Abstract
In a screen for gene copy-number changes in mouse mammary tumors, we identified a tumor with a small 350-kb amplicon from a region that is syntenic to a much larger locus amplified in human cancers at chromosome 11q22. The mouse amplicon contains only one known gene, Yap, encoding the mammalian ortholog of Drosophila Yorkie (Yki), a downstream effector of the Hippo(Hpo)–Salvador(Sav)–Warts(Wts) signaling cascade, recently identified in flies as a critical regulator of cellular proliferation and apoptosis. In nontransformed mammary epithelial cells, overexpression of human YAP induces epithelial-to-mesenchymal transition, suppression of apoptosis, growth factor-independent proliferation, and anchorage-independent growth in soft agar. Together, these observations point to a potential oncogenic role for YAP in 11q22-amplified human cancers, and they suggest that this highly conserved signaling pathway identified in Drosophila regulates both cellular proliferation and apoptosis in mammalian epithelial cells.
Keywords: breast, mammary, transformation, Yorkie
Genomewide analysis of tumors for gene copy gains and losses by using array comparative genomic hybridization (array CGH) enables a detailed characterization of loci implicated in tumorigenesis (1). Whereas human cancers frequently show extensive chromosomal instability, mouse tumor models may provide a more stable baseline from which to dissect essential tumor-related alterations. This approach may be particularly powerful when used to search for somatically acquired genetic lesions in the background of Brca1/Trp-53 inactivation, a genotype associated with somatic oncogene amplification. We have recently shown that as many as 73% of mouse Brca1/Trp-53-driven mammary tumors have amplification of the gene encoding the Met protein, pointing to gross overexpression of this growth factor receptor as a common secondary event in tumors with this genetic background (2). In analyzing these mammary tumors, we also observed a tumor with a selective amplification of a small region of mouse chromosome 9, syntenic with the 11q22 amplicon commonly observed in human cancers (3–11).
Amplification of 11q22 is evident in glioblastomas; oral squamous-cell carcinomas; and in cancers of the pancreas, lung, ovary, and cervix (3–11). The human amplicon tends to be large [0.7–2.6 megabases (Mb)], including a cluster of matrix metalloproteinase (MMP) genes, two members of the BIRC family (BIRC2 and BIRC3, also known as the cIAP family), and YAP (3–5, 8, 10). Most analyses of this amplicon have focused on the role of BIRC (cIAP) proteins, whose antiapoptotic functions are well described (12). The possible contribution of the YAP gene in driving this amplicon has not been explored.
The YAP protein was initially isolated by virtue of its binding to the Src family member nonreceptor tyrosine kinase YES (Yes kinase-associated protein) (13). Additional YAP-interacting proteins have been described more recently, including a number of transcription factors [phosphatidylethanolamine-binding protein 2α (PEBP2α), p73, and TEA domain/transcription enhancer factor (TEAD/TEF) family members], with which YAP acts as a transcriptional coregulator (14–16). The Drosophila ortholog of YAP, Yorkie (Yki), also functions as a transcriptional coactivator, and it was recently described as a target of the Hippo(Hpo)–Salvador(Sav)–Warts(Wts) pathway that negatively regulates growth by simultaneously inhibiting proliferation and promoting apoptosis (17, 18). Yki activates proliferation by inducing the expression of cyclin E and inhibits apoptosis by induction of the caspase-inhibitor protein DIAP1 (Drosophila inhibitor of apoptosis) (17). The upstream Hpo–Wts kinase cascade negatively regulates these activities (18–25).
In this study, we report that a mouse tumor-derived amplicon defined by high-density array CGH excludes the MMP and BIRC (cIAP) genes, pointing to YAP as a critical gene-amplification “driver.” Overexpression of YAP in human nontransformed mammary epithelial cells results in phenotypic alterations that are hallmarks of tumorigenic transformation, including epithelial-to-mesenchymal transition (EMT), suppression of apoptosis, growth factor-independent proliferation, and anchorage-independent growth in soft agar. Collectively, these findings suggest that YAP contributes to malignant transformation in cancers harboring the 11q22 amplicon, and they support the potential significance of this pathway in human cancer.
Results
Mapping of the Yap-Containing Amplicon.
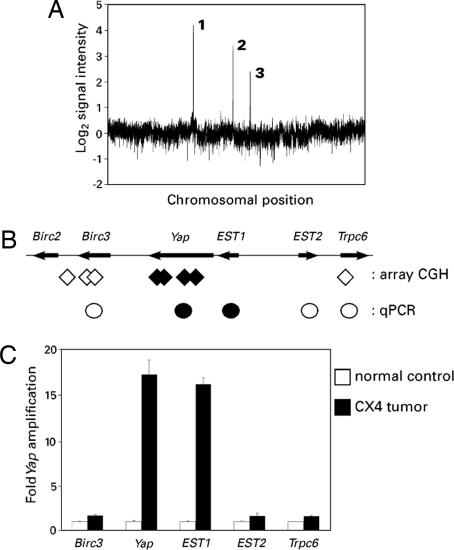
Mammary tumors arising in mice with a tissue-specific knockout of Brca1, engineered on a Trp-53-heterozygous background (Brca1Δ11/co Trp-53+/− MMTV-Cre) (26), were subjected to whole-genome array CGH analysis for gene copy-number alterations. One of 15 tumors analyzed, CX4, harbored three distinct high-level amplifications (Fig. 1A). The first was centered on the Met protooncogene, a recurrent and specific genetic abnormality that is present in the majority of Brca1/Trp-53-driven mouse mammary tumors (2). Whereas the amplification on chromosome 10 encompassed a region of >4 Mb with a large number of genes, the amplification on chromosome 9 was centered on a single known gene, Yap (Fig. 1B). The Yap amplicon was of particular interest because amplification of the syntenic locus on human chromosome 11q22 is found in diverse cancers, but the large size of the human amplicons has precluded identification of the key oncogene(s) driving this amplification. In contrast, the CX4 tumor amplicon was small (350 kb) and restricted to Yap and a neighboring uncharacterized EST. The array CGH data were confirmed by using real-time quantitative PCR (qPCR), precisely defining the boundaries of the amplicon (Fig. 1 B and C).
