Abstract
The chromatin-assembly factor I (CAF-I) and the replication-coupling assembly factor (RCAF) complexes function in chromatin assembly during DNA replication and repair and play a role in the maintenance of genome stability. Here, we have investigated their role in checkpoints and S-phase progression. FACS analysis of mutants lacking Asf1 or Cac1 as well as various checkpoint proteins indicated that normal rates of S-phase progression in asf1 mutants have a strong requirement for replication checkpoint proteins, whereas normal S-phase progression in cac1 mutants has only a weak requirement for either replication or DNA-damage checkpoint proteins. Furthermore, asf1 mutants had high levels of Ddc2.GFP foci that were further increased in asf1 dun1 double mutants consistent with a requirement for checkpoint proteins in S-phase progression in asf1 mutants, whereas cac1 mutants had much lower levels of Ddc2.GFP foci that were not increased by a dun1 mutation. Our data suggest that RCAF defects lead to unstable replication forks that are then stabilized by replication checkpoint proteins, whereas CAF-I defects likely cause different types of DNA damage.
Keywords: chromatin assembly, DNA-damage response, DNA replication, genome instability
DNA replication and repair require new chromatin assembly, during which the four core histones must be assembled into a histone octamer containing two H2A/H2B dimers and a H3/H4 tetramer wrapped in DNA to form a nucleosome. In the absence of proper chromatin assembly in human cells, DNA synthesis cannot be completed (1, 2). Many histone chaperones are known to be involved in nucleosome and chromatin assembly, of which chromatin assembly factor 1 (CAF-I) and anti-silencing function 1 (Asf1) are among the most studied (reviewed in ref. 3). CAF-I is a nucleosome assembly factor complex that functions during DNA replication and repair and deposits histone H3/H4 tetramers onto DNA (4–7). Asf1 binds histones H3 and H4, forming the replication-coupling assembly factor (RCAF) complex, and is thought to play the role of a histone donor, functioning synergistically with CAF-I (8–11). Among other factors involved in chromatin assembly are NAP-1 (12), which functions as a cytoplasmic-nuclear histone H2A/H2B transfer factor during DNA synthesis, and HIRA (Hir1 and Hir2 in Saccharomyces cerevisiae) proteins, which are histone H2A/H2B chaperones that act in the assembly of chromatin independently of DNA synthesis (13, 14).
CAF-I is evolutionarily conserved and in most species contains three subunits. In S. cerevisiae, these three subunits are Cac1, Cac2, and Cac3, of which Cac3 directly interacts with histones H3 and H4, and the largest subunit, Cac1, binds proliferating cell nuclear antigen (PCNA) (15). CAF-I facilitates nucleosome assembly preferentially onto newly synthesized DNA (4, 16). In S. cerevisiae, deletion of any of the subunits leads to increased UV sensitivity (7) and silencing defects at telomeres as well as at the mating type loci (7, 17, 18). Furthermore, a synergistic decrease in silencing was observed in S. cerevisiae cells lacking both CAF-I and Asf1 (8), and there appears to be a synergistic interaction between CAF-I and Asf1 during chromatin assembly linked to DNA replication in vitro (10). Consistent with these observations, CAF-I has been shown to physically interact with Asf1 in both human (10) and Drosophila (11) cells.
The RCAF complex in S. cerevisiae and Drosophila consists of the Asf1 histone chaperone protein and histones H3 and H4 (11, 19). As opposed to CAF-I, the RCAF complex cannot promote chromatin assembly coupled to DNA replication on its own; however, it appears to synergize with CAF-I in this role in vitro (8, 11). S. cerevisiae mutants lacking Asf1 exhibit sensitivity to a wider range of DNA-damaging agents and have a slow growth phenotype compared with CAF-I-defective mutants (8), suggesting that CAF-I and RCAF may have some distinct functions. Asf1 has also been implicated in the buffering of free histones during DNA-damage-induced cell cycle arrest (20) as well as chromatin disassembly at certain loci (21).
Several studies have linked both CAF-I and RCAF to checkpoint regulation. In S. cerevisiae, Asf1 and the checkpoint protein Rad53 are known to interact, and Asf1 is dissociated from phosphorylated Rad53 after cells are treated with the DNA damaging agent methyl methane sulfonate (MMS) or hydroxyurea (HU) (22, 23). However, a recent analysis of physiological levels of Asf1 has indicated that Asf1 can interact with phosphorylated Rad53 in response to MMS (24). In humans and Drosophila, Asf1 has also been implicated in checkpoint related signaling due to being the only known substrate for Tlk (tousled-like kinase), which is inhibited by the ATM/Chk1 DNA-damage-checkpoint pathway (25, 26). Furthermore, asf1 mutant cells have been previously shown to be partially defective in HU-induced Rad53 phosphorylation (22), suggesting that asf1 mutants may be partially checkpoint-defective. Consistent with this idea, the apparent inability of asf1 mutants to recover from HU arrest as suggested by FACS (8) is similar to the behavior of mutants that have a defect in the replication checkpoint (27). In addition, expression of a dominant-negative Cac1 protein, the largest component of CAF-I, has been shown to cause DNA damage and activate the S-phase checkpoint in human cells (2). In our previous work, we showed that mutations in the genes encoding the CAF-I and RCAF complexes caused increased rates of accumulation of gross chromosomal rearrangements in S. cerevisiae (28). Our genetic analysis suggested that Asf1 defects could result in DNA damage that activated both the replication and DNA-damage checkpoints, whereas CAF-I defects might result in activation of the DNA-damage checkpoint. Consistent with this, a recent study has demonstrated activation of the DNA-damage checkpoint in an asf1 mutant (29).
