Abstract
In this study, 83 clinical isolates purified from biofilms colonizing 18 silicone gastrostomy devices (12 “buttons” and six tubes converted to skin level devices) were selected for subtype characterization utilizing genetic analysis. The tubes, previously used for feeding, remained in place for 3 to 47 months (mean, 20.0 months) in children ranging in age from 6 months to 17 years. Classification of specific microbes using random amplified polymorphic DNA (RAPD) analysis revealed genetic similarities and differences among isolates belonging to the same genus. Both gram-positive and -negative bacteria were investigated, including 2 isolates of Bacillus brevis, 4 isolates of Bacillus licheniformis, 2 isolates of Bacillus pumilus, 3 isolates of Enterococcus durans, 19 isolates of Enterococcus faecalis, 8 isolates of Enterococcus faecium, 2 isolates of Enterococcus hirae, 7 isolates of Escherichia coli, 8 isolates of Lactobacillus plantarum, 19 isolates of Staphylococcus aureus, 2 isolates of Staphylococcus epidermidis, and 7 isolates of Staphylococcus saprophyticus. Amplified DNA fragments (amplicons) provided species-specific fingerprints for comparison by agarose gel electrophoresis. A total of 62 distinct RAPD types were categorized from the five genera studied. Typing analysis suggested cross acquisition of E. coli, E. faecalis, and S. aureus in three patient pairs. Genomic polymorphism detection proved efficient and reliable for classifying bacterial subtypes isolated from biofilms adhering to various portions of commonly employed enteral access tubes.
Long-term enteral access is a common practice associated with modern medicine, especially for direct gastric access via gastrostomy. Percutaneous endoscopic gastrostomy (PEG), first described in 1980, has facilitated direct gastric access, and skin level devices have removed many of the disadvantages associated with long enteral catheters (11). It is estimated that between 180,000 and 200,000 PEGs are performed annually in the United States (10). However, problems associated with microbial adhesion and related to patient health and device failure require further investigation.
Several studies have looked at microbial attachment on medical devices, such as catheters and prosthetic implants, while other investigations have researched fungal contamination on silicone gastrostomy tubes. The following organisms have been found associated with gastrostomy devices: Candida tropicalis, Candida albicans, Torulopsis glabrata, Engyodontium album, Wangiella dermatitidis, Pseudomonas aeruginosa, Candida krusei, Escherichia coli, Enterobacter cloacae, alpha-hemolytic streptococci, Lactobacillus sp., Bacteriodes sp., Bacillus brevis, Bacillus licheniformis, Bacillus pumilus, Enterococcus durans, Enterococcus faecalis, Enterococcus faecium, Enterococcus hirae, Lactobacillus plantarum, Micrococcus sedentarius, Staphylococcus aureus, Staphylococcus epidermidis, and Staphylococcus saprophyticus (6, 13, 14, 16, 19). Tube deterioration in association with attached fungi, as well as gastrostomy site wound infections, were found related to these organisms.
Bacterial biofilms, defined as a structured community of sessile bacterial cells enclosed in a self-produced extracellular polysaccharide matrix adhering to inert or living surfaces, occur in a wide variety of environments (5). Biofilms preferentially develop on inert surfaces or dead tissue, which is problematic in immunocompromised individuals requiring some form of medical implant. In addition, biofilms produce localized environments of corrosive molecules and/or protons in excessive concentrations as well as enzymes in direct contact with the substratum (4). Molecules are produced faster than they can diffuse through the matrix, leading to device deterioration. It has also been reported that species of the genera Torulopsis, Candida, and Aspergillus can catabolize intermediate-chain-length hydrocarbons for cellular growth (7, 17). Based on these findings, fungi in association with biofilm bacteria could enhance device destruction via the metabolism of silicone components.
Microbial typing is most accurately determined by genomic fingerprinting methods (21). Several methodologies are commonly employed, including pulsed-field electrophoresis (15), ribotyping (8), restriction endonuclease analysis (18), multilocus enzyme electrophoresis (12), and PCR-based procedures (9). Random amplified polymorphic DNA (RAPD) analysis can be utilized to determine variations in DNA sequences among closely related species, thus allowing subtype differentiation (20). Single primers, usually 10 bases long, are used to amplify random domains of purified DNA, producing fingerprints characteristic of a particular strain. Single-base substitutions, or insertions and deletions, will alter primer annealing, causing differing RAPD profiles that can be used to estimate nucleotide diversity and divergence (3). An advantage of RAPD analysis is that it can be applied to any strain or species of a bacterial group without previous knowledge of that isolate (26). In addition, much smaller quantities of DNA are required, which is especially important when dealing with gram-positive species. PCR-based technologies of this type produce high concentrations of DNA composing the amplicons, which eliminates the need for expensive data-imaging software. RAPD typing has been used successfully for the characterization of numerous organisms, including P. aeruginosa (1, 21), S. aureus, Staphylococcus intermedius (25), Streptococcus mutans, Streptococcus sobrinus, Streptococcus rattus (22), Salmonella enterica subsp. enterica (2), Lactobacillus casei, Lactobacillus curvatus, Lactobacillus fermentum, Lactobacillus halotolerans, Lactobacillus pentosus, L. plantarum, Lactobacillus rhamnosus, Lactobacillus sake (26), and E. coli (23).
RAPD analysis was used in this study to classify 83 isolates into 62 distinct subtypes. RAPD analysis is inexpensive, efficient, and well suited for investigations incorporating large sample numbers (1). In the present study, isolates associated with pediatric feeding tubes were compared to determine the numerous subtypes capable of proliferating throughout the biofilm as well as proliferation of single organisms throughout multiple areas of PEG tubes. In addition, potentially common organisms were studied in an effort to link a general localized source of contamination leading to cross acquisition of species.
