Abstract
Current methods for identification of Mycobacterium spp. rely upon time-consuming phenotypic tests, mycolic acid analysis, and narrow-spectrum nucleic acid probes. Newer approaches include PCR and sequencing technologies. We evaluated the MicroSeq 500 16S ribosomal DNA (rDNA) bacterial sequencing kit (Applied Biosystems, Foster City, Calif.) for its ability to identify Mycobacterium isolates. The kit is based on PCR and sequencing of the first 500 bp of the bacterial rRNA gene. One hundred nineteen mycobacterial isolates (94 clinical isolates and 25 reference strains) were identified using traditional phenotypic methods and the MicroSeq system in conjunction with separate databases. The sequencing system gave 87% (104 of 119) concordant results when compared with traditional phenotypic methods. An independent laboratory using a separate database analyzed the sequences of the 15 discordant samples and confirmed the results. The use of 16S rDNA sequencing technology for identification of Mycobacterium spp. provides more rapid and more accurate characterization than do phenotypic methods. The MicroSeq 500 system simplifies the sequencing process but, in its present form, requires use of additional databases such as the Ribosomal Differentiation of Medical Microorganisms (RIDOM) to precisely identify subtypes of type strains and species not currently in the MicroSeq library.
Mycobacterium species are a group of acid-fast, aerobic, slow-growing bacteria. The genus comprises more than 70 different species, of which about 30 have been associated with human disease (23). The most important species is Mycobacterium tuberculosis, the causative agent of tuberculosis. Organisms in the M. avium complex (MAC), opportunists found in soil and water, often infect immunocompromised patients (2). Many other species referred to as atypical or nontuberculous mycobacteria have also been associated previously with disease (1, 8, 28, 29).
Nucleic acid probes and high-performance liquid chromatography (HPLC) are two rapid methods that have replaced the traditionally slow biochemical tests (1) for the identification of mycobacterial isolates. Commercial nonradiolabeled probes are available for identifying isolates of M. tuberculosis, M. gordonae, M. kansasii, and M. avium-intracellulare complex. When performed correctly, they can be highly sensitive and specific but do require approximately 105 to 106 CFU to determine conclusive results (10, 24). With the use of isolates from solid media, HPLC examines the mycolic acid fingerprint patterns that differ among most species or complexes of mycobacteria. A small number of species (complexes) have not been separable by HPLC, including many of the pathogenic rapidly growing mycobacterial species (5; unpublished data).
The use of sequencing techniques for identification of Mycobacterium species can replace the conventional methods mentioned above. At least three gene targets have been reported elsewhere to be useful for sequencing to distinguish Mycobacterium species: the 16S rRNA gene (2, 7, 14, 18, 22, 26), the hsp65 gene (21), and the recA gene (3). The gene encoding the small subunit of rRNA (16S rDNA) is highly conserved but contains genus- or species-specific sequence variations in certain positions. Access to monitored sequence databases is scarce for the hsp65 and recA genes. While the turnaround time remains similar to that of HPLC, depending upon the volume of testing, identification by sequencing is more accurate and provides more information. This paper describes our experience with the commercially available MicroSeq 500 16S rDNA bacterial sequencing kit (Applied Biosystems, Foster City, Calif.) used for identification of Mycobacterium spp. in our laboratory. PCR primers anneal to DNA extracted from pure bacterial isolates, allowing amplification of a 500-bp product from the 5" end of the 16S rRNA gene to be used for sequencing. As part of the system, a sequence database is included to determine the genus and species of bacteria. The software allows for exporting of sequences to be compared with other databases, a task that we found to be a necessary part of our protocol. Tools for phylogenetic analysis of bacteria are also included with the software package.
MATERIALS AND METHODS
Bacterial isolates.
All mycobacterial isolates were grown on Lowenstein-Jensen medium (Hardy Diagnostics, Santa Maria, Calif.). Twenty-five frequently isolated mycobacterial strains were purchased from the American Type Culture Collection (ATCC) (Table 1). Ninety-four mycobacterial species either recovered from patient clinical material or sent to the Associated Regional and University Pathologists (ARUP) Mycobacteriology Laboratory directly for identification were characterized. These strains comprised isolates from various geographical regions within the continental United States and Hawaii.
TABLE 1.