In the present study, we have investigated whether RCAF and CAF-I play a role in checkpoint regulation. Our results show that defects in these chromatin assembly factors do not cause checkpoint defects. In contrast, asf1 mutants were found to depend highly on the S-phase checkpoints for normal progression through S phase. Furthermore, cells that are defective for both CAF-I and RCAF appear to have increased S-phase progression defects resulting in the accumulation of cells arrested in G2/M consistent with the accumulation of DNA damage during S phase. These results are interpreted in terms of models in which RCAF mutants are partially defective in maintaining replication fork structure and that this defect is exacerbated by both checkpoint and CAF-I defects.
Results
asf1 Mutants Are Sensitive to Killing by MMS but Not HU.
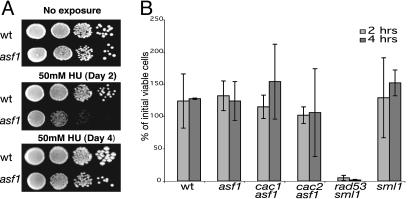
Many checkpoint-defective mutants, such as a rad53Δ mutant, are sensitive to killing by both HU and MMS. We therefore tested all possible single, double, and triple combinations of CAF-I subunit mutations, none of which caused sensitivity to either 50 mM HU or 0.01% or 0.02% MMS (Fig. 7, which is published as supporting information on the PNAS web site) (data not shown); note that in an S288c strain (our background) CAF-I defects did not cause sensitivity to 0.02% MMS, whereas in a W303 strain sensitivity to 0.02% MMS was seen (30). In contrast, the asf1 single mutant and the cac1 asf1, cac2 asf1, and cac3 asf1 double mutants were all sensitive to killing by MMS (Fig. 8, which is published as supporting information on the PNAS web site). The asf1 single mutant and the cac1 asf1, cac2 asf1, and cac3 asf1 double mutants appeared to be sensitive to killing by HU when the plates were incubated at 30°C for up to 3 days, but upon longer incubation all four mutant strains exhibited wild-type levels of survival (Fig. 1A) (data not shown). Thus, these mutants appear to grow much more slowly in the presence of HU but are not killed.
Fig. 1.
asf1 mutants are not killed by either chronic or acute HU treatment. (A) Cells were plated on YPD and 50 mM HU plates and incubated at 30°C, as indicated. (B) Cell survival after acute treatment of cells with 200 mM HU for 2- or 4-h periods is shown as the percent of viable cells present before treatment.
Mutants that are defective for the replication checkpoint cannot recover from acute treatment with concentrations of HU (200 mM) that result in depletion of nucleotide pools and subsequent DNA replication arrest. Consistent with this, the checkpoint-defective rad53 sml1 mutant was unable to recover from either 2- or 4-h treatment with HU. In contrast, the asf1 single mutant and the cac1 asf1, cac2 asf1, and cac3 asf1 double mutants all showed the same full recovery as the wild-type and sml1 single mutant control strains (Fig. 1B). This result confirms that the asf1 single mutant and the cac1 asf1, cac2 asf1, and cac3 asf1 double mutants are not sensitive to killing by HU.
Analysis of Cell Morphology Suggests That RCAF Single Mutants and RCAF CAF-I Double Mutants Have a Cell Cycle Progression Defect.
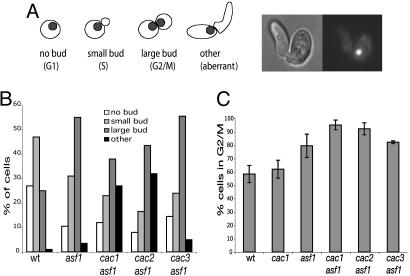
The asf1 single mutant and the cac1 asf1, cac2 asf1, and cac3 asf1 double mutants all appear to have a modest reduction in growth rate compared with wild-type control strains or cac1, cac2, or cac3 single mutants. To better understand this, the morphology of cells from log-phase cultures of these mutants and control strains were analyzed by fluorescence microscopy (Fig. 2A and B). The largest class of wild-type cells had small buds and a single nucleus, and thus appeared to be in early S phase. The cac1, cac2, and cac3 single mutants were all similar to wild-type cells (see legend of Fig. 2). Compared with the wild-type strain, asf1 mutants had a large increase in the number of large-budded cells along with a decrease in the proportion of cells with either no bud and a single nucleus or a small bud and a single nucleus; the cac3 asf1 double mutant showed a distribution of cells that was similar to that of the asf1 mutant. Likewise, in comparison with the wild-type cells, the cac1 asf1 and cac2 asf1 double mutants showed an increase in large-budded cells with DNA staining in both buds along with a decrease in the proportion of cells with either no bud and a single nucleus or a small bud and a single nucleus. However, in contrast to asf1 mutants, these two mutants exhibited a large increase in the number of cells having aberrant bud morphologies in which buds were elongated and misshapen, with a single nucleus at the bud neck (Fig. 2A).
Fig. 2.
asf1 caf-1 double mutants exhibit a higher proportion of cells in G2/M and accumulate aberrant buds indicative of DNA damage. (A) Log-phase cells were stained with DAPI and analyzed by fluorescent microscopy. Each strain was scored for the percentage of cells with no buds, small buds, large buds, and aberrant buds. The distribution for CAF-I mutants, in the order stated above, were cac1 (18%, 48%, 30%, and 4%), cac2 (19%, 48%, 29%, and 3%), and cac3 (15%, 55%, 28%, and 2%). (B) The DNA content of the same series of mutants was determined by FACS analysis of cells from log-phase unsynchronized cultures.