MATERIALS AND METHODS
Isolation and identification of biofilm microorganisms.
Low-profile PEG tubes, composed of silicone rubber, were collected from 18 pediatric patients from The Children's Hospital of Greenville Hospital System (Greenville, S.C.). Areas of the gastrostomy tubes, including the inner and outer portions of the internal stabilizer, the inner and outer portions of the shaft, and the valve, if present, were scraped with a sterile scalpel to remove the viable biofilm (6). The biofilm cells were placed in brain heart infusion broth (Difco, Detroit, Mich.) with 0.01 g of cycloheximide (Sigma, St. Louis, Mo.)/ml (BHI-C). Aliquots were also plated in duplicate on chocolate agar (Difco) plates containing 0.01 g of cycloheximide/ml. One set of tubes and plates was incubated under 5% CO2 for 1 week at 37°C, and the other set was incubated in an anaerobic Gas-Pak 100 jar (BBL Biological Systems, Cockeysville, Md.) for 1 week at 37°C. Colonies from chocolate agar plates containing 0.01 g of cycloheximide/ml were inoculated into BHI-C and incubated either aerobically or anaerobically as previously described. Tubes of BHI-C from the initial biofilm inoculation were diluted (10−5) in phosphate-buffered saline (pH 7.2) (Sigma), plated on brain heart infusion agar (Difco) plates containing 0.01 g of cycloheximide/ml, and incubated aerobically or anaerobically as previously described. Well-isolated, morphologically distinct colonies from brain heart infusion agar plates containing 0.01 g of cycloheximide/ml were inoculated into BHI/C and cultured as described above. Genus and species determinations were obtained with the BBL CRYSTAL identification systems (Becton Dickinson, Sparks, Md.). Gram-positive isolates were identified with a BBL CRYSTAL Gram-Positive identification kit (Becton Dickinson), while gram-negative isolates were identified with a BBL CRYSTAL Enteric/Nonfermenter identification kit (Becton Dickinson). Both types of panels were inoculated according to the manufacturer's specifications.
Chromosomal DNA preparation.
For chromosomal DNA preparations, a 1.5-ml sample of the desired strain was grown in brain heart infusion broth (Difco), pelleted with a microcentrifuge (Eppendorf model 5415C; Brinkmann Instruments, Westbury, N.Y.) at 14,000 rpm for 2 to 3 min, and extracted using methods described by Ulrich and Hughes (24). The preparations were analyzed on a 1% agarose gel containing 0.5 μg of ethidium bromide/ml.
RAPD analysis.
RAPD reactions were performed in duplicate using Ready-to-Go RAPD analysis beads (Amersham Pharmacia Biotech, Piscataway, N.J.). Multiple primers were tested for the capacity to amplify the target DNA from each isolate. A single primer (5"-GTTTCGCTCC-3") was chosen for its ability to produce a distinguishable RAPD profile with a limited number of amplicons visible under UV light. Reactions were performed by combining 2 μl (15 pmol/μl) of primer, 1 μl of template DNA (approximately 50 ng), and 22 μl of sterile distilled water with the reaction beads for a total volume of 25 μl. Amplification was performed in a Genius thermal cycler (Techne Ltd., Cambridge, United Kingdom) programmed for one cycle of 5 min at 95°C followed by 45 cycles of 1 min at 95°C, 1 min at 36°C, and 2 min at 72°C. The amplicons were analyzed on a 1.5% agarose gel containing 0.5 μg of ethidium bromide/ml at 200 V for 1 h and 30 min.
RAPD data analysis.
The RAPD patterns of individual strains were compared based on the index of similarity (IOS) between samples (2). The following formula was used for calculations: Fxy = 2nxy/(nx + ny), where nxy is the number of RAPD bands both samples share and nx and ny are the number of RAPD bands in each sample. The IOS provides a mathematical model to study genetic variation, especially with difficult or closely related RAPD patterns. Computational analysis of this type allows for direct comparisons without the need to count bands, which is especially important after loss of resolution resulting from manuscript duplication via photocopying.
RESULTS
Eighty-three isolates from 15 patients were classified into 62 subtypes as determined by unique banding patterns obtained from RAPD analysis and IOS. The patients ranged in age from 6 months to 17 years (mean, 93.4 months), and the implantation time ranged from 3 to 47 months (mean, 20.0 months). All RAPD reactions were performed in duplicate to determine the consistency and reproducibility of the RAPD methodology. No deviations in RAPD patterns between the duplicate reactions were encountered using the selected primer discussed in Materials and Methods. Isolate codes were based on organism classification, patient number, and location of the tube (IIS, inner portion of the internal stabilizer; OIS, outer portion of the internal stabilizer; IS, inner portion of the shaft; OS, outer portion of the shaft; and V, valve) and were designated with a number if multiple isolates were found in the same location. IOS values were determined by comparing two isolates and ranged from 0.000 (showing no similarity) to 1.000 (indicating identical subtypes).