ATCC strains used to evaluate the sequencing system
| Mycobacterium sp. | ATCC no. | Sequence identity | Distance scorea (%) |
|---|---|---|---|
| M. asiaticum | 25276T | M. asiaticum | 0.00 |
| M. avium | 25291T | M. avium | 0.00 |
| M. celatum | 51131T | M. celatum | 0.19 |
| M. flavescens | 14474T | M. flavescens | 0.20 |
| M. fortuitum | 6841T | M. fortuitum | 0.00 |
| M. genavense | 51233 | M. genavensec | 0.00 |
| M. gordonae | 14470T | M. gordonae | 0.00 |
| M. haemophilum | 29548T | M. haemophilumc | 0.00 |
| M. kansasii | 19478T | M. kansasii/gastri | 0.00 |
| M. lentiflavum | 51985T | M. lentiflavumc | 0.00 |
| M. mageritense | 700351T | M. mageritensec | 0.00 |
| M. marinum | 927T | M. marinum/ulceransc | 0.00 |
| M. microti | 19422T | M. tuberculosis complexb | 0.00 |
| M. nonchromogenicum | 19530T | M. nonchromogenicum | 0.00 |
| M. peregrinum | 14467T | M. peregrinum | 0.00 |
| M. phlei | 11758T | M. phlei | 0.00 |
| M. porcinum | 3376T | M. porcinum | 0.00 |
| M. scrofulaceum | 19981T | M. scrofulaceum | 0.00 |
| M. simiae | 15275T | M. simiae | 0.00 |
| M. szulgai | 35799T | M. szulgai | 0.00 |
| M. terrae | 15755T | M. terrae | 0.00 |
| M. tuberculosis | 27294T | M. tuberculosis complexb | 0.00 |
| M. tuberculosis | 25177 | M. tuberculosis complexb | 0.00 |
| M. ulcerans | 19423T | M. marinum/ulceransc | 0.00 |
| M. xenopi | 19250T | M. xenopi | 0.10 |
Sequence divergence from the strain closest in the database (0.00% is a perfect match).
Includes M. tuberculosis, M. microti, M. bovis, M. africanum, and M. bovis BCG Pasteur.
Absent from MicroSeq database but identified by RIDOM database.
Conventional identification.
Prior to adoption of sequencing technology, identification of Mycobacterium spp. in our laboratory included observation of growth characteristics, nucleic acid probes, and mycolic acid analysis by HPLC. Our identification algorithm begins with the observation of each acid-fast isolate for growth rate, chromogenicity, and colony morphology. Slowly growing nonchromogenic isolates were submitted for nucleic acid probe assays (AccuProbe; Gen-Probe Inc., San Diego, Calif.) targeting the M. tuberculosis complex and M. avium-intracellulare complex. Probe assays targeting M. gordonae and M. kansasii were performed on chromogenic colonies. Probe hybridizations were performed according to the manufacturer's protocol. AccuProbe results were considered positive when the relative light units (RLU) were greater than 80,000. Repeat analysis was warranted if results were between 10,000 and 80,000 RLU.
Probe-negative isolates were submitted for mycolic acid analysis by HPLC (HP series 1050; Hewlett-Packard Company, Wilmington, Del.). For HPLC, rapidly growing mycobacteria were harvested after 7 days of incubation while slow-growing organisms were harvested after 2 weeks. The equivalent of one generous loopful of cells was suspended in 2 ml of saponification reagent (20% potassium hydroxide in 50% methanol). Suspensions were capped and incubated at 100°C for 2 h. After cooling, 2 ml of chloroform was added, followed by 1.5 ml of acidification reagent (50% hydrochloric acid). After vigorous mixing, the bottom chloroform layer was removed and evaporated to dryness. Samples were evaporated again after the addition of 100 μl of potassium bicarbonate reagent (2% KHCO3 in 50% methanol). After the addition of 1 ml of chloroform and 50 μl of derivatization reagent (0.1 mM p-bromo-phenacyl bromide and 0.005 mM dicyclohexyl-18-crown-6 ether in acetonitrile), samples were heated at 100°C for 20 min. Cooled samples were mixed with 1 ml of clarification reagent (equal parts of acidification reagent and methanol). After vigorous mixing, the bottom layer was removed and evaporated to dryness. A standard solution in methylene chloride was prepared using high (8 μg/100 μl)- and low (4 μg/μl)-molecular-weight standards (Ribi ImmunoChem Research, Inc., Hamilton, Mont.). Samples were suspended in approximately 100 μl of the standard solution prior to application to the HPLC equipped with a C18, reverse-phase analytical cartridge column, 4.6 mm by 7.5 cm, packed with 3-μm silica (Beckman Coulter, Inc., Fullerton, Calif.) at 35°C. Solvent concentrations (methanol:methylene chloride [vol/vol]) were set at 98:2 at 0 min, 80:20 at 1 min, and 35:65 at 10 min, changing linearly between time points at a flow rate of 2.5 ml/min. Chromatograms were analyzed using the Sherlock Microbial Identification System software, version 2.95 (MIDI, Inc., Newark, Del.). If the software did not produce conclusive results, chromatograms were manually compared to figures 1 to 23 of the “Standardized Method for HPLC Identification of Mycobacteria” printed by the U.S. Department of Health and Human Services, 1996.