Analysis of the G2/M DNA content in the same cultures by FACS was consistent with the cell morphology analysis (summarized in Fig. 2C). One interpretation of the FACS results in combination with the cell morphology analysis is that the asf1 single and double mutant cells are accumulating DNA damage during DNA replication resulting in slower progression through the G2/M phase of the cell cycle, whereas this may not be the case with CAF-I single mutants.
Chromatin Assembly Mutants Are Proficient for the Replication and Intra-S Checkpoints.
There appeared to still be some question as to whether asf1 mutants are checkpoint-defective (31), leading us to further study the replication and intra-S checkpoint proficiency of chromatin assembly mutants. We examined chromatin assembly mutants, as well as appropriate controls, by FACS to monitor their ability to resume the cell cycle after arrest with 200 mM HU. We found that all of the chromatin assembly mutants showed the same kinetics and extent of recovery from HU arrest as the wild-type control (Fig. 9, which is published as supporting information on the PNAS web site). Intra-S checkpoint proficiency was examined by releasing α-factor arrested cells into 0.03% MMS and observing the ability of mutant and control cells to sense the DNA damage, demonstrated by significant slowing of S-phase progression. Again, all of the chromatin assembly mutants behaved like the wild-type control (Fig. 10, which is published as supporting information on the PNAS web site). Thus, these mutants have fully functional replication and intra-S checkpoints.
S-Phase Progression in asf1 Mutants Depends on Functional Checkpoints.
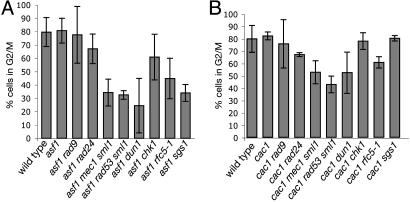
We have previously suggested that defects in the genes encoding CAF-I appear to cause some type of DNA damage that is processed by the DNA-damage checkpoints, whereas a defect in the ASF1 gene causes some type of DNA damage that is processed by both the DNA-damage and the replication checkpoints (28). Therefore, it was of interest to determine whether inactivation of different checkpoints has an effect on S-phase progression in these chromatin assembly mutants. We monitored S-phase progression of asf1 and cac1 mutants containing mutations that inactivate DNA-damage and intra-S DNA-damage checkpoint sensors (rad24 and rad9), replication checkpoint sensors (rfc5-1 and dpb11-1), replication and intra-S checkpoint sensors (sgs1), and checkpoint transducers (mec1, rad53, dun1, and chk1) after release from G1 arrest (31, 32). The rate of progression through S phase was evaluated by determining the proportion of cells that accumulated in G2/M phase at 40 min after release from α-factor arrest (Fig. 3).
Fig. 3.
asf1 and cac1 mutants require the function of different checkpoints for normal S-phase progression. FACS was used to monitor the rate of S-phase progression of various chromatin assembly/checkpoint double mutants after release from α-factor arrest. The proportion of cells in G2/M was determined at the 40-min time point, at which at least 80% of wild-type cells have reached G2/M. Error bars represent the SD. The percent of cells in G2/M for the control strains was as follows: rad9, 81 ± 4%; rad24, 86 ± 6%; mec1 sml1, 85 ± 5%; rad53 sml1, 81 ± 4%; dun1, 80 ± 2%; chk1, 82 ± 10%; rfc5-1, 74 ± 7%; sgs1, 83 ± 7%.
Despite initiating S phase slightly later than wild-type cells and growing slightly slower, asf1 mutants accumulated the same proportion of cells in G2/M by 40 min after release as the wild-type cells (81% vs. 80%). The asf1 rad9 and asf1 rad24 mutants did not demonstrate a significant S-phase progression delay compared with wild-type cells or asf1 mutants as evidenced by their proportion of G2/M cell cells at 40 min (78% and 67%, respectively). In contrast, the asf1 dun1, asf1 mec1 sml1, asf1 rad53 sml1, and asf1 sgs1 mutants exhibited a significant delay in S-phase progression (24%, 34%, 32%, and 34% G2/M cells at 40 min, respectively) and also had a higher proportion of S-phase cells at this time point. The asf1 rfc5-1 mutants also showed a reproducible decrease in G2/M content cells (45%), although not as large as in asf1 dun1, asf1 mec1 sml1, asf1 rad53 sml1, and asf1 sgs1 mutants. It was not possible to analyze the asf1 dbp11-1 double mutant because it had a severe growth defect (data not shown). Unlike asf1 dun1 or asf1 rad53 sml1 mutants, the asf1 chk1 mutant did not appear to exhibit a significant S-phase delay (61%). Cells with a cac1 mutation did not appear to exhibit slowed progression through S phase and had an average G2/M content of 80% of the cells at 40 min after α-factor release (Fig. 3B). The rad9, rad24, rfc5-1, and sgs1 mutations did not appear to cause a significant slowing of S-phase progression in a cac1 mutant. However, cac1 mec1 sml1, cac1 dun1, and cac1 rad53 sml1 mutants did appear to show a small but significant reduction in S-phase progression (53%, 52%, and 43% G2/M cells at 40 min, respectively) as compared with the cac1 single mutant.
The analysis presented here indicates that asf1 and cac1 mutations cause different effects in the S-phase progression assay. The data are consistent with the idea that asf1 mutants depend on replication checkpoint proteins to transit S phase, as evidenced by the S-phase progression phenotype of the asf1 rfc5-1, asf1 mec1 sml1, asf1 dun1, asf1 rad53 sml1, and asf1 sgs1 mutants and the lack of a slowed phenotype of the asf1 rad9 and asf1 rad24 mutants. In contrast, cac1 mutants show little if any checkpoint protein dependence for progression through S phase as evidenced by the weak phenotypes seen only with the cac1 mec1 sml1, cac1 rad53 sml1, and cac1 dun1 mutants.