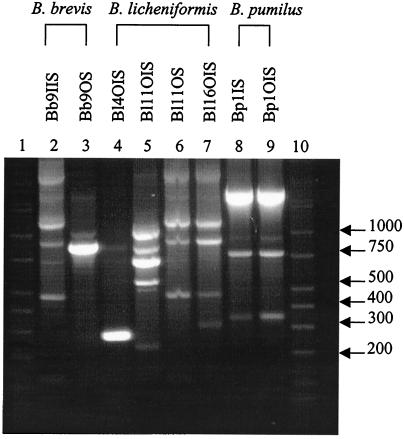
Two B. brevis (Bb), four B. licheniformis (Bl), and one B. pumilus (Bp) subtypes were discerned based on unique banding patterns (Fig. 1) and IOS analysis of each isolate. The IOS for the B. brevis isolates, Bb9IIS and Bb9OS, was 0.308. A comparison of B. licheniformis isolates can be seen in Table 1. None of the B. licheniformis isolates were genetically identical, and the largest IOS value was between isolates Bl11OS and Bl16OIS. The B. pumilus isolates, Bp1IS and Bp1OIS, had an IOS of 1.000.
FIG. 1.
RAPD fingerprints of Bacillus isolates. Lanes 1 and 10, DirectLoad wide-range DNA markers ranging from 50 to 10,000 bp (D7058; Sigma); lanes 2 to 9, B. brevis, B. licheniformis, and B. pumilus isolates.
TABLE 1.
IOS for B. licheniformis
| Isolate | IOS
|
||
|---|---|---|---|
| Bl11OIS | Bl11OS | Bl16OIS | |
| Bl4OIS | 0.600 | 0.222 | 0.000 |
| Bl11OIS | 0.000 | 0.000 | |
| Bl11OS | 0.800 | ||
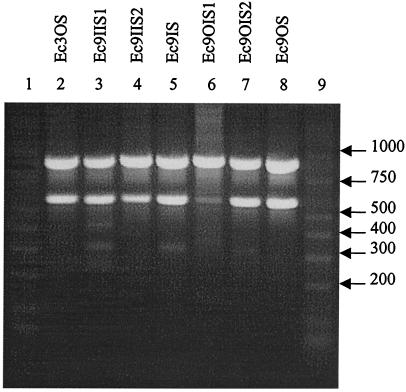
Three E. coli (Ec) types were also distinguished and categorized as seen in Fig. 2 and Table 2. Isolates Ec3OS, Ec9IIS2, Ec9OIS1, Ec9OIS2, and Ec9OS produced identical amplicon patterns and IOS values. The least related isolates based on amplified DNA were Ec9IIS1 and Ec9IS, with an IOS of 0.500.
FIG. 2.
RAPD fingerprints of E. coli isolates. Lanes 1 and 9, DirectLoad wide-range DNA markers ranging from 50 to 10,000 bp (D7058; Sigma); lanes 2 to 8, E. coli isolates.
TABLE 2.
IOS for E. coli
| Isolate | IOS
|
|||||
|---|---|---|---|---|---|---|
| Ec9IIS1 | Ec9IIS2 | Ec9IS | Ec9OIS1 | Ec9OIS2 | Ec9OS | |
| Ec3OS | 0.571 | 1.000 | 0.800 | 1.000 | 1.000 | 1.000 |
| Ec9IIS1 | 0.571 | 0.500 | 0.571 | 0.571 | 0.571 | |
| Ec9IIS2 | 0.800 | 1.000 | 1.000 | 1.000 | ||
| Ec9IS | 0.800 | 0.800 | 0.800 | |||
| Ec9OIS1 | 1.000 | 1.000 | ||||
| Ec9OIS2 | 1.000 | |||||
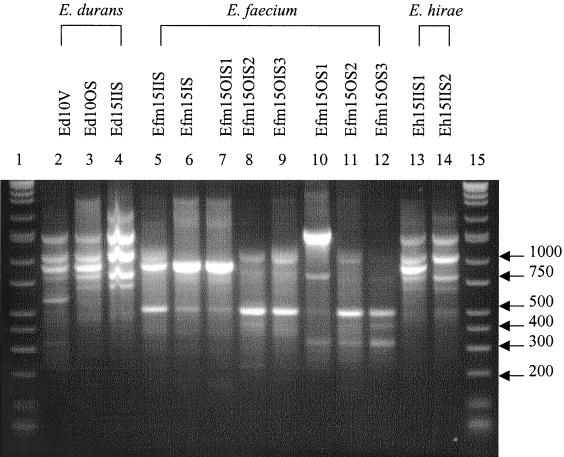
Three E. durans (Ed), seven E. faecium (Efm), and two E. hirae (Eh) subtypes were also grouped using RAPD profiles (Fig. 3) and IOS values. The IOS for the E. durans isolates Ed10V and Ed10OS was 0.727, that for Ed10V and Ed15IIS was 0.546, and that for Ed10OS and Ed15IIS was 0.800. E. faecium IOS results are reported in Table 3. Isolates Efm15IS and Efm15OIS1 produced homologous RAPD patterns. The IOS for the E. hirae isolates, Eh15IIS1 and Eh15IIS2, was 0.833.
FIG. 3.
RAPD fingerprints of Enterococcus isolates. Lanes 1 and 15, DirectLoad wide-range DNA markers ranging from 50 to 10,000 bp (D7058; Sigma); lanes 2 to 14, E. durans, E. faecium, and E. hirae isolates.
TABLE 3.