Fig. 1.
Yap is a candidate “driver” gene in the mouse chromosome 9 amplicon. (A) Whole-genome profile of an individual tumor (CX4) showing ratio of tumor DNA signal vs. normal DNA control from the same mouse; x-axis coordinates represent oligonucleotide probes ordered by genomic map position, with the whole-genome filtered median (three nearest neighbors) data set plotted. High-level amplifications are labeled 1–3 and correspond to: 1, 2.6-Mb amplicon on chromosome 6, centering on Met (2); 2, 350-kb amplicon on chromosome 9, centering on Yap; 3, 4.6-Mb amplicon on chromosome 10, containing a large number of genes. (B) Mouse chromosome 9 amplicon. Filled diamonds denote the positions of probes detecting genomic amplification in CX4. Open diamonds indicate the positions of closest-neighbor probes detecting normal DNA copy number. Circles represent data from C. Filled circles denote amplification, and open circles indicate normal DNA content. (C) Independent confirmation of the amplicon boundaries by using qPCR.
Induction of EMT in Mammary Epithelial Cells.
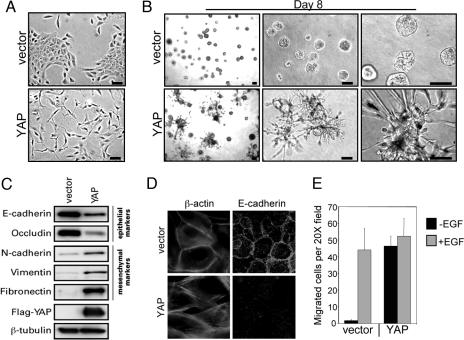
To examine the function of YAP in mammalian cells, we introduced this gene by retroviral infection into the immortalized, but nontumorigenic, human mammary epithelial cell line MCF10A. We have previously used this cell line in a three-dimensional culture model to investigate the biological activities of known and candidate oncogenes within an architecture that mimics mammary acini in vivo (27, 28). To avoid clonal-selection effects, all experiments were performed with short-term cultures of drug-selected but uncloned pools of cells, stably expressing YAP (MCF10A-YAP). Whereas control MCF10A cells grow in epithelial-type islands on monolayer cultures, cells overexpressing YAP displayed a loss of cell–cell contacts and cell scattering (Fig. 2A). YAP expression also disrupted the morphogenesis of MCF10A cells in three-dimensional cultures of reconstituted basement membrane (Matrigel). MCF10A-YAP cells failed to form spherical acinar-like structures similar to the vector control cells (Fig. 2B). Instead, these cells formed structures characterized by spike-like projections and cords of cells that invaded the basement-membrane gel. This invasive phenotype was evident as early as day 4, and it was detectable in ≈50% of the structures by day 8 (Fig. 2B). These morphological changes in monolayer and three-dimensional cultures, i.e., a spindled morphology with cell scattering, and invasion in Matrigel, suggested that MCF10A-YAP cells had undergone EMT. EMT was evaluated by examining the expression patterns of epithelial and mesenchymal markers. The mesenchymal markers fibronectin, vimentin, and N-cadherin were up-regulated, and the epithelial markers E-cadherin and occludin were down-regulated in MCF10A-YAP cells, as demonstrated by the immunoblotting analysis in Fig. 2C. MCF10A-YAP cells also displayed disorganization of adherens junctions, another hallmark of EMT, as shown by immunofluorescence analyses of E-cadherin and actin localization (Fig. 2D). Finally, there was a 20- to 30-fold increase in the migration of MCF10A-YAP cells compared with control cells in Transwell assays (Fig. 2E). Interestingly, the increased migration was evident only in the absence of EGF. Collectively, these morphological, biochemical, and cell-biological observations suggest that YAP was able to induce EMT in MCF10A cells.
Fig. 2.
YAP induces an EMT. (A) YAP induces a morphology change on monolayer cultures. Representative phase-contrast images of MCF10A-YAP and vector control cells growing in monolayer cultures are shown. (Scale bars, 100 μm.) (B) YAP induces an invasive three-dimensional morphology. MCF10A-YAP and vector control cells were cultured on Matrigel for 8 days. Representative phase-contrast images are shown, increasing in magnification from left to right. (Scale bars, 100 μm.) (C) Expression of YAP results in loss of epithelial markers and gain of mesenchymal markers. Immunoblotting analysis reveals a decrease in E-cadherin and occludin (epithelial markers) and concomitant increase in N-cadherin, vimentin, and fibronectin (mesenchymal markers). β-Tubulin is used to show equal loading. (D) YAP overexpression results in loss of membrane E-cadherin and cortical actin. Immunofluorescence analysis shows loss of plasma membrane E-cadherin (magnification, ×50) and loss of localization of actin (stained with phalloidin; magnification, ×100) at cortical sites adjacent to cell–cell interfaces in MCF10A-YAP cells. (E) YAP induces Transwell migration. Control and YAP-expressing MCF10A cells were plated onto 8-μm Transwell filters and allowed to migrate for 24 h either in the presence (+EGF) or in the absence (−EGF) of EGF. Data are the mean number of migrated cells per ×20 field of four fields. Error bars equal ±SD of three independent experiments.