Analysis of Spontaneous Checkpoint Protein Assembly in asf1 and cac1 Mutants.
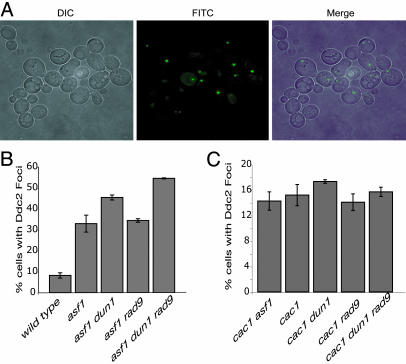
Previously published results showing that both asf1 and cac1 mutants have elevated levels of gross chromosomal rearrangements and that asf1 and cac1 mutations showed synergistic interactions with different checkpoint defects (28) raised the hypothesis that chromatin assembly mutants accumulate DNA damage that activates checkpoints during replication. The findings presented here that asf1 mutants, and to a small extent cac1 mutants, depend on checkpoint proteins for normal S-phase progression is consistent with this idea. To further investigate this hypothesis, we examined the formation of DNA-damage foci in log-phase cultures of key mutant strains using a functional GFP-tagged version of the Ddc2 protein (Fig. 4). Ddc2 is recruited to sites of DNA damage during checkpoint activation (33); thus, formation of Ddc2.GFP foci is indicative of endogenous DNA damage being sensed by the checkpoint machinery.
Fig. 4.
Ddc2.GFP checkpoint protein complexes accumulate in asf1 and cac1 mutants. Live cells were analyzed in log phase by deconvolution microscopy to visualize Ddc2.GFP expression from an endogenous functional DDC2.GFP fusion gene resulting in Ddc2.GFP foci. (A) Cells are shown in Left (DIC), GFP fluorescence is shown in Center (FITC), and the two images are merged in Right (Merge). The experimental results quantified for asf1 (B) and cac1 (C) mutant cells are expressed as the percentage of cells containing Ddc2.GFP foci. The error bars presented are SDs. The control mutants rad9 DDC2.GFP and dun1 DDC2.GFP had 13% and 7% cells with Ddc2.GFP foci, respectively.
We found that in the wild-type strain, 8% of the cells had Ddc2.GFP foci, consistent with previously published results (33). There was a slight, but significant, increase in the cac1 mutant cells, with 15% containing Ddc2.GFP foci (Fig. 4C). In contrast, asf1 mutants exhibited an ≈4-fold increase over wild-type cells, with 33% of cells containing Ddc2.GFP foci (Fig. 4B), similar to a recent study (29). The Ddc2.GFP foci were only seen in budding cells. These data suggest that there are varying degrees of endogenous DNA damage in asf1 and cac1 mutant cells, and this could explain the dependency of S-phase progression on the Mec1–Rad53–Dun1 pathway. Interestingly, the cac1 asf1 double mutant had a similar proportion of cells with Ddc2.GFP foci (14%) to that seen in a cac1 mutant (Fig. 4C), despite exhibiting a more severe growth phenotype than either of the single mutants, suggesting that the growth and morphological defects in cac1 asf1 mutants may not be due to replication defects.
We also examined Ddc2.GFP foci formation in both the asf1 dun1 and cac1 dun1 mutants. A dun1 mutation would be expected to inactivate the checkpoint downstream from assembly of Ddc2.GFP foci, allowing us to monitor checkpoint activation in the absence of checkpoint execution (31, 34, 35). We found a significant increase in Ddc2.GFP foci in asf1 dun1 mutants, with 45% of cells containing one or more Ddc2.GFP focus (Fig. 4B). In contrast, the cac1 dun1 cells exhibited a smaller, but significant increase in damage foci formation compared with the cac1 single mutant, with 17% of cells containing Ddc2.GFP foci (Fig. 4C). These findings are consistent with the results of the FACS experiments suggesting that asf1 mutants, more so than cac1 mutants, require Mec1–Rad53–Dun1 checkpoint pathway proteins for progression through S phase. These findings also support the view that in the absence of a functional checkpoint, chromatin assembly mutants, especially asf1 mutants, have an exacerbated S-phase progression defect that results in increased levels of Ddc2 foci.
We also examined the formation of Ddc2.GFP foci in asf1 rad9 and cac1 rad9 cells. If asf1 or cac1 mutants indeed activated a Rad9 DNA-damage checkpoint, then it is possible that in the absence of a Rad9 pathway asf1 and cac1 mutants would accumulate more damage resulting in more recruitment of Ddc2 to DNA. However, consistent with the FACS analysis, there was no increase in Ddc2.GFP foci formation in either asf1 rad9 (34%) or cac1 rad9 (14%) mutants (Fig. 4 B and C). Interestingly, the asf1 dun1 rad9 triple mutant had a modestly higher proportion of cells with Ddc2-GFP foci (55%) compared with both the asf1 and asf1 dun1 mutants. In contrast, the cac1 dun1 rad9 triple mutant did not have an increased proportion of cells with Ddc2.GFP foci (15%) compared with either the cac1 or cac1 dun1 mutants. These data support the view that asf1 defects primarily result in a requirement for replication checkpoint proteins, in the absence of which a secondary requirement for the DNA-damage checkpoint might occur. In contrast, cac1 defects cause little requirement for checkpoint proteins.