IOS for E. faecium
| Isolate | IOS
|
||||||
|---|---|---|---|---|---|---|---|
| Efm15IS | Efm15OIS1 | Efm15OIS2 | Efm15OIS3 | Efm15OS1 | Efm15OS2 | Efm15OS3 | |
| Efm15IIS | 0.667 | 0.667 | 0.600 | 0.750 | 0.500 | 0.400 | 0.444 |
| Efm15IS | 1.000 | 0.364 | 0.444 | 0.444 | 0.364 | 0.400 | |
| Efm15OIS1 | 0.182 | 0.222 | 0.444 | 0.546 | 0.400 | ||
| Efm15OIS2 | 0.800 | 0.400 | 0.333 | 0.546 | |||
| Efm15OIS3 | 0.500 | 0.400 | 0.667 | ||||
| Efm15OS1 | 0.600 | 0.667 | |||||
| Efm15OS2 | 0.909 | ||||||
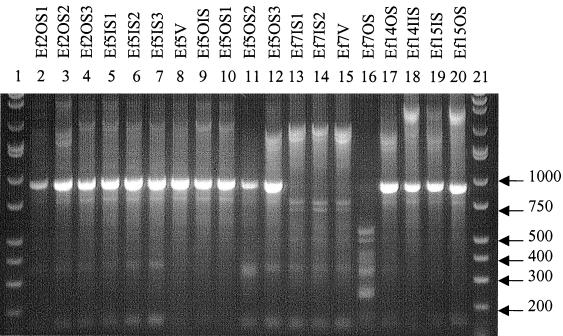
Twelve E. faecalis (Ef) types were differentiated and classified as reported in Fig. 4 and Table 4. Isolates Ef2OS2 and Ef5OS3 were identical in amplicon patterns and IOS values. Also, isolates Ef2OS3, Ef5IS1, Ef5IS2, and Ef5IS3; Ef5OIS and Ef5OS1; and Ef7IS1, Ef7IS2, and Ef7V were identical.
FIG. 4.
RAPD fingerprints of E. faecalis isolates. Lanes 1 and 21, DirectLoad wide-range DNA markers ranging from 50 to 10,000 bp (D7058; Sigma); lanes 2 to 20, E. faecalis isolates.
TABLE 4.
IOS for E. faecalis
| Isolate | IOS
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ef2OS2 | Ef2OS3 | Ef5IS1 | Ef5IS2 | Ef5IS3 | Ef5V | Ef5OIS | Ef5OS1 | Ef5OS2 | Ef5OS3 | Ef7IS1 | Ef7IS2 | Ef7V | Ef7OS | Ef14OS | Ef14IIS | Ef15IS | Ef15OS | |
| Ef2OS1 | 0.333 | 0.400 | 0.400 | 0.400 | 0.400 | 0.667 | 0.500 | 0.500 | 0.667 | 0.333 | 0.000 | 0.000 | 0.000 | 0.000 | 0.400 | 0.400 | 0.333 | 0.500 |
| Ef2OS2 | 0.667 | 0.667 | 0.667 | 0.667 | 0.571 | 0.500 | 0.500 | 0.571 | 1.000 | 0.800 | 0.800 | 0.800 | 0.200 | 0.889 | 0.667 | 0.800 | 0.500 | |
| Ef2OS3 | 1.000 | 1.000 | 1.000 | 0.667 | 0.857 | 0.857 | 0.667 | 0.667 | 0.667 | 0.667 | 0.667 | 0.222 | 0.500 | 0.500 | 0.444 | 0.571 | ||
| Ef5IS1 | 1.000 | 1.000 | 0.667 | 0.857 | 0.857 | 0.667 | 0.667 | 0.667 | 0.667 | 0.667 | 0.222 | 0.500 | 0.500 | 0.444 | 0.571 | |||
| Ef5IS2 | 1.000 | 0.667 | 0.857 | 0.857 | 0.667 | 0.667 | 0.667 | 0.667 | 0.667 | 0.222 | 0.500 | 0.500 | 0.444 | 0.571 | ||||
| Ef5IS3 | 0.667 | 0.857 | 0.857 | 0.667 | 0.667 | 0.667 | 0.667 | 0.667 | 0.222 | 0.500 | 0.500 | 0.444 | 0.571 | |||||
| Ef5V | 0.800 | 0.800 | 0.500 | 0.571 | 0.286 | 0.286 | 0.286 | 0.000 | 0.667 | 0.667 | 0.571 | 0.800 | ||||||
| Ef5OIS | 1.000 | 0.400 | 0.500 | 0.500 | 0.500 | 0.500 | 0.000 | 0.571 | 0.571 | 0.500 | 0.667 | |||||||
| Ef5OS1 | 0.400 | 0.500 | 0.500 | 0.500 | 0.500 | 0.000 | 0.571 | 0.571 | 0.500 | 0.667 | ||||||||
| Ef5OS2 | 0.571 | 0.286 | 0.286 | 0.286 | 0.286 | 0.333 | 0.333 | 0.286 | 0.400 | |||||||||
| Ef5OS3 | 0.600 | 0.600 | 0.600 | 0.200 | 0.889 | 0.667 | 0.600 | 0.500 | ||||||||||
| Ef7IS1 | 1.000 | 1.000 | 0.200 | 0.444 | 0.444 | 0.400 | 0.250 | |||||||||||
| Ef7IS2 | 1.000 | 0.200 | 0.444 | 0.444 | 0.400 | 0.250 | ||||||||||||
| Ef7V | 0.200 | 0.444 | 0.444 | 0.400 | 0.250 | |||||||||||||
| Ef7OS | 0.000 | 0.000 | 0.000 | 0.000 | ||||||||||||||
| Ef14OS | 0.750 | 0.889 | 0.571 | |||||||||||||||
| Ef14IIS | 0.667 | 0.857 | ||||||||||||||||
| Ef15IS | 0.750 | |||||||||||||||||
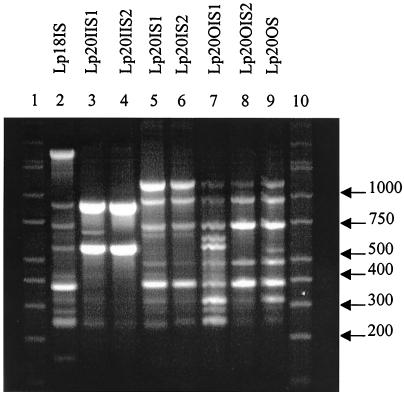
Eight L. plantarum (Lp) types were also distinguished (Fig. 5) and categorized (Table 5). None of the isolates had identical banding patterns; however, Lp20IS2 and Lp20OS had the highest IOS value of 0.875, while Lp20IIS2 and Lp20OS did not share any common amplicons.