YAP Overexpression Induces a Proliferative Advantage.
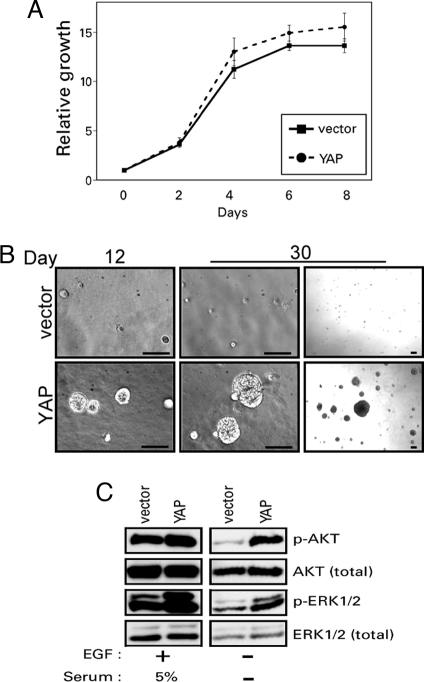
Overexpression of the Drosophila YAP ortholog yki causes an overgrowth phenotype resulting from both the activation of proliferation and the inhibition of cell death (17). Because YAP disrupted morphogenesis of MCF10A cells and induced highly invasive three-dimensional structures, we were unable to evaluate the effects of YAP expression on proliferation of outer cells or cell death of the center cells of acini in this model. To investigate further the biological activities of YAP in mammalian cells, we examined MCF10A-YAP cells more directly for proliferative and antiapoptotic phenotypes in other assays. To assess the effects of YAP on cell proliferation, we took advantage of the stringent requirement of MCF10A cells for EGF to support proliferation, and we assayed MCF10A-YAP cells in both the presence and absence of this growth factor. MCF10A-YAP cells did not display an increased rate of proliferation in monolayer cultures in the presence of EGF (Fig. 3A). However, these cells were able to proliferate three-dimensionally in the absence of EGF, in contrast to vector control cells, which failed to proliferate under these conditions (Fig. 3B). By 12 days in culture, MCF10A-YAP cells had formed three-dimensional structures in the absence of EGF that continued to grow larger until the assay was stopped at day 30. Approximately 30% of the total input of MCF10A-YAP cells were able to form structures after 30 days in culture, whereas no control cells were able to proliferate in this assay. Interestingly, these EGF-independent three-dimensional structures did not display the invasive morphology that was observed in the presence of EGF (Fig. 2B), suggesting that EGF is required for the YAP-induced invasive activity.
Fig. 3.
YAP overexpression promotes proliferation. (A) YAP overexpression does not affect growth rate in the presence of EGF. MCF10A-vector cells were grown in parallel to MCF10A-YAP cells, and their growth was assessed over an 8-day time course. (B) YAP overexpression activates EGF-independent growth. MCF10A-YAP cells and vector control cells were grown on Matrigel in medium without EGF for 30 days. Representative phase-contrast images are shown from one of three independent experiments on day 12 on the Left, and both a high-power (Center) and low-power (Right) magnification of day 30. (Scale bars, 100 μm.) (C) YAP overexpression results in activation of ERK1/2 and AKT pathways. Immunoblotting analysis with antibodies to phosphorylated ERK1/2 (Thr-202/Tyr-204) and AKT (Ser-473) shows increased activation in MCF10A-YAP cells in the absence of EGF and serum.
To gain insight into the mechanism responsible for the ability of MCF10A-YAP cells to proliferate in the absence of EGF, we examined whether YAP expression could activate signaling through either ERK or AKT, two of the major signaling pathways that can contribute to EGF-independent growth of MCF10A cells (29), by immunoblotting with activation-sensitive, phospho-specific antibodies. Whereas exogenous growth factors were required for activation of ERK and AKT in vector control cells, both of these proteins displayed strong activation in the absence of growth factors in MCF10A-YAP cells (Fig. 3C). Thus, the activation of AKT and ERK could contribute to the ability of YAP to promote proliferation of MCF10A cells in the absence of EGF.
Inhibition of Apoptosis by YAP Overexpression.