Analysis of Spontaneous Rad53 Phosphorylation in asf1 and cac1 Mutants.
To confirm the results obtained by examining Ddc2.GFP foci formation, we evaluated checkpoint activation by monitoring Rad53 phosphorylation using quantitative mass spectrometry (24). The relative abundance of both unphosphorylated peptides and phosphopeptides of Rad53 purified from wild-type cells and either cac1 or asf1 cells was quantified by stable isotope labeling and mass spectrometry. As shown in Tables 1 and 2 (which are published as supporting information on the PNAS web site), the relative abundance of Rad53 in wild-type, cac1 and asf1 cells was similar, as measured by quantitative analysis of multiple unphosphorylated peptides.
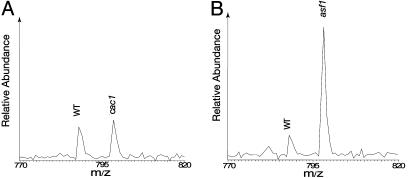
We next identified and quantified the relative abundance of phosphopeptides of Rad53 in wild-type and cac1 cells. Five different phosphopeptides were found (see Table 3, which is published as supporting information on the PNAS web site), representing both proline-directed and non-proline-directed phosphorylation of Rad53. As shown previously (24), the non-proline-directed phosphorylation is due to autophosphorylation of Rad53, which accompanies its activation. The detection of the autophosphorylation of Rad53 is not unexpected and it indicates a basal level of Rad53 activation in the unperturbed cells. Comparison of the phosphopeptides of Rad53 between wild-type and cac1 cells shows that there was little change in their abundance (Table 3). As an example, a phosphopeptide of Rad53 (LLHS*NNTENVK), containing phosphorylation of Ser-560, is shown in Fig. 5A. Phosphorylation of Ser-560 is an autophosphorylation event that serves as an indicator for Rad53 activation, demonstrating that there was no detectable change in the activation of Rad53 in the cac1 mutants.
Fig. 5.
Levels of Rad53 phosphorylation in cac1 and asf1 cells compared with that in wild-type cells. A phosphopeptide (LLHS*NNTENVK; the asterisk indicates the phosphorylated Ser to its right, i.e., Ser-560) of Rad53 is used as an example. (A) The abundance of this phosphopeptide was unaffected by deletion of CAC1. (B) The abundance of this phosphopeptide increased to almost 5-fold in asf1 cells, compared with wild-type cells. In both cases, Rad53 peptides from wild-type cells were labeled by d0-leucine, whereas those from cac1 or asf1 cells were labeled by d10-leucine.
In contrast, when the phosphopeptides of Rad53 in asf1 cells were compared with those in wild-type cells, the non-proline-directed autophosphorylation of Rad53 was clearly enhanced. As shown in Fig. 5B, the level of phosphorylation of Ser-560 in asf1 mutants was almost 5-fold higher than that in wild-type cells. Similarly, phosphorylation of Ser-789, an initial event in Rad53 autophosphorylation, and phosphorylation of Ser-747–748 and Ser-774 were also more abundant in asf1 cells (see Table 4, which is published as supporting information on the PNAS web site). Previous analysis of MMS-treated wild-type cells identified many more autophosphorylation sites of Rad53 with a much higher level of induction (24) than seen here in asf1 mutants. Thus, the level of Rad53 activation resulting from deletion of ASF1 appears to be higher than in wild-type cells but below the levels of a full checkpoint response, which is consistent with Western blot analysis of Rad53 phosphorylation in asf1 mutants (22, 29).
Discussion
In the present study, we have examined the genetic interplay between the RCAF and CAF-I chromatin assembly factors and different checkpoints. Our results demonstrate that defects in the genes encoding the RCAF and CAF-I complexes do not cause defects in the replication or intra-S DNA-damage checkpoints. We further found that normal progression of asf1 mutants through S phase depended on replication checkpoint proteins, whereas S-phase progression of CAF-I-defective mutants only weakly depended on checkpoint proteins. Consistent with these checkpoint protein requirements, asf1 mutants exhibited increased levels of Ddc2.GFP foci, which were further increased when checkpoint function was inactivated downstream of Ddc2 by a dun1 mutation. In contrast, CAF-I-defective mutants exhibited a much smaller increase in Ddc2.GFP foci in these assays. However, quantitative analysis of Rad53 phosphorylation indicated that asf1 mutants showed only low levels of spontaneous checkpoint activation similar to previous results (22, 29), and cac1 mutants did not show any checkpoint activation. The fact that a rad9 mutation did not increase the level of Ddc2.GFP foci in asf1 mutants and that rad9 and rad24 mutations did not affect the rate of S-phase progression in an asf1 mutant again points to a role for replication, rather than damage checkpoint proteins, in the stabilization of damaged DNA structures that may arise in asf1 mutants. Finally, although CAF-I defects did not cause a cell cycle progression defect, RCAF CAF-I double mutants showed a more severe cell cycle progression defect with lower levels of Ddc2.GFP foci than that seen in an asf1 mutant.
