscopy.
NMR samples (≈0.5 mM) were prepared as described in ref. 11. Spectra were acquired at 280 K on DRX-600 and DRX-800 NMR spectrometers (Bruker, Karlsruhe, Germany), processed by using xwinnmr 3.5, and analyzed by using xeasy (33). The following spectra were acquired: double-quantum-filtered COSY, total correlation spectroscopy (TOCSY; τm = 70 ms), NOESY (τm = 150 ms), ct-HNCA, HNHA, HNCACB, CBCA(CO)NH, HNCO, HCCH-TOCSY, HCCH-COSY, 13C-separated NOESY, and 15N-separated NOESY. Assignments were made by using standard methodology. NOE-derived distance restraints were obtained from the 2D NOESY and 3D 13C-edited NOESY. φ-Angle restraints based on the 3JHNHα coupling constants were measured from an HNHA (34). Additional φ and ψ restraints were included based on the use of talos (10).
Structure Calculations.
NOESY spectra were peak picked and used as input into aria 1.2 (35) implemented in cns (Version 1.1) (36). A total of 63 φ and 27 ψ angle constraints were also used (with a precision of ±40° for HNHA-derived values and twice the reported standard errors for talos-derived restraints). Four pairs of disulfide bonds together with 10 backbone H-bond restraints also were incorporated into the calculations. Backbone H-bond restraints were included in regions of regular secondary structure where both an unambiguous donor–acceptor pair was identified in preliminary structures and the amide proton exchanged slowly with solvent (see Supporting Textand Fig. 9, which are published as supporting information on the PNAS web site). The calculation protocol comprised eight cycles of 20 structures each and a final cycle of 500 structures. Manually assigned NOEs were included in iteration zero as soft restraints. The cut-off value for an ambiguous assignment was reduced from 1.01 for the first iteration to 0.80 in iteration 8. A total of 1,547 unambiguous and 188 ambiguous restraints were identified by aria, and these restraints were checked and corrected manually where necessary. The 100 lowest-energy structures from iteration 8 were refined in a 9-Å shell of water molecules and the 20 conformers with the lowest value of Etot were visualized and analyzed by using molmol (37) and procheck-nmr (38).
X-Ray Fiber Diffraction.
X-ray fiber diffraction data were collected from samples of EAS rodlets prepared in either the presence or the absence of a 0.5-T magnetic field. Data were collected on an in-house source and examined by using standard methods. More details are available in Supporting Text.
Modeling.
Residues on EAS monomers that appeared to be capable of forming intermolecular H-bonds with neighboring EAS molecules were identified by inspection. Calculations were carried out in aria 1.2 using the restraint lists used in the monomer structure calculations together with putative intermolecular H-bonds. H-bonds incorporating the disordered loops also were included to incorporate these regions into the rodlet structure. In each trial, a single iteration of 100 structures was carried out (incorporating the parameters used in the final iteration of a normal aria calculation), and all restraints were included as fixed restraints. The 10 conformers with lowest energy from each set of structure calculations were examined by using molmol and the Protein–Protein Interaction Server (www.biochem.ucl.ac.uk/bsm/PP/server).
Supplementary Material
Acknowledgments
We thank Ross Beever for providing the initial inspiration for this work, Molly Clifton for assistance with mass spectrometry, and Ian Kaplin from the Electron Microscope Unit at the University of Sydney for assistance with EM. This work was supported in part by the Marsden Fund, administered by the Royal Society of New Zealand, Contract HRT 701 (to M.D.T.).
Abbreviation
- NOE
nuclear Overhauser effect.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The family of 20 low-energy structures has been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2FMC).
Butko, P., Goodwin, J. S., Bufford, J. P., Stroud, P., McCormick, C. L. & Cannon, G. C. (2001) Biophys. J. 80, 404A (abstr.).
References
- 1.Wösten H. A. B., van Wetter M. A., Lugones L. G., van der Mei H. C., Busscher H. J., Wessels J. G. H. Curr. Biol. 1999;9:85–88. doi: 10.1016/s0960-9822(99)80019-0. [DOI] [PubMed] [Google Scholar]
- 2.Wösten H. A. B., Richter M., Willey J. M. Fungal Genet. Biol. 1999;27:153–160. doi: 10.1006/fgbi.1999.1130. [DOI] [PubMed] [Google Scholar]
- 3.Wösten H. A. B. Annu. Rev. Microbiol. 2001;55:625–646. doi: 10.1146/annurev.micro.55.1.625. [DOI] [PubMed] [Google Scholar]
- 4.Wessels J. G. H. Adv. Microb. Physiol. 1997;38:1–45. doi: 10.1016/s0065-2911(08)60154-x. [DOI] [PubMed] [Google Scholar]
- 5.Wessels J. G. H. Annu. Rev. Phytopathol. 1994;32:413–437. [Google Scholar]
- 6.Beever R. E., Dempsey G. Nature. 1978;272:608–610. doi: 10.1038/272608a0. [DOI] [PubMed] [Google Scholar]