FIG. 1.
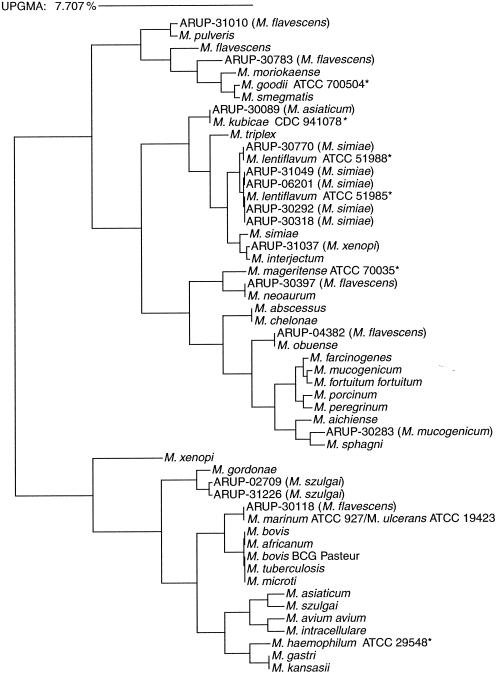
Phylogenetic tree relating sequence-based identification of Mycobacterium species discordant from conventional identification. The algorithm used to construct the tree is the unweighted pair group method using averages (UPGMA). Asterisks indicate species absent from the MicroSeq database but included in the RIDOM database. The numbers following “ARUP-” are designated patient isolates and are followed by the identification provided by conventional methods (see text for full discussion).
Sequencing of 16S rDNA.
The person performing the sequencing protocol was blinded to the conventional identification of the strains. The MicroSeq 500 system includes a kit with PCR and cycle sequencing reagents, bacterial identification and analysis software, and a 16S rDNA sequence database library. DNA extracts were made from pure cultures of mycobacteria using the Prepman protocol for gram-positive bacteria described by Applied Biosystems. The DNA extracts were stored frozen (−20°C) until PCR was performed. A 500-bp 16S rDNA fragment was amplified in a reaction volume of 50 μl (25 μl of MicroSeq PCR master mix, 23.5 μl of molecular-grade water, 0.5 μl of uracil-N-glycosylase, and 1 μl of DNA extract). Prior to sequencing, amplified products were purified by Microcon-100 microconcentrator columns (Amicon, Beverly, Mass.). Forward and reverse sequencing reactions were performed for each amplified product according to instructions supplied with the MicroSeq 500 kit. The kit includes reagents for dRhodamine Dye Terminator chemistry. Sequencing reaction mixtures were purified using premade columns of Sephadex G-50 (Amersham Pharmacia, Piscataway, N.J.) in wells of a multiscreen HV plate (Millipore, Bedford, Mass.). Briefly, the Sephadex was hydrated with 300 μl of water for 3 h and centrifuged for 2 min at 900 × g to remove the interstitial water. The sequencing reaction mixtures were applied to the columns and spun for 2 min at 900 × g, the purified product being collected in a 96-well plate. The extension products were then dehydrated in a vacuum centrifuge and sequenced using the ABI PRISM 377 DNA sequencer (Applied Biosystems) according to the manufacturer's instructions.
Sequence data analysis.
Sequences were assembled and edited using the MicroSeq software, version 1.36. The software was used to assemble each forward and reverse sequence into a consensus sequence, which was edited to resolve base pair ambiguities between the two strands by evaluation of the electropherograms. Early in the validation, it became apparent that important clinical species such as M. marinum, M. genavense, and M. lentiflavum, among others, were absent from the MicroSeq library. Because of this, it was necessary to seek an alternative database for sequence analysis. Each consensus sequence was compared to two different libraries: (i) the MicroSeq 500 bacterial database using the Full Alignment Tool of the software package and (ii) the Ribosomal Differentiation of Medical Microorganisms (RIDOM) database from the University of Würzburg, Würzburg, Germany (11), accessible via the internet (www.RIDOM.de). The final result from the MicroSeq database search was a list of the closest matches with a distance score describing the percent difference between the unknown sequence and the database sequence. The species from the MicroSeq and/or RIDOM database giving a perfect match (0.00% distant) was used to determine the final identification. As suggested by Patel et al. (18), if the distance score was greater than 0.00% and less than 0.80%, the unknown organism was said to be most closely related to the species giving the closest match. Sequences resulting in distance scores of 0.80% or greater from both the MicroSeq and RIDOM databases were considered unique species or subtypes of species.