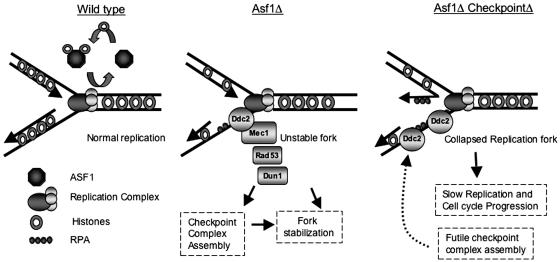
Initial studies indicated that RCAF functions in the assembly of chromatin during DNA replication, although subsequent studies have implicated RCAF in additional processes including disassembly of chromatin and assembly of selected replication factors at replication forks (3, 36, 37). A model interpreting the results of our analysis of asf1 mutants, as well as prior results, is presented in Fig. 6. We propose that in the absence of Asf1, replication forks are somewhat unstable, explaining the observation that asf1 mutants appear to enter S phase slightly later and progress through S phase slightly more slowly than wild-type cells. Fork instability could result from a deficit of histones on newly replicated DNA near replication forks, a lack of chromatin remodeling by the Asf1 chromatin disassembly function during DNA replication, or decreased assembly of replication factors at replication forks (21, 37, 38). However, these modestly unstable replication forks are primarily stabilized by proteins that function in the replication checkpoint and not the damage checkpoints (39, 40). Because a number of replication checkpoint proteins are known to be normal components of the replication machinery (reviewed in ref. 31), stabilization could be mediated either directly by assembly of these proteins at replication forks or through checkpoint activation implied by the observed increase in Ddc2.GFP foci when Dun1 is inactivated in an asf1 mutant. These mechanisms would explain the much slower S-phase progression and synergistic increase in genomic instability of mutants defective for both Asf1 and the replication checkpoint as well as the increased assembly of checkpoint protein foci in asf1 mutants containing downstream checkpoint defects (28). Although the DNA-damage checkpoint proteins appear to play little role in S-phase progression of asf1 mutants, the increase in Ddc2.GFP foci in the asf1 dun1 rad9 triple mutant could reflect a possible conversion of replicating DNA in the asf1 dun1 double mutant into structures that activate the DNA-damage checkpoint.
Fig. 6.
A model for the role of Asf1 in DNA replication and checkpoint activation. Asf1 plays an important role at replication forks during replication and repair. In the absence of Asf1, replication forks become unstable, leading to slower progression through S phase. Although somewhat unstable, these replication forks remain functional because of the presence of various replication checkpoint proteins, such as Ddc2, Rad53, and others, which play roles in replication forks stabilization. In the absence of a functional replication checkpoint, these somewhat unstable replication forks may collapse, thus leading to even slower S-phase progression, as well as conversion of the replication forks into structures that activate the DNA-damage checkpoint.
It has been suggested that RCAF and CAF-I function in the same chromatin assembly process (8, 11), leading us to perform parallel analyses of RCAF and CAF-I defects in the present study. Consistent with other studies indicating that RCAF and CAF-I have at least some distinct cellular roles (36, 41), we observed that defects in CAF-I did not cause the same effects on HU or MMS sensitivity, S-phase progression, Ddc2.GFP foci formation, or checkpoint activation as caused by an asf1 mutation. The aberrant bud phenotype seen in cac1 asf1 and cac2 asf1 double mutants is particularly interesting in that it correlates with a strong growth defect which is much more severe than in the respective single mutants. Furthermore, the double mutants are similar to asf1 single mutants in regard to S-phase progression and damaging agent sensitivity and similar to cac1 single mutants in regard to Ddc2.GFP foci phenotypes. Interestingly, the cac3 asf1 double mutant does not show either the increased aberrant bud or the slow growth phenotype, suggesting that these cac1 asf1 and cac2 asf1 phenotypes may be independent of histone deposition defects. It is tempting to speculate that these cac1 asf1 and cac2 asf1 defects are due to a defect in cell division and budding rather than DNA replication and chromatin assembly. Consistent with this, CAF-I has been implicated in kinetochore maintenance (42, 43) and has recently been linked to the control of the anaphase-promoting complex (APC) (41), a process thought not to involve Cac3. Given this, it is possible that cac1- and cac2-specific DNA damage leading to genome instability is the result of errors occurring during cell division, such as chromosome mis-segregation due to inappropriate kinetochore assembly, rather than errors occurring during DNA replication. This might explain the high gross chromosomal rearrangement rate seen in cac1 mutants even though cac1 mutants do not accumulate high levels of Ddc2.GFP.
This study identifies differences between the RCAF and CAF-I complexes in vivo, providing further evidence that although both complexes act as histone chaperones for histones H3 and H4, their functions during DNA replication and cell division may be quite different. Although further work is needed to investigate whether the high gross chromosomal rearrangement rates and low levels of checkpoint complex assembly in cac1 mutants actually reflect a role for CAF-I during cell division, it is apparent that Asf1 plays a critical role during DNA replication, specifically at replication forks. Future experiments will be required to investigate the role of Asf1 in replication fork stabilization and progression.
Materials and Methods
Strains and Media.
S. cerevisiae strains were grown in yeast extract/peptone/dextrose (YPD) medium or synthetic complete medium lacking the appropriate amino acid. G418 resistant colonies were selected on YPD plates containing 200 mg/liter geneticin. All of the strains used were derived from the S288c strain RDKY3615 (MATa, ura3-52, leu2Δ1, trp1Δ63, his3Δ200, lys2ΔBgl, hom3-10, ade2Δ1, ade8, hxt13::URA3) by either crossing with other RDKY3615 derivatives or by standard gene disruptions. The RDKY5764 strain background (MATa, ura3-52, trp1Δ63, his3Δ200, DDC2.GFP) used for analysis of Ddc2.GFP foci was created by Kristina S. Schmidt (Ludwig Institute for Cancer Research); it was derived from the same S288c parent strain as RDKY3615. The detailed genotypes of these strains are listed in Table 5, which is published as supporting information on the PNAS web site.
Sensitivity to HU and MMS.
Cells were grown in YPD medium to log phase. Serial dilutions were made and spotted onto previously prepared sensitivity plates (50 mM HU, 0.01% MMS, and 0.02% MMS) and grown for 2 (or more, as indicated) days at 30°C.