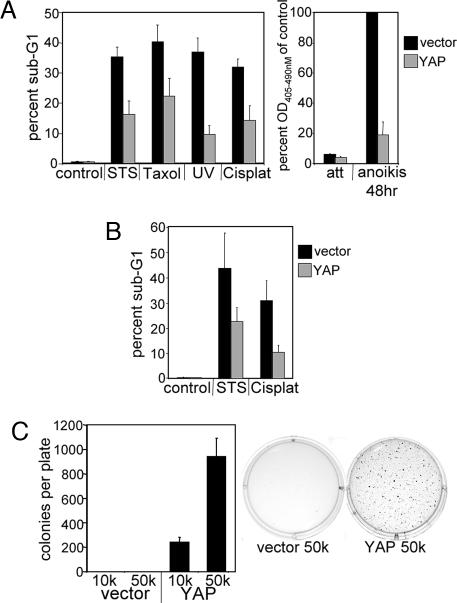
Although yki expression inhibits apoptosis in Drosophila (17), previous reports of YAP function in mammalian cells indicate that expression of this gene activates apoptosis in several tumor cell lines (30, 31). To assess the effect of YAP expression on apoptosis in MCF10A cells, we exposed MCF10A-YAP and control cells to a variety of apoptosis-inducing stresses, including the chemotherapeutic agents Taxol (paclitaxel) and cisplatin, the pan-kinase inhibitor staurosporine (STS), UV irradiation, and loss of matrix attachment (anoikis). Surprisingly, expression of YAP conferred protection from apoptosis induced by each of these stresses as measured by DNA fragmentation (Fig. 4A). Thus, in contrast to reports of YAP function in tumor cell lines, overexpression of YAP broadly inhibits cell death in MCF10A cells. To evaluate whether the effects YAP were specific to MCF10A cells, we examined YAP-induced effects on apoptosis in another immortalized, but nontumorigenic cell line, HMECtert. Immortalized human mammary epithelial cells (HMEC) were generated by infection with a retrovirus encoding the catalytic subunit of telomerase (hTert) (32). YAP was subsequently overexpressed in these cells by retroviral infection, and the stable cell pool (HMECtert-YAP) was assayed for apoptosis after exposure to STS and cisplatin. As with MCF10A cells, YAP expression in HMEC conferred resistance to cell death induced by both apoptotic inducers (Fig. 4B). Thus, in contrast to the proapoptotic effect of YAP reported in some cancer cell lines (30, 31), YAP displayed antiapoptotic activity in two nontransformed epithelial cell lines.
Fig. 4.
YAP overexpression inhibits apoptosis and transforms MCF10A cells. (A) YAP overexpression inhibits apoptosis in MCF10A cells. Control and MCF10A-YAP monolayer cells were treated with STS, Taxol, UV light, or cisplatin (Cisplat), or they were detached from matrix (anoikis) to induce apoptosis. (A Left) Data are the mean percentages of sub-G1 DNA content cells determined by flow-cytometric analysis of propidium iodide-stained samples collected after the indicated treatments. (A Right) Cell-death data were quantified by using the cell-death detection ELISA kit, which measures DNA fragmentation; the data are the mean absorbance readings at 405–490 nm relative to vector control cells after 48 h of anoikis. att, attached control monolayer cells. All error bars equal ±SD of three independent experiments. (B) YAP overexpression inhibits apoptosis in HMEC. HMECtert-vector cells and HMECtert-YAP cells were treated with STS and cisplatin to induce apoptosis. Data are the mean percentages of the sub-G1 content cells determined by flow cytometry as in A. Error bars equal ±SD of three independent experiments. (C) YAP overexpression induces anchorage-independent growth in soft agar. MCF10A-YAP cells and vector control cells were plated in soft-agar assays and allowed to grow for 21 days. (Left) Data are the mean number of colonies per six-well culture of either 10,000 cells (10k) or 50,000 cells (50k). Error bars equal ±SD of three independent experiments. (Right) Representative wells stained with 0.02% iodonitrotetrazolium chloride are shown.
YAP Induction of Colony Formation in Soft Agar.
To evaluate a more stringent parameter of oncogenic transformation, we examined the effect of YAP on the ability of MCF10A cells to form colonies in soft agar, a property that frequently correlates with tumorigenicity. As expected, MCF10A-vector control cells failed to produce anchorage-independent colonies in soft agar. In marked contrast, MCF10A-YAP cells formed large colonies after 3 weeks in soft agar (Fig. 4C), demonstrating that YAP is able to induce a fully transformed phenotype.
Discussion
In this work, we demonstrate that overexpression of YAP in MCF10A cells induces phenotypic alterations that are commonly associated with potent transforming oncogenes, that is, induction of anchorage-independent growth, EMT, growth factor-independent proliferation and activation of AKT and ERK, and inhibition of apoptosis. Notably, most other oncogenes that display activities similar to YAP are typically constitutively activated mutant variants of cellular proteins, such as the smGTPase H-Ras (33), the tyrosine kinase Src (34, 35), and phosphatidylinositol 3-kinase (36–39). Thus, the ability of wild-type YAP to induce transformation of immortalized mammary epithelial cells by mere overexpression indicates that this gene has potent oncogenic potential. This oncogenic activity of YAP in mammalian cells is consistent with the described functions of the Drosophila YAP ortholog yki, whose overexpression causes an overgrowth phenotype resulting from both increased proliferation and reduced cell death (17). In parallel studies, Lowe and coworkers (40) have also demonstrated oncogenic activity for Yap in a mouse model of hepatocellular carcinoma where Yap amplification contributes to the development of tumors.
In Drosophila, the activity of Yki is negatively regulated by an upstream kinase cascade in which the Hpo kinase, together with its binding partner Sav, activates the Wts kinase–Mats complex, which, in turn, inactivates Yki. Members of this upstream pathway were identified before yki in genetic screens for inhibitors of cell growth in the Drosophila eye and wing (18–25, 41, 42). The ability of human YAP, like yki, to rescue pupal lethality induced by overexpression of hpo and wts in Drosophila had previously suggested that the growth-promoting functions of yki are conserved in the human YAP ortholog (17). The YAP-induced phenotypes described here in mammalian cells support the notion that YAP, like Yki, can both activate proliferation and inhibit apoptosis. The combination of these YAP-driven phenotypes is sufficient to transform the nontumorigenic human epithelial cell line MCF10A.