- 7.Wösten H. A. B., Devries O. M. H., Wessels J. G. H. Plant Cell. 1993;5:1567–1574. doi: 10.1105/tpc.5.11.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beever R. E., Redgewell R. J., Dempsey G. J. Bacteriol. 1979;140:1063–1070. doi: 10.1128/jb.140.3.1063-1070.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Vries O. M. H., Fekkes M. P., Wösten H. A. B., Wessels J. G. H. Arch. Microbiol. 1993;159:330–335. [Google Scholar]
- 10.Cornilescu G., Delaglio F., Bax A. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 11.Mackay J. P., Matthews J. M., Winefield R. D., Mackay L. G., Haverkamp R. G., Templeton M. D. Structure (London) 2001;9:83–91. doi: 10.1016/s0969-2126(00)00559-1. [DOI] [PubMed] [Google Scholar]
- 12.Paananen A., Vuorimaa E., Torkkeli M., Penttila M., Kauranen M., Ikkala I., Lemmetyinen H., Serimaa R., Linder M. B. Biochemistry. 2003;42:5253–5258. doi: 10.1021/bi034031t. [DOI] [PubMed] [Google Scholar]
- 13.Torkkeli M., Serimaa R., Ikkala O., Linder M. Biophys. J. 2002;83:2240–2247. doi: 10.1016/S0006-3495(02)73984-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssen M. I., van Leeuwen M. B. M., van Kooten T. G., de Vries J., Dijkhuizen L., Wosten H. A. B. Biomaterials. 2004;25:2731–2739. doi: 10.1016/j.biomaterials.2003.09.060. [DOI] [PubMed] [Google Scholar]
- 15.Linder M. B., Qiao M. Q., Laumen F., Selber K., Hyytia T., Nakari-Setala T., Penttila M. E. Biochemistry. 2004;43:11873–11882. doi: 10.1021/bi0488202. [DOI] [PubMed] [Google Scholar]
- 16.Wu J., Watson J. T. Protein Sci. 1997;6:391–398. doi: 10.1002/pro.5560060215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murzin A. G., Brenner S. E., Hubbard T., Chothia C. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 18.Hakanpaa J., Paananen A., Askolin S., Nakari-Setala T., Parkkinen T., Penttila M., Linder M. B., Rouvinen J. J. Biol. Chem. 2004;279:534–539. doi: 10.1074/jbc.M309650200. [DOI] [PubMed] [Google Scholar]
- 19.Gebbink M. F. B. G., Claessen D., Bouma B., Dijkhuizen L., Wösten H. A. B. Nature Rev. Microbiol. 2005;3:333–341. doi: 10.1038/nrmicro1127. [DOI] [PubMed] [Google Scholar]
- 20.de Vocht M. Ph.D. thesis. Groningen, The Netherlands: University of Groningen; 2001. [Google Scholar]
- 21.Wösten H. A. B., Schuren F. H. J., Wessels J. G. H. EMBO J. 1994;13:5848–5854. doi: 10.1002/j.1460-2075.1994.tb06929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu J., Thomas R., Penfold J. Adv. Coll. Inter. Sci. 2000;84:143–304. doi: 10.1016/s0001-8686(99)00019-6. [DOI] [PubMed] [Google Scholar]
- 23.de Vocht M. L., Reviakine I., Ulrich W. P., Bergsma-Schutter W., Wosten H. A. B., Vogel H., Brisson A., Wessels J. G. H., Robillard G. T. Protein Sci. 2002;11:1199–1205. doi: 10.1110/ps.4540102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X., Permentier H. P., Rink R., Kruijtzer J. A., Liskamp R. M., Wosten H. A., Poolman B., Robillard G. T. Biophys. J. 2004;87:1919–1928. doi: 10.1529/biophysj.104.041616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wosten H. A. B., de Vocht M. L. Biochim. Biophys. Acta Rev. Biomembr. 2000;1469:79–86. doi: 10.1016/s0304-4157(00)00002-2. [DOI] [PubMed] [Google Scholar]
- 26.Wang X., Shi F. X., Wosten H. A. B., Hektor H., Poolman B., Robillard G. T. Biophys. J. 2005;88:3434–3443. doi: 10.1529/biophysj.104.057794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yaguchi M., Pusztai-Cary M., Roy C., Surewicz W. K., Carey P. R., Stevenson K. J., Richards W. C., Takai S. In: Dutch Elm Disease Research: Cellular and Molecular Approaches. Sticklen M. B., Sherald J. L., editors. New York: Springer; 1993. pp. 152–170. [Google Scholar]
- 28.Kishimoto A., Hasegawa K., Suzuki H., Taguchi H., Namba K., Yoshida M. Biochem. Biophys. Res. Comm. 2004;315:739–745. doi: 10.1016/j.bbrc.2004.01.117. [DOI] [PubMed] [Google Scholar]
- 29.Perutz M. F., Finch J. T., Berriman J., Lesk A. Proc. Natl. Acad. Sci. USA. 2002;99:5591–5595. doi: 10.1073/pnas.042681399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson R., Sawaya M. R., Balbirnie M., Madsen A. O., Riekel C., Grothe R., Eisenberg D. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Templeton M. D., Greenwood D. R., Beever R. E. Exp. Mycol. 1995;19:166–169. doi: 10.1006/emyc.1995.1020. [DOI] [PubMed] [Google Scholar]
- 32.Catanzariti A. M., Soboleva T. A., Jans D. A., Board P. G., Baker R. T. Protein Sci. 2004;13:1331–1339. doi: 10.1110/ps.04618904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartels C., Xia T., Billeter M., Guntert P., Wuthrich K. J. Biomol. NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 34.Vuister G. W., Bax A. J. Biomol. NMR. 1994;4:193–200. doi: 10.1007/BF00175247. [DOI] [PubMed] [Google Scholar]
- 35.Nilges M., Macias M. J., O’Donoghue S. I., Oschkinat H. J. Mol. Biol. 1997;269:408–422. doi: 10.1006/jmbi.1997.1044. [DOI] [PubMed] [Google Scholar]
- 36.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta. Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Koradi R., Billeter M., Wuthrich K. J. Mol. Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. 29–32. [DOI] [PubMed] [Google Scholar]
- 38.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.