HU-Survival Assay.
Cells were first grown to log phase in YPD, and serial dilutions of the culture were plated onto YPD plates to determine the concentration of viable cells. HU was then added to the culture at a final concentration of 200 mM for periods of 2 and 4 h, after which serial dilutions of the cultures were plated onto YPD plates. The plates were incubated for 2–3 days at 30°C. Survival was calculated by determining the percentage of viable cells present after HU treatment compared with the untreated culture. These experiments were performed in triplicate.
α-Factor Arrest.
Cells were grown in YPD medium to log phase. α-factor (Sigma) was added to the culture at a concentration of 5 μg/ml. The culture was then incubated for 2 h at 30°C with shaking, and arrest was monitored by phase-contrast light microscopy. The cells were then pelleted, washed with water, and resuspended in fresh YPD.
FACS Analysis.
Cells were fixed in 70% ethanol for 1 h at room temperature, harvested by centrifugation, and resuspended in 50 mM sodium citrate buffer (pH 7.0). After sonication and centrifugation, the cells were once again resuspended in sodium citrate buffer and treated with 250 μg/ml RNase A (United States Biochemical) and 1 mg/ml proteinase K (Sigma) overnight at 37°C. The cells were then harvested by centrifugation, resuspended in 1 ml of sodium citrate buffer containing 1 μM Sytox Green (Molecular Probes), and incubated at room temperature for 2 h. Samples were analyzed by using a FACSVantage SE machine (BD Immunocytometry Systems).
Calculating G2/M Content of Cells.
FACS profiles were analyzed by using winmdi 2.8 software (http://facs.scripps.edu). G2/M content was calculated by measuring the number of events in the later half of the 2N DNA content peaks. This number was then multiplied by 2 and expressed as the percentage of the total of gated events. Experiments were performed in duplicate.
Quantitation of Bud Morphology.
Cells were grown in YPD medium to log phase, sonicated, stained with DAPI, and examined by fluorescence microscopy. Four hundred to 600 cells were counted for each mutant. Experiments were done in duplicate, and the percentage of cells in indicated morphology classes was determined.
Quantitation of Ddc2.GFP Foci.
Cells were grown in YPD medium to log phase and examined live by using a DeltaVision Restoration confocal microscope (Applied Precision). Images were collected in 0.2-μm z-sections to allow viewing of the entire content of the cells. Two hundred to 300 cells were imaged and counted for each experiment. softworx software (Applied Precision) was used for image analysis. Experiments were done in duplicate.
Determination of Rad53 Phosphorylation by Mass Spectrometry.
Purification of Rad53-TAP in wild-type, cac1, and asf1 background cells was performed as described in ref. 24. The purified Rad53 was digested by trypsin and labeled by an N-isotag reagent with d0- or d10-leucine, instead of d0- or d6-γ-amino butyric acid (GABA). There is no difference in protein quantification when using different amino acid based N-isotag reagents (unpublished observations). Leucine is used here because of its simplicity in synthesis, and the d0 form is commercially available. Boc-d0-Leu-NHS was purchased from NovaBiochem, and Boc-d10-Leu-NHS was synthesized from d10-leucine (Sigma-Aldrich), using the same protocol as for synthesis of Boc-d6-GABA-NHS (24). Mass spectrometry analysis and protein quantification were performed as described in ref. 24.
Supplementary Material
Acknowledgments
We thank Scarlet Shell, Jorrit Enserink, and Kristina Schmidt for comments on the manuscript; Kyungjae Myung (National Institutes of Health, Bethesda) and members of the Kolodner and Zhou laboratories for helpful discussions; Dennis Young (Flow Cytometry Resource, Moores University of California at San Diego Cancer Center) for FACS analysis; Stefanie Ness for DNA sequencing; and Jim Feramisco and his laboratory for help with fluoresence microscopy. We are particularly grateful to Kristina Schmidt for providing the Ddc2.GFP-expressing strain and Paul Maddox for advice about fluorescence microscopy. This work was supported by National Institutes of Health Grant GM26017 (to R.D.K.) and a K22 award from the National Human Genome Research Institute (to H.Z.).