FIG. 5.
RAPD fingerprints of L. plantarum isolates. Lanes 1 and 10, DirectLoad wide-range DNA markers ranging from 50 to 10,000 bp (D7058; Sigma); lanes 2 to 9, L. plantarum isolates.
TABLE 5.
IOS for L. plantarum
| Isolate | IOS
|
||||||
|---|---|---|---|---|---|---|---|
| Lp20IIS1 | Lp20IIS2 | Lp20IS1 | Lp20IS2 | Lp20OIS1 | Lp20OIS2 | Lp20OS | |
| Lp18IS | 0.667 | 0.615 | 0.444 | 0.471 | 0.700 | 0.400 | 0.588 |
| Lp20IIS1 | 0.800 | 0.533 | 0.571 | 0.471 | 0.333 | 0.286 | |
| Lp20IIS2 | 0.308 | 0.167 | 0.400 | 0.200 | 0.000 | ||
| Lp20IS1 | 0.824 | 0.700 | 0.800 | 0.706 | |||
| Lp20IS2 | 0.737 | 0.857 | 0.875 | ||||
| Lp20OIS1 | 0.706 | 0.737 | |||||
| Lp20OIS2 | 0.857 | ||||||
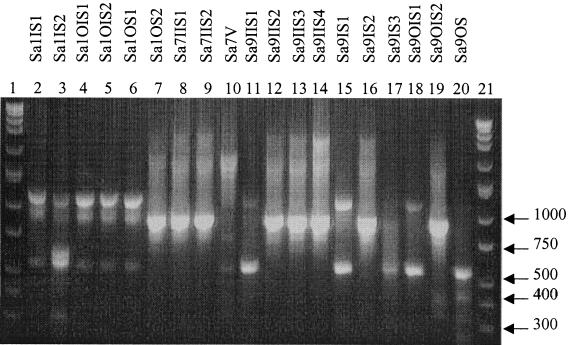
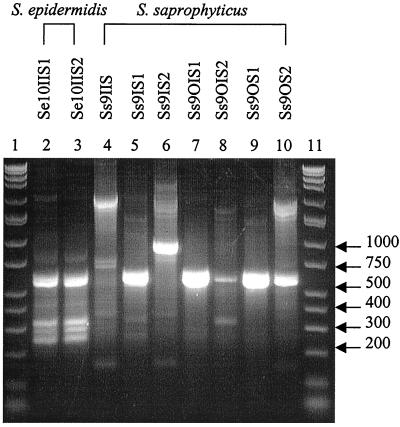
Twelve S. aureus (Sa) types were determined based on banding patterns (Fig. 6) and IOS (Table 6). The identical isolates were as follows: Sa1IS1, Sa1OIS1, Sa1OIS2, and Sa1OS1; Sa1OS2 and Sa9IIS2; Sa7IIS1 and Sa7IIS2; and Sa9IIS1, Sa9IS1, and Sa9OIS1. The two S. epidermidis species analyzed produced the same RAPD profile (Fig. 7). Seven S. saprophyticus types were classified and produced different RAPD fingerprints (Fig. 7) and IOS values (Table 7).
FIG. 6.
RAPD fingerprints of S. aureus isolates. Lanes 1 and 21, DirectLoad wide-range DNA markers ranging from 50 to 10,000 bp (D7058; Sigma); lanes 2 to 20, S. aureus isolates.
TABLE 6.
IOS for S. aureus
| Isolate | IOS
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sa1IS2 | Sa1OIS1 | Sa1OIS2 | Sa1OS1 | Sa1OS2 | Sa7IIS1 | Sa7IIS2 | Sa7V | Sa9IIS1 | Sa9IIS2 | Sa9IIS3 | Sa9IIS4 | Sa9IS1 | Sa9IS2 | Sa9IS3 | Sa9OIS1 | Sa9OIS2 | Sa9OS | |
| Sa1IS1 | 0.750 | 1.000 | 1.000 | 1.000 | 0.400 | 0.333 | 0.333 | 0.286 | 0.800 | 0.400 | 0.333 | 0.286 | 0.800 | 0.400 | 0.500 | 0.800 | 0.333 | 0.333 |
| Sa1IS2 | 0.750 | 0.750 | 0.750 | 0.286 | 0.250 | 0.250 | 0.222 | 0.571 | 0.286 | 0.250 | 0.222 | 0.571 | 0.286 | 0.333 | 0.571 | 0.250 | 0.500 | |
| Sa1OIS1 | 1.000 | 1.000 | 0.400 | 0.333 | 0.333 | 0.286 | 0.800 | 0.400 | 0.333 | 0.286 | 0.800 | 0.400 | 0.500 | 0.800 | 0.333 | 0.333 | ||
| Sa1OIS2 | 1.000 | 0.400 | 0.333 | 0.333 | 0.286 | 0.800 | 0.400 | 0.333 | 0.286 | 0.800 | 0.400 | 0.500 | 0.800 | 0.333 | 0.333 | |||
| Sa1OS1 | 0.400 | 0.333 | 0.333 | 0.286 | 0.800 | 0.400 | 0.333 | 0.286 | 0.800 | 0.400 | 0.500 | 0.800 | 0.333 | 0.333 | ||||
| Sa1OS2 | 0.800 | 0.800 | 0.333 | 0.000 | 1.000 | 0.800 | 0.667 | 0.000 | 0.500 | 0.000 | 0.000 | 0.400 | 0.000 | |||||
| Sa7IIS1 | 1.000 | 0.286 | 0.000 | 0.800 | 0.667 | 0.857 | 0.000 | 0.400 | 0.500 | 0.000 | 0.333 | 0.000 | ||||||
| Sa7IIS2 | 0.286 | 0.000 | 0.800 | 0.667 | 0.857 | 0.000 | 0.400 | 0.500 | 0.000 | 0.333 | 0.000 | |||||||
| Sa7V | 0.333 | 0.333 | 0.571 | 0.500 | 0.333 | 0.333 | 0.400 | 0.333 | 0.000 | 0.286 | ||||||||
| Sa9IIS1 | 0.000 | 0.000 | 0.000 | 1.000 | 0.000 | 0.667 | 1.000 | 0.000 | 0.400 | |||||||||
| Sa9IIS2 | 0.800 | 0.667 | 0.000 | 0.500 | 0.667 | 0.000 | 0.400 | 0.000 | ||||||||||
| Sa9IIS3 | 0.857 | 0.000 | 0.800 | 0.000 | 0.000 | 0.667 | 0.000 | |||||||||||
| Sa9IIS4 | 0.000 | 0.667 | 0.400 | 0.000 | 0.571 | 0.000 | ||||||||||||
| Sa9IS1 | 0.000 | 0.667 | 1.000 | 0.000 | 0.400 | |||||||||||||
| Sa9IS2 | 0.000 | 0.000 | 0.800 | 0.000 | ||||||||||||||
| Sa9IS3 | 0.667 | 0.000 | 0.500 | |||||||||||||||
| Sa9OIS1 | 0.000 | 0.400 | ||||||||||||||||
| Sa9OIS2 | 0.333 | |||||||||||||||||
FIG. 7.
RAPD fingerprints of Staphylococcus isolates. Lanes 1 and 11, DirectLoad wide-range DNA markers ranging from 50 to 10,000 bp (D7058; Sigma); lanes 2 to 10, S. epidermidis and S. saprophyticus isolates.
TABLE 7.
IOS for S. saprophyticus
| Isolate | IOS
|
|||||
|---|---|---|---|---|---|---|