The National Reference Centre for Mycobacteriology (Christine Turenne, Canadian Science Centre for Human and Animal Health, Health Canada, Winnipeg, Manitoba, Canada) used their library to analyze the sequences that gave results discrepant from our conventional methods of identification. Their database consists of collections of confirmed isolates from patients and chosen GenBank submissions as well as numerous reference strains (i.e., ATCC strains) which have been sequenced in their laboratory. Their database also includes unique sequences belonging to uncharacterized Mycobacterium species obtained from submitted clinical strains.
RESULTS
All 25 ATCC strains evaluated were correctly identified using the MicroSeq system (Table 1). An additional 94 clinical isolates recovered from primary cultures or sent to ARUP for identification were evaluated by sequencing as well as by conventional methods. The following species were included in the RIDOM database but not included in the MicroSeq 500 database: M. marinum, M. haemophilum, M. lentiflavum, M. genavense, M. kubicae, and M. mageritense (Fig. 1). A compilation of the results is listed in Table 2. Fifteen discrepancies between conventional identification methods and the sequencing strategy employed were found. Nine of the discrepancies were perfect matches (0.00% distance score) to strains from either the MicroSeq database or the RIDOM database, with two being near-perfect matches (0.59 and 0.20% distant). The distance scores of the remaining discrepant sequences were higher (1.37, 1.17, 2.19, and 1.40%), suggesting species absent from either database. Two of the species with higher divergence, identified as M. szulgai by HPLC, were most closely related to M. gordonae by sequencing. An M. gordonae AccuProbe (Gen-Probe) assay was performed for each isolate. Both isolates were repeatedly negative with RLU values of <11,000. The Canadian National Reference Centre for Mycobacteriology also matched the sequence as most closely related to M. gordonae. A phylogenetic tree (Fig. 1) shows that it is definitely not M. szulgai and is most likely a variant of M. gordonae that is not similar enough to react with the probe. Previously published results have also illustrated that HPLC can misidentify some M. gordonae strains as M. szulgai (27). It has been observed elsewhere that, unlike other mycobacteria, which show a conservation of the rRNA sequence at the species level, M. gordonae exhibits rDNA variation (13).
TABLE 2.
Comparison of sequencing with conventional identification methods for Mycobacterium species
| Species by conventional identification | Conventional identification
|
Sequence-based identification by:
|
|||||
|---|---|---|---|---|---|---|---|
| Final method of identification | No. of isolates | MicroSeq 500 database
|
RIDOM database
|
||||
| Species | % Divergencea (no. of isolates) | Species | % Divergencea (no. of isolates) | ||||
| M. avium/intracellulare | AccuProbe | 11 | M. avium | 0.00 (5) | M. avium | 0.00 (5) | |
| M. intracellulare | 0.00 (6) | M. intracellulare | 0.00 (6) | ||||
| M. gordonae | AccuProbe | 11 | M. gordonae | 0.00 (4) | M. gordonae | 0.00 (4) | |
| 0.20 (1) | M. gordonae | 0.00 (1) | |||||
| 0.39 (1) | M. gordonae | 0.00 (1) | |||||
| 0.59 (1) | M. gordonae | 0.00 (1) | |||||
| 0.78 (3) | M. gordonae | 0.00 (3) | |||||
| 0.98 (1) | M. gordonae | 0.00 (1) | |||||
| M. tuberculosis | AccuProbe | 11 | M. tuberculosis complexb | 0.00 (10) | M. tuberculosis complexb | 0.00 (10) | |
| 0.39 (1) | M. tuberculosis complexb | 0.00 (1) | |||||
| M. kansasii | AccuProbe | 11 | M. kansasii/gastri | 0.00 (10) | M. kansasii/gastri | 0.00 (10) | |