Similar to the conservation between yki and YAP, the human orthologs of wts, hpo, and mats can rescue their corresponding Drosophila mutants, suggesting that the entire upstream Yki-regulating pathway might be conserved in mammalian cells (18, 22, 43). In support of this hypothesis, the human Hpo ortholog, MST2, can phosphorylate and activate the human Wts orthologs LATS1 and LATS2 (44). Hints that this pathway might be tumor-suppressive in mammalian cells have also been reported, including a tumor-predisposition phenotype (soft-tissue sarcomas and ovarian tumors) in mice lacking one of the two wts orthologs, Lats1 (45), and suppression of RasV12-driven transformation of NIH 3T3 cells by the second wts ortholog, Lats2 (46). Furthermore, the Hpo–Sav–Wts–Yki pathway in Drosophila was recently reported to lie downstream of signaling from Merlin (47), which is the product of the NF2 tumor-suppressor gene that is mutated in humans with neurofibromatosis type II (48, 49). Whether any of these tumor-suppressor functions in mammalian cells could be mediated by the inhibition of YAP activity is not known. In screening for intragenic mutations in human cancer-derived cell lines, we detected homozygous deletions in SAV in two renal cancer cell lines (24), suggesting that this pathway could be targeted by the inactivation of upstream regulators in addition to amplification of YAP.
In Drosophila, Yki functions as a transcriptional coregulator that activates proliferation and inhibits apoptosis by increasing the expression of the cyclin E gene and diap1 (17). We examined the expression levels of cyclin E, cIAP1, and cIAP2 proteins in MCF10A-YAP cells, and we found them to be similar to the expression levels in control cells (data not shown). Thus, although YAP appears to promote phenotypes in mammalian cells similar to those promoted by Yki in Drosophila, the mechanism of YAP action may be different. Previous studies of YAP in mammalian cells have uncovered a variety of seemingly nonoverlapping functions. Since its discovery as a YES-binding protein (13), other cytoplasmic functions for YAP have been revealed, including the recruitment of Smad7 to TGF-β receptor I (50), as well as nuclear functions, where YAP is a binding partner and cotranscriptional regulator of a variety of transcription factors, including PEBP2α, TEAD/TEF, and p73 family proteins (14–16). Whether any of these reported YAP functions or binding partners might contribute to the phenotypes reported here remains to be determined. The description of YAP acting as an inhibitor of apoptosis is in direct contrast to previous reports in which YAP was an activator of cell death in mammalian cells. YAP was previously shown to activate apoptosis in response to DNA damage by interacting with p73 in several cancer cell lines (30, 31). Suppressive interactions between p73 and endogenously high levels of p63 isoforms expressed in the nontransformed cells used here may have modulated the observed effects (51).
Finally, although these data predict a role for YAP in human cancer, this role remains to be clearly defined. As discussed above, amplification of the 11q22 chromosomal locus including YAP is observed in multiple cancer types. The Brca1/Trp-53-driven mouse mammary tumor with selective amplification of YAP led us to focus on this proposed oncogene by using mammary epithelial transformation assays. However, using qPCR analysis of microscopically dissected specimens, we did not detect YAP amplification in >100 sporadic human breast cancers (data not shown). Thus, it is possible that the physiological significance of YAP amplification may be more relevant for other cancers that are more commonly known to have amplification of the 11q22 locus, such as oral squamous-cell carcinomas, where it is present in 5–15% of primary tumors (3, 10). In addition, although the specific YAP amplification in the mouse model emphasized the unique contribution of this gene to tumorigenesis, the larger size of the common human amplicon points to additional genes that may jointly contribute to malignancy. For example, in some contexts the BIRC2 and BIRC3 genes encoding the apoptotic inhibitors cIAP1 and cIAP2 might also be relevant targets of this amplicon. This indeed appears to be the case in parallel work from a hepatocellular carcinoma model, where the Yap and cIAP1 genes are coamplified and jointly contribute to the development of tumors in mice (40). Together, our studies point to potent oncogenic effects of YAP, a component of a highly conserved pathway regulating proliferation and apoptosis in Drosophila. The precise mechanisms underlying the oncogenic effects of YAP itself and the cellular contexts in which it can contribute to malignancy remain to be defined.
Materials and Methods
Mammary Tumor Analysis.
Experimental mice (26) and array CGH screening of tumors (2) have been previously described.
qPCR.
The sequences of the PCR primer pairs and fluorogenic MGB probes (Applied Biosystems, Foster City, CA) (all listed from 5′ to 3′) used for DNA copy number analyses were: Mm.Yap_F, CCTATGACCTCGCAGCATTCT; Mm.Yap_R, GGAAACCTCCTCCCGTGTCT; Mm.Yap_probe, VIC-CCCCAGGGTCCACTC-MGBNFQ; Mm.Birc3_F, CCCCTGAGCCTTCCAACA; Mm.Birc3_R, ATTGCACAAAATTGAGGGCTTT; Mm.Birc3_probe, VIC-ACAGCAGATTTTAAACACTT-MGBNFQ; Mm.EST1_F, GGCAAGGAATGACGGTCACT; Mm.EST1_R, AATGTTGCCTCCTACCCAACA; Mm.EST1_probe, VIC-CACAAAACTGAACACTTTACCTA-MGBNFQ; Mm.EST2_F, TCTGTTGTATTCTGTTGCTGATGCT; Mm.EST2_R, AACACCGGAGATAGAGACCCTAGA; Mm.EST2_probe, VIC-TGTATGGTTCTAGATTTC-MGBNFQ; Mm.Trpc6_F, AAGTACCTGAACGCCCATTTTC; Mm.Trpc6_R, GAATGATGGCGTCTTTCAAGTG; Mm.Trpc6_probe, VIC-TCCTGAGTCTAATGCCT-MGBNFQ; Mm.Edem_F, GTTTCCACACCACCTTTGATTCT; Mm.Edem_R, GTCAGGAGGAACACCTGTCTTCA; and Mm.Edem_probe, VIC-CCCACTGCAGGTGAA-MGBNFQ.