| Ss9IS1 | Ss9IS2 | Ss9OIS1 | Ss9OIS2 | Ss9OS1 | Ss9OS2 | |
| Ss9IIS | 0.154 | 0.667 | 0.250 | 0.546 | 0.222 | 0.546 |
| Ss9IS1 | 0.143 | 0.286 | 0.600 | 0.500 | 0.400 | |
| Ss9IS2 | 0.000 | 0.333 | 0.000 | 0.500 | ||
| Ss9OIS1 | 0.400 | 0.667 | 0.400 | |||
| Ss9OIS2 | 0.667 | 0.750 | ||||
| Ss9OS1 | 0.667 | |||||
In several species, all isolates tested shared major amplicons. The two B. brevis isolates shared one band, the seven tested E. coli strains shared two amplicons, three common bands were found in the three E. durans isolates, one amplicon was shared by all eight E. faecium samples, and the two isolated E. hirae strains shared four bands.
Based on the RAPD fingerprints and the calculated IOS values, 62 distinct types were discerned among the 83 total isolates investigated in this study. Isolates with the same amplicon pattern were observed in multiple patients as follows: one E. coli type was seen in isolates Ec3OS, Ec9IIS2, Ec9OIS1, Ec9OIS2, and Ec9OS; one S. aureus profile was seen in isolates Sa1OS2 and Sa9IIS2; and two E. faecalis profiles were seen, one in strains Ef2OS2 and Ef5OS3 and the other in strains Ef2OS3, Ef5IS2, and Ef5IS3. Patients 3 and 9 had identical E. coli RAPD profiles and were seen in the hospital for tube removal 1 month and 25 days apart. RAPD profiles in patients 2 and 5 showed identical E. faecalis types, and these patients were seen 28 days apart for device replacement. The third pair, patients 1 and 9, seen 2 months and 20 days apart, shared the same S. aureus type.
DISCUSSION
It has been shown that RAPD fingerprints produced from arbitrarily primed PCR can be used to compare bacterial strains (27). In the present study, RAPD analysis was applied to clinical isolates for subtype differentiation and to possibly link common isolates to specific patients. The results showed that this methodology could be used to distinguish different subtypes based on the numerous fingerprints generated within a microbial genus and species group. Not only did the RAPD profiles allow for type distinction, the degree of relatedness between isolates was calculated based on band similarities. This was apparent with B. brevis, E. coli, E. durans, E. faecium, E. hirae, and S. epidermidis, all of which had at least one band in common within each species, indicating a potential species-specific probe for strain identification.
RAPD profiles from the genus Bacillus led to several interpretations. None of the isolates from the three species B. brevis, B. licheniformis, and B. pumilus showed identical RAPD profiles. The B. brevis isolates were purified from two specific locations on the same tube from one patient, the inner portion of the internal stabilizer and the outer portion of the shaft. A hypothesis, supported by several reports (1, 22), can be drawn that the isolates came from different environmental sources and were not from one organism proliferating through various locations of the tube. B. licheniformis was found in biofilms isolated from three different patients, and all of the RAPD profiles were different, leading to the hypothesis that each patient encountered different environmental sources of the contaminant that produced biofilm proliferation. Patient 11 had two different B. licheniformis types in two different areas of the tube, reinforcing the conclusion that multiple sources of bacterial contamination exist. In addition to these observations, the two B. pumilus isolates had identical RAPD profiles and were found on two areas of the same patient's device, implying a second characteristic related to biofilm proliferation. Based on this fact, it is also possible for microorganisms to spread to multiple areas of the tube from a single source.