Abbreviations
- HU
hydroxyurea
- MMS
methyl methane sulfonate
- YPD
yeast extract/peptone/dextrose.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Nelson D. M., Ye X., Hall C., Santos H., Ma T., Kao G. D., Yen T. J., Harper J. W., Adams P. D. Mol. Cell. Biol. 2002;22:7459–7472. doi: 10.1128/MCB.22.21.7459-7472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ye X., Franco A. A., Santos H., Nelson D. M., Kaufman P. D., Adams P. D. Mol. Cell. 2003;11:341–351. doi: 10.1016/s1097-2765(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 3.Loyola A., Almouzni G. Biochim. Biophys. Acta. 2004;1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 4.Smith S., Stillman B. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 5.Gaillard P. H., Martini E. M., Kaufman P. D., Stillman B., Moustacchi E., Almouzni G. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 6.Kamakaka R. T., Bulger M., Kaufman P. D., Stillman B., Kadonaga J. T. Mol. Cell. Biol. 1996;16:810–817. doi: 10.1128/mcb.16.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaufman P. D., Kobayashi R., Stillman B. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 8.Tyler J. K., Adams C. R., Chen S. R., Kobayashi R., Kamakaka R. T., Kadonaga J. T. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 9.Sharp J. A., Fouts E. T., Krawitz D. C., Kaufman P. D. Curr. Biol. 2001;11:463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- 10.Mello J. A., Sillje H. H., Roche D. M., Kirschner D. B., Nigg E. A., Almouzni G. EMBO Rep. 2002;3:329–334. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyler J. K., Collins K. A., Prasad-Sinha J., Amiott E., Bulger M., Harte P. J., Kobayashi R., Kadonaga J. T. Mol. Cell. Biol. 2001;21:6574–6584. doi: 10.1128/MCB.21.19.6574-6584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishimi Y., Hirosumi J., Sato W., Sugasawa K., Yokota S., Hanaoka F., Yamada M. Eur. J. Biochem. 1984;142:431–439. doi: 10.1111/j.1432-1033.1984.tb08305.x. [DOI] [PubMed] [Google Scholar]
- 13.Mosammaparast N., Ewart C. S., Pemberton L. F. EMBO J. 2002;21:6527–6538. doi: 10.1093/emboj/cdf647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park Y. J., Chodaparambil J. V., Bao Y., McBryant S. J., Luger K. J. Biol. Chem. 2005;280:1817–1825. doi: 10.1074/jbc.M411347200. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z., Shibahara K., Stillman B. Nature. 2000;408:221–225. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- 16.Verreault A., Kaufman P. D., Kobayashi R., Stillman B. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 17.Enomoto S., McCune-Zierath P. D., Gerami-Nejad M., Sanders M. A., Berman J. Genes Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- 18.Enomoto S., Berman J. Genes Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le S., Davis C., Konopka J. B., Sternglanz R. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 20.Groth A., Ray-Gallet D., Quivy J. P., Lukas J., Bartek J., Almouzni G. Mol. Cell. 2005;17:301–311. doi: 10.1016/j.molcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 21.Adkins M. W., Howar S. R., Tyler J. K. Mol. Cell. 2004;14:657–666. doi: 10.1016/j.molcel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 22.Hu F., Alcasabas A. A., Elledge S. J. Genes Dev. 2001;15:1061–1066. doi: 10.1101/gad.873201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emili A., Schieltz D. M., Yates J. R., III, Hartwell L. H. Mol. Cell. 2001;7:13–20. doi: 10.1016/s1097-2765(01)00150-2. [DOI] [PubMed] [Google Scholar]
- 24.Smolka M. B., Albuquerque C. P., Chen S. H., Schmidt K. H., Wei X. X., Kolodner R. D., Zhou H. Mol. Cell. Proteomics. 2005;4:1358–1369. doi: 10.1074/mcp.M500115-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groth A., Lukas J., Nigg E. A., Sillje H. H., Wernstedt C., Bartek J., Hansen K. EMBO J. 2003;22:1676–1687. doi: 10.1093/emboj/cdg151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrera P., Moshkin Y. M., Gronke S., Sillje H. H., Nigg E. A., Jackle H., Karch F. Genes Dev. 2003;17:2578–2590. doi: 10.1101/gad.276703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desany B. A., Alcasabas A. A., Bachant J. B., Elledge S. J. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myung K., Pennaneach V., Kats E. S., Kolodner R. D. Proc. Natl. Acad. Sci. USA. 2003;100:6640–6645. doi: 10.1073/pnas.1232239100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramey C. J., Howar S., Adkins M., Linger J., Spicer J., Tyler J. K. Mol. Cell. Biol. 2004;24:10313–10327. doi: 10.1128/MCB.24.23.10313-10327.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linger J. G., Tyler J. K. Genetics. 2005;171:1513–1522. doi: 10.1534/genetics.105.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lambert S., Carr A. M. Biochimie. 2005;87:591–602. doi: 10.1016/j.biochi.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 32.Cobb J. A., Shimada K., Gasser S. M. Curr. Opin. Genet. Dev. 2004;14:292–300. doi: 10.1016/j.gde.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 33.Melo J. A., Cohen J., Toczyski D. P. Genes Dev. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allen J. B., Zhou Z., Siede W., Friedberg E. C., Elledge S. J. Genes Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 35.Gardner R., Putnam C. W., Weinert T. EMBO J. 1999;18:3173–3185. doi: 10.1093/emboj/18.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adkins M. W., Tyler J. K. J. Biol. Chem. 2004;279:52069–52074. doi: 10.1074/jbc.M406113200. [DOI] [PubMed] [Google Scholar]
- 37.Franco A. A., Lam W. M., Burgers P. M., Kaufman P. D. Genes Dev. 2005;19:1365–1375. doi: 10.1101/gad.1305005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pulm W., Knippers R. Adv. Exp. Med. Biol. 1984;179:127–141. doi: 10.1007/978-1-4684-8730-5_13. [DOI] [PubMed] [Google Scholar]
- 39.Lopes M., Cotta-Ramusino C., Pellicioli A., Liberi G., Plevani P., Muzi-Falconi M., Newlon C. S., Foiani M. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 40.Lucca C., Vanoli F., Cotta-Ramusino C., Pellicioli A., Liberi G., Haber J., Foiani M. Oncogene. 2004;23:1206–1213. doi: 10.1038/sj.onc.1207199. [DOI] [PubMed] [Google Scholar]
- 41.Harkness T. A., Arnason T. G., Legrand C., Pisclevich M. G., Davies G. F., Turner E. L. Eukaryot. Cell. 2005;4:673–684. doi: 10.1128/EC.4.4.673-684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharp J. A., Franco A. A., Osley M. A., Kaufman P. D. Genes Dev. 2002;16:85–100. doi: 10.1101/gad.925302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharp J. A., Krawitz D. C., Gardner K. A., Fox C. A., Kaufman P. D. Genes Dev. 2003;17:2356–2361. doi: 10.1101/gad.1131103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.