All samples were analyzed in triplicate, and the relative copy number was derived by standardizing the input DNA to the control signal (Edem1, a gene on chromosome 6 that is genomically stable based on the array CGH analysis).
Plasmid Construction.
The human YAP ORF was cloned into the pBABEpuro vector as an EcoRI–BamHI fragment. A single FLAG tag was added to the N terminus during PCR with the following primers: Hs.YAP.F, CCGGGATCCACCATGGATTACAAGGATGACGACGATAAGATGGACCCCGGGCAGCAGCCCGCCGC; and Hs.YAP.R, CCGGAATTCCTATAACCATGTAAGAAAGCTTTC.
Immunoblotting Analysis.
Cells were lysed in NETN lysis buffer [150 mM NaCl/1 mM EDTA/20 mM Tris, pH 8/0.5% Nonidet P-40/1× protease inhibitor mixture (Roche, Indianapolis, IN)]. Lysates were run on an SDS/10% polyacrylamide gel and transferred onto PVDF membranes (Millipore, Bedford, MA), and immunoblots were visualized with a Western Lightning Plus chemiluminescence kit (PerkinElmer, Boston, MA).
Immunofluorescence Analysis.
Immunofluorescence was analyzed as described in ref. 52.
Antibodies.
Phospho-AKT (Ser-473), AKT, phospho-ERK1/2 (Thr-202/Tyr-204), and ERK1/2 antibodies were purchased from Cell Signaling Technology (Beverly, MA). Anti-occludin (His-279) was from Santa Cruz Biotechnology (Santa Cruz, CA). The β-tubulin monoclonal antibody was from Upstate Biotechnology (Lake Placid, NY). The fibronectin and FLAG (M2) antibodies were from Sigma (St. Louis, MO). The E-cadherin, N-cadherin, and vimentin antibodies were from BD Biosciences (San Jose, CA).
Cell Culture.
MCF10A cells were cultured as described in ref. 53. HMEC were obtained from Cambrex (East Rutherford, NJ) and cultured in MEGM (Cambrex). HMEC were immortalized by retroviral introduction of hTert (HMECtert). A retroviral construct encoding hTert (pBABEhygro-hTert) was a kind gift from W. C. Hahn (Dana–Farber Cancer Institute, Boston, MA). Vesicular stomatitis virus glycoprotein-pseudotyped retroviruses were generated as described in ref. 53 and used to infect MCF10A and HMEC. Cell lines were selected with 2 μg/ml puromycin for MCF10A and 1 μg/ml puromycin or 50 μg/ml hygromycin for HMEC.
Three-Dimensional Morphogenesis Assays.
Cells were cultured in growth factor-reduced reconstituted basement membrane (Matrigel; BD Biosciences) as described in ref. 53. Cell lines were assayed in three independent experiments.
Transwell Migration Assays.
Transwell migration assays were performed essentially as described in ref. 54. Cells (2 × 105) were plated without EGF, or 2.5 × 104 cells were plated with 5 ng/ml EGF on Transwell filters (8-μm pore size; Corning, Corning, NY) in three-dimensional medium (described above). Assays were stained and quantified after cells migrated for 24 h.
Two-Dimensional Cell Proliferation.
Two-dimensional cell proliferation was measured by using the fluorescent nuclear stain Syto 60 (Molecular Probes, Eugene, OR) as described in ref. 52.
EGF-Independent Proliferation.
Cells were plated on Matrigel as for three-dimensional morphogenesis assays but without EGF. Cells were fed every 4 days with medium lacking EGF for the duration of the experiment.
Cell Death Assays.
Monolayer MCF10A cultures were assayed for DNA fragmentation after treatment with 0.5 μM STS (Sigma) for 18 h, 100 nM Taxol (Sigma) for 48 h, 100 μM cisplatin for 24 h, or UV light [45-s exposure of UV-C (254 nm) from a 30-W G30T8 bulb (VWR, Bridgeport, NJ] for 24 h. HMECtert were assayed after treatment with 0.5 μM STS or 50 μM cisplatin for 24 h. Floating cells were collected and combined with trypsinized cells, fixed in 75% ethanol, treated with RNase A (0.25 mg/ml), stained with propidium iodide (10 μg/ml), and analyzed on a FACSCalibur flow cytometer (BD Biosciences) for percentage of cells with sub-G1 DNA content. Data were analyzed by using CellQuest (BD Biosciences). For anoikis, MCF10A cells were plated in growth medium on tissue-culture plates pretreated with poly(2-hydroxyethyl methacrylate) [poly-HEMA from Sigma–Aldrich (St. Louis, MO) (6 mg/ml in 95% ethanol at 37°C until dry)] to prevent adherence. After 48 h, cells were collected and analyzed for DNA fragmentation by using the cell-death detection ELISA kit (Roche Diagnostics, Mannheim, Germany). All cell-death assays were performed in three independent experiments.
Soft-Agar Assays.
Cells (1 × 104 or 5 × 104) were added to 1.5 ml of growth medium with 0.4% agar and layered onto 2 ml of 0.5% agar beds in six-well plates. Cells were fed with 1 ml of medium with 0.4% agar every 7 days for 3 weeks, after which colonies were stained with 0.02% iodonitrotetrazolium chloride (Sigma–Aldrich) and photographed. Colonies larger than 50 μm in diameter were counted as positive for growth. Assays were conducted in duplicate in three independent experiments.