Of the seven E. coli isolates, five had the same amplicon distribution but were not from the same patient. One was from patient 3, while the other four were from patient 9. A finding of this nature could show cross acquisition between patients via direct patient-to-patient interaction or contact with a common environmental source. Accessible records indicate that 1 month and 25 days elapsed between office visits and a different doctor cared for each patient. This information provides insight into a potential source, but further testing was restricted, so a definitive source linking the two could not be determined.
Comparisons of enterococcus profiles support the previously stated findings that biofilm formation occurs due to multiple sources as well as proliferation from a single contaminant. The two E. durans isolates Ed10V and Ed10OS were from the same patient's tube but different locations. Two E. faecium isolates from the inner portion of the shaft and the outer portion of the internal stabilizer from the same patient produced the same pattern by RAPD analysis, supporting the finding of biofilm proliferation from a single bacterium. All other isolates (Efm15IIS, Efm15OIS2, Efm15OIS3, Efm15OS1, Efm15OS2, and Efm15OS3) from the same patient were genetically different, showing that numerous microorganisms were involved in the formation of the biofilm. Two E. hirae isolates from the same location were genetically different, supporting the previously stated finding. E. faecalis was found in five patients, but two of them showed multiple RAPD types with an IOS of 1.000. Samples Ef2OS2 and Ef5OS3 were identical, further supporting the statement that cross acquisition between patients could occur. Interestingly, these two patients shared another subtype seen in Ef2OS3, Ef5IS2, and Ef5IS3.
L. plantarum was found in only two patients, but all eight of the isolates were genetically different based on RAPD profiles. Many of the samples were similar (Lp20IIS1 and Lp20IIS2, Lp20IS1 and Lp20IS2, and Lp20OIS2 and Lp20OS), but one or two bands that were present in one were absent in the other (Fig. 5), indicating similarity in the genetic template due to the amplicons produced in each sample while the missing bands show the isolates were not identical. Interestingly, Lp20IIS2 and Lp20OS did not share any amplicons.
Results from the genus Staphylococcus RAPD analysis supported all three findings stated so far: that multiple sources contribute to biofilm composition, proliferation of a specific organism occurs, and patient cross acquisition is possible. A single S. aureus type was found associated with two patients seen in the same office 2 months and 20 days apart, indicating the possibility of direct patient-to-patient contact or the presence of a common environmental source. In patient 1, 4 of the S. aureus isolates were identical, showing proliferation of one type over the surface of the device, as did 2 of the 3 isolates in patient 7 and 3 of the 10 S. aureus strains in patient 9. The remaining isolates of S. aureus as well as S. saprophyticus species were genetically different based on RAPD profiles.
RAPD profiles, using the primer described in Materials and Methods, produced a limited number of bands, which facilitated fingerprint analysis without the need for computer intervention. In addition, the relative prevalence of microbial colonization throughout various locations of the device and potential cross acquisition among patients was determined. Identical isolates, based on RAPD profiles, were found on multiple areas of the tube, suggesting the spread of the biofilm from the initial point of attachment. In addition, numerous subtypes were found associated with a single tube, leading to the observation that multiple bacterial subtypes are involved in the formation of gastrostomy tube-associated biofilms. Identical RAPD fingerprints for E. coli, E. faecalis, and S. aureus were identified in three patient pairs, suggesting a potential transfer of microorganisms from patient to patient via direct interaction or contact with a common source.
Investigations of large numbers of isolates by RAPD would benefit from computer-assisted discrimination by generating a database of patterns for the comparison of present and future isolates, including antibiotic sensitivity and resistance, isolate source, and patient-associated disease. Additional studies are using plasmid analysis to investigate the transfer of antibiotic resistance genes in order to link RAPD fingerprints to antibiotic resistance and sensitivity. This investigation supports previous reports that RAPD analysis is efficient, reproducible, and capable of detecting genomic polymorphisms among various microbial species without previous knowledge of the nucleotide sequence on the target DNA (22), and the technique has been shown to be valuable in studies dealing with biofilm formation on enteral access tubes. RAPD technology is an inexpensive way to type organisms without specialized equipment not readily available to general molecular microbiology laboratories. Although RAPD is sensitive to annealing temperatures, reproducibility has been achieved previously as well as in this study (22, 25). RAPD is also preferable for gram-positive species, where chromosomal preparations are difficult and low concentrations of nucleic acid are achieved. RAPD typing has proved to be efficient and cost-effective while maintaining reproducible and accurate results for analyzing large numbers of gram-positive organisms.