Acknowledgments
We thank Scott Lowe for sharing unpublished results and Lynda Chin for array CGH analysis. This work was supported by National Cancer Institute/National Institutes of Health Grants CA080111 and CA089393 and the Breast Cancer Research Foundation (to J.S.B.); National Institutes of Health Grant P01 95281, the Doris Duke Foundation Distinguished Clinical Investigator Award, and a National Foundation for Cancer Research grant (to D.A.H.); National Institutes of Health Grant F32 CA117737 (to G.A.S.); and National Cancer Institute/National Institutes of Health Institutional Training Grant T32CA09361 (to M.O.).
Glossary
Abbreviations
- CGH
comparative genomic hybridization
- EMT
epithelial-to-mesenchymal transition
- HMEC
human mammary epithelial cells
- hTert
catalytic subunit of telomerase
- Mb
megabase(s)
- qPCR
quantitative PCR
- STS
staurosporine
- YAP
Yes kinase-associated protein.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Pinkel D., Segraves R., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W. L., Chen C., Zhai Y., et al. Nat. Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 2.Smolen G. A., Muir B., Mohapatra G., Barmettler A., Kim W. J., Rivera M. N., Haserlat S. M., Okimoto R. A., Kwak E., Dahiya S., et al. Cancer Res. 2006;66:3452–3455. doi: 10.1158/0008-5472.CAN-05-4181. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin C., Garnis C., Zhang L., Rosin M. P., Lam W. L. Cancer Res. 2005;65:7561–7567. doi: 10.1158/0008-5472.CAN-05-1513. [DOI] [PubMed] [Google Scholar]
- 4.Bashyam M. D., Bair R., Kim Y. H., Wang P., Hernandez-Boussard T., Karikari C. A., Tibshirani R., Maitra A., Pollack J. R. Neoplasia. 2005;7:556–562. doi: 10.1593/neo.04586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai Z., Zhu W. G., Morrison C. D., Brena R. M., Smiraglia D. J., Raval A., Wu Y. Z., Rush L. J., Ross P., Molina J. R., et al. Hum. Mol. Genet. 2003;12:791–801. doi: 10.1093/hmg/ddg083. [DOI] [PubMed] [Google Scholar]
- 6.Hermsen M., Alonso Guervos M., Meijer G., van Diest P., Suarez Nieto C., Marcos C. A., Sampedro A. Cell. Oncol. 2005;27:191–198. doi: 10.1155/2005/407216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imoto I., Tsuda H., Hirasawa A., Miura M., Sakamoto M., Hirohashi S., Inazawa J. Cancer Res. 2002;62:4860–4866. [PubMed] [Google Scholar]
- 8.Imoto I., Yang Z. Q., Pimkhaokham A., Tsuda H., Shimada Y., Imamura M., Ohki M., Inazawa J. Cancer Res. 2001;61:6629–6634. [PubMed] [Google Scholar]
- 9.Lambros M. B., Fiegler H., Jones A., Gorman P., Roylance R. R., Carter N. P., Tomlinson I. P. J. Pathol. 2005;205:29–40. doi: 10.1002/path.1681. [DOI] [PubMed] [Google Scholar]
- 10.Snijders A. M., Schmidt B. L., Fridlyand J., Dekker N., Pinkel D., Jordan R. C., Albertson D. G. Oncogene. 2005;24:4232–4242. doi: 10.1038/sj.onc.1208601. [DOI] [PubMed] [Google Scholar]
- 11.Weber R. G., Sommer C., Albert F. K., Kiessling M., Cremer T. Lab. Invest. 1996;74:108–119. [PubMed] [Google Scholar]
- 12.Deveraux Q. L., Reed J. C. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 13.Sudol M. Oncogene. 1994;9:2145–2152. [PubMed] [Google Scholar]
- 14.Yagi R., Chen L. F., Shigesada K., Murakami Y., Ito Y. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strano S., Munarriz E., Rossi M., Castagnoli L., Shaul Y., Sacchi A., Oren M., Sudol M., Cesareni G., Blandino G. J. Biol. Chem. 2001;276:15164–15173. doi: 10.1074/jbc.M010484200. [DOI] [PubMed] [Google Scholar]
- 16.Vassilev A., Kaneko K. J., Shu H., Zhao Y., DePamphilis M. L. Genes Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J., Wu S., Barrera J., Matthews K., Pan D. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Wu S., Huang J., Dong J., Pan D. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 19.Harvey K. F., Pfleger C. M., Hariharan I. K. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- 20.Jia J., Zhang W., Wang B., Trinko R., Jiang J. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kango-Singh M., Nolo R., Tao C., Verstreken P., Hiesinger P. R., Bellen H. J., Halder G. Development (Cambridge, U.K.) 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- 22.Lai Z. C., Wei X., Shimizu T., Ramos E., Rohrbaugh M., Nikolaidis N., Ho L. L., Li Y. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 23.Pantalacci S., Tapon N., Leopold P. Nat. Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- 24.Tapon N., Harvey K. F., Bell D. W., Wahrer D. C., Schiripo T. A., Haber D. A., Hariharan I. K. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 25.Udan R. S., Kango-Singh M., Nolo R., Tao C., Halder G. Nat. Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- 26.Brodie S. G., Xu X., Qiao W., Li W. M., Cao L., Deng C. X. Oncogene. 2001;20:7514–7523. doi: 10.1038/sj.onc.1204929. [DOI] [PubMed] [Google Scholar]
- 27.Debnath J., Brugge J. S. Nat. Rev. Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 28.Witt A. E., Hines L. M., Collins N. L., Hu Y., Gunawardane R. N., Moreira D., Raphael J., Jepson D., Koundinya M., Rolfs A., et al. J. Proteome Res. 2006;5:599–610. doi: 10.1021/pr050395r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Debnath J., Walker S. J., Brugge J. S. J. Cell Biol. 2003;163:315–326. doi: 10.1083/jcb.200304159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basu S., Totty N. F., Irwin M. S., Sudol M., Downward J. Mol. Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 31.Strano S., Monti O., Pediconi N., Baccarini A., Fontemaggi G., Lapi E., Mantovani F., Damalas A., Citro G., Sacchi A., et al. Mol. Cell. 2005;18:447–459. doi: 10.1016/j.molcel.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Elenbaas B., Spirio L., Koerner F., Fleming M. D., Zimonjic D. B., Donaher J. L., Popescu N. C., Hahn W. C., Weinberg R. A. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taparowsky E., Suard Y., Fasano O., Shimizu K., Goldfarb M., Wigler M. Nature. 1982;300:762–765. doi: 10.1038/300762a0. [DOI] [PubMed] [Google Scholar]
- 34.Iba H., Takeya T., Cross F. R., Hanafusa T., Hanafusa H. Proc. Natl. Acad. Sci. USA. 1984;81:4424–4428. doi: 10.1073/pnas.81.14.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levy J. B., Iba H., Hanafusa H. Proc. Natl. Acad. Sci. USA. 1986;83:4228–4232. doi: 10.1073/pnas.83.12.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samuels Y., Wang Z., Bardelli A., Silliman N., Ptak J., Szabo S., Yan H., Gazdar A., Powell S. M., Riggins G. J., et al. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 37.Kang S., Bader A. G., Vogt P. K. Proc. Natl. Acad. Sci. USA. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikenoue T., Kanai F., Hikiba Y., Obata T., Tanaka Y., Imamura J., Ohta M., Jazag A., Guleng B., Tateishi K., et al. Cancer Res. 2005;65:4562–4567. doi: 10.1158/0008-5472.CAN-04-4114. [DOI] [PubMed] [Google Scholar]
- 39.Isakoff S. J., Engelman J. A., Irie H. Y., Luo J., Brachmann S. M., Pearline R. V., Cantley L. C., Brugge J. S. Cancer Res. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 40.Zender L., Spector M., Xue W., Flemming P., Cordon-Cardo C., Silke J., Fan S., Luk J., Wigler M., Hannon G., et al. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Justice R. W., Zilian O., Woods D. F., Noll M., Bryant P. J. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 42.Xu T., Wang W., Zhang S., Stewart R. A., Yu W. Development (Cambridge, U.K.) 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 43.Tao W., Zhang S., Turenchalk G. S., Stewart R. A., St. John M. A. R., Chen W., Xu T. Nat. Genet. 1999;21:177–181. doi: 10.1038/5960. [DOI] [PubMed] [Google Scholar]
- 44.Chan E. H., Nousiainen M., Chalamalasetty R. B., Schafer A., Nigg E. A., Sillje H. H. Oncogene. 2005;24:2076–2086. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- 45.St. John M. A. R., Tao W., Fei X., Fukumoto R., Carcangiu M. L., Brownstein D. G., Parlow A. F., McGrath J., Xu T. Nat. Genet. 1999;21:182–186. doi: 10.1038/5965. [DOI] [PubMed] [Google Scholar]
- 46.Li Y., Pei J., Xia H., Ke H., Wang H., Tao W. Oncogene. 2003;22:4398–4405. doi: 10.1038/sj.onc.1206603. [DOI] [PubMed] [Google Scholar]
- 47.Hamaratoglu F., Willecke M., Kango-Singh M., Nolo R., Hyun E., Tao C., Jafar-Nejad H., Halder G. Nat. Cell. Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 48.Rouleau G. A., Merel P., Lutchman M., Sanson M., Zucman J., Marineau C., Hoang-Xuan K., Demczuk S., Desmaze C., Plougastel B., et al. Nature. 1993;363:515–521. doi: 10.1038/363515a0. [DOI] [PubMed] [Google Scholar]
- 49.Trofatter J. A., MacCollin M. M., Rutter J. L., Murrell J. R., Duyao M. P., Parry D. M., Eldridge R., Kley N., Menon A. G., Pulaski K., et al. Cell. 1993;72:791–800. doi: 10.1016/0092-8674(93)90406-g. [DOI] [PubMed] [Google Scholar]
- 50.Ferrigno O., Lallemand F., Verrecchia F., L’Hoste S., Camonis J., Atfi A., Mauviel A. Oncogene. 2002;21:4879–4884. doi: 10.1038/sj.onc.1205623. [DOI] [PubMed] [Google Scholar]
- 51.Rocco J. W., Leong C. O., Kuperwasser N., DeYoung M. P., Ellisen L. W. Cancer Cell. 2006;9:45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 52.Smolen G. A., Sordella R., Muir B., Mohapatra G., Barmettler A., Archibald H., Kim W. J., Okimoto R. A., Bell D. W., Sgroi D. C., et al. Proc. Natl. Acad. Sci. USA. 2006;103:2316–2321. doi: 10.1073/pnas.0508776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Debnath J., Muthuswamy S. K., Brugge J. S. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 54.Gunawardane R. N., Sgroi D. C., Wrobel C. N., Koh E., Daley G. Q., Brugge J. S. Cancer Res. 2005;65:11572–11580. doi: 10.1158/0008-5472.CAN-05-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]