REFERENCES
- 1.Campbell, M., E. Mahenthiralingam, and D. P. Speert. 2000. Evaluation of random amplified polymorphic DNA typing of Pseudomonas aeruginosa. J. Clin. Microbiol. 38:4614-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chansiripornchai, N., P. Ramasoota, A. Bangtrakulnonth, J. Sasipreeyajan, and S. B. Svenson. 2000. Application of randomly amplified polymorphic DNA (RAPD) analysis for typing avian Salmonella enterica subsp. enterica. FEMS Immunol. Med. Microbiol. 29:221-225. [DOI] [PubMed] [Google Scholar]
- 3.Clark, A. G., and C. M. S. Lanigan. 1993. Prospects for estimating nucleotide divergence with RAPDs. Mol. Biol. Evol. 10:1096-1111. [DOI] [PubMed] [Google Scholar]
- 4.Costerton, J. W. 1992. The pivotal role of biofilms in the focused attack of bacteria on soluble substrates. Int. Biodeterior. Biodegradation 30:123-133. [Google Scholar]
- 5.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilm: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 6.Dautle, M. P. 1999. Isolation and identification of biofilm microorganisms from silicone gastrostomy devices. M.S. thesis. Clemson University, Clemson, S.C. [DOI] [PubMed]
- 7.Davies, J. S., and D. W. Westlake. 1979. Crude oil utilization by fungi. Can. J. Microbiol. 25:146-156. [DOI] [PubMed] [Google Scholar]
- 8.Denamur, E., B. Picard, P. Goullet, E. Bingen, N. Lambert, and J. Elion. 1991. Complexity of Pseudomonas aeruginosa infection in cystic fibrosis: combined results from esterase electrophoresis and rDNA restriction fragment length polymorphism analysis. Epidemiol. Infect. 106:531-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elaichouni, A., A. Vershraegen, G. Claeys, M. Devleeschouwer, C. Godard, and M. Vaneechoute. 1994. Pseudomonas aeruginosa serotpye O12 outbreak studied by arbitrary primer PCR. J. Clin. Microbiol. 32:666-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gauderer, M. 1999. Twenty years of percutaneous endoscopic gastrostomy: origin and evolution of a concept and its expanded applications. Gastrointest. Endosc. 50:879-883. [DOI] [PubMed] [Google Scholar]
- 11.Gauderer, M. W. L., R. S. Abrams, and J. H. Hammond. 1998. Initial experience with the changeable skin-level port-valve: a new concept for long-term gastrointestinal access. J. Pediatr. Surg. 33:73-75. [DOI] [PubMed] [Google Scholar]
- 12.Gilmour, M. N., T. S. Whittam, M. Kilian, and R. K. Selander. 1987. Genetic relationships among the oral streptococci. J. Bacteriol. 169:5247-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottlieb, K., M. DeMeo, and P. Borton. 1992. Gastrostomy tube deterioration and fungal colonization. Am. J. Gastroenterol. 87:1683.. [PubMed] [Google Scholar]
- 14.Gottlieb, K., J. Leya, P. M. Kruss, S. Mobarhan, and F. L. Iber. 1993. Intraluminal fungal colonization of gastrostomy tubes. Gastrointest. Endosc. 39:413-415. [DOI] [PubMed] [Google Scholar]
- 15.Grothues, D., V. Koopmann, H. van der Hardt, and B. Tümmler. 1988. Genome fingerprinting of Pseudomonas aeruginosa indicates colonization of cystic fibrosis siblings with closely related strains. J. Clin. Microbiol. 26:1973-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hull, M. A., J. Rawlings, F. E. Murray, J. Field, A. S. McIntyre, and Y. R. Mahida. 1993. Audit of outcome of long-term enteral nutrition by percutaneous endoscopic gastrostomy. Lancet 341:869-872. [DOI] [PubMed] [Google Scholar]
- 17.Klug, M. J., and A. J. Markovetz. 1971. Utilization of aliphatic hydrocarbons by microorganisms. Adv. Microb. Physiol. 5:1-43. [DOI] [PubMed] [Google Scholar]
- 18.Kulkarni, G. V., K. H. Chan, and H. J. Sandham. 1989. Investigation into the use of restriction endonuclease analysis for the study of transmission of mutans streptococci. J. Dent. Res. 68:1155-1161. [DOI] [PubMed] [Google Scholar]
- 19.Marcuard, S. P., J. L. Finley, and K. G. MacDonald. 1993. Large-bore feeding tube occlusion by yeast colonies. J. Parenter. Enteral Nutr. 17:187-190. [DOI] [PubMed] [Google Scholar]
- 20.Mileham, A. J. 1997. Identification of microorganisms using random primed PCR. Mol. Biotechnol. 8:139-145. [DOI] [PubMed] [Google Scholar]
- 21.Speijer, H., P. H. M. Sauelkoul, M. J. Bonten, E. E. Stobberingh, and J. H. T. Tjhie. 1999. Application of different genotyping methods for Pseudomonas aeruginosa in a setting of endemicity in an intensive care unit. J. Clin. Microbiol. 37:3654-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Truong, T. L., C. Menard, C. Mouton, and L. Trahan. 2000. Identification of mutans and other oral streptococci by random amplified polymorphic DNA analysis. J. Med. Microbiol. 49:63-71. [DOI] [PubMed] [Google Scholar]
- 23.Tseng, C., E. Ting, D. Johnson, M. Saluta, and R. Dunst. 2001. Use of RAPD fingerprinting for differentiating E. coli isolates from human and animal sources. Life Sci. News 7:10-11. [Google Scholar]
- 24.Ulrich, R. L., and T. A. Hughes. 2000. A rapid procedure for the isolation of chromosomal DNA from Gram-positive bacteria. Lett. Appl. Microbiol. 32:52-56. [DOI] [PubMed] [Google Scholar]
- 25.Van Leeuwen, W., M. Sijmons, J. Sluijs, H. Verbrugh, and A. van Belkum. 1996. On the nature and use of randomly amplified DNA from Staphylococcus aureus. J. Clin. Microbiol. 34:2770-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veyrat, A., M. C. Miralles, and G. Pérez-Martinez. 1999. A fast method for monitoring the colonization rate of lactobacilli in a meat model system. J. Appl. Microbiol. 87:49-61. [DOI] [PubMed] [Google Scholar]
- 27.Welsh, J., and M. McClelland. 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 18:7213-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]