| 1.17 (1) | M. kansasii | 0.23 (1) | |||||
| M. fortuitum | HPLC | 9 | M. fortuitum | 0.00 (6) | M. fortuitum | 0.00 (6) | |
| M. peregrinum | 0.00 (1) | M. peregrinum/septicum | 0.00 (1) | ||||
| M. farcinogenes | 0.00 (1) | M. farcinogenes/senegalense/fortuitum | 0.00 (1) | ||||
| M. porcinum | 0.00 (1) | M. porcinum/fortuitumc | 0.00 (1) | ||||
| M. szulgai | HPLC | 4 | M. szulgai | 0.00 (2) | M. szulgai | 0.00 (2) | |
| M. gordonae | 1.37 (1) | M. gordonae | 1.36 (1) | ||||
| 1.17 (1) | M. gordonae | 1.14 (1) | |||||
| M. marinum | HPLC | 3 | M. asiaticum | 1.56 (3) | M. marinum/ulcerans | 0.00 (3) | |
| M. xenopi | HPLC | 5 | M. xenopi | 0.10 (2) | M. xenopi | 0.00 (2) | |
| 0.29 (2) | M. xenopi | 0.00 (2) | |||||
| M. interjectum | 0.20 (1) | M. interjectum | 0.24 (1) | ||||
| M. flavescens | HPLC | 5 | M. obuense | 0.00 (1) | M. obuense | 0.00 (1) | |
| M. asiaticum | 1.56 (1) | M. marinum/ulcerans | 0.00 (1) | ||||
| M. moriokaense | 2.19 (1) | M. moriokaense | 3.01 (1) | ||||
| M. pulveris | 0.59 (1) | M. pulveris | 0.59 (1) | ||||
| M. neoaurum | 0.00 (1) | M. neoaurum | 0.00 (1) | ||||
| M. asiaticum | HPLC | 1 | M. simiae, M. triplex, or M. interjectum | 1.79 (1) | M. kubicae | 0.00 (1) | |
| M. simiae | HPLC | 5 | M. simiae | 1.00 (4) | M. lentiflavumd | 0.00 (4) | |
| 1.39 (1) | M. lentiflavume | 0.00 (1) | |||||
| M. mucogenicum | HPLC | 4 | M. mucogenicum | 0.00 (3) | M. mucogenicum | 0.00 (3) | |
| M. sphagni or M. aichiense | 1.40 (1) | M. sphagni or M. aichiense | 1.64 (1) | ||||
| M. chelonae/abscessus | HPLC | 11 | M. chelonae/abscessus | 0.00 (11) | M. chelonae/abscessus | 0.00 (11) | |
| M. bovis BCG | HPLC | 3 | M. tuberculosis complexb | 0.00 (3) | M. tuberculosis complexb | 0.00 (3) | |
Sequence divergence from the strain closest in the database (0.00% is a perfect match).
Includes M. tuberculosis, M. bovis, M. microti, M. africanum, and M. bovis BCG Pasteur.
Third biovariant.
DSM 44418, ATCC 51985.
ATCC 51988.
Three isolates, each a close relative of M. fortuitum as illustrated in the dendrogram of Fig. 1, were identified by sequencing as M. peregrinum, M. farcinogenes, and M. porcinum but were considered M. fortuitum by HPLC. These are all closely related members of the M. fortuitum complex (15). M. avium-intracellulare complex is identified by the MAC AccuProbe used in our lab and does not reveal differences between the two species of the complex. The MicroSeq 500 system provides species differentiation between M. avium and M. intracellulare (Table 2).
Sequencing of 16S rDNA does not distinguish M. kansasii from M. gastri (22); however, chromogenicity will differentiate the two. M. chelonae is not distinguished from M. abscessus by HPLC, colony morphology, or 16S rDNA sequencing of the 5" end. The full 16S rDNA sequence will distinguish M. chelonae from M. abscessus. The full gene sequence will not distinguish the type strains of M. kansasii and M. gastri. Furthermore, the first 500 bases as well as the full 16S rRNA gene sequence result in identical sequences for each member of the M. tuberculosis complex (M. tuberculosis, M. bovis, M. microti, M. africanum, and M. bovis BCG Pasteur). AccuProbe targets the 16S rRNA and does not provide this distinction either. HPLC, on the other hand, can distinguish M. bovis BCG from the other members of the M. tuberculosis complex (4).
Each of five isolates identified as M. simiae by HPLC resulted in a distance score of 1.00 or 1.36% by the MicroSeq 500 database, which first led us to believe that the identifications were concordant. Unlike the patient isolates, the M. simiae ATCC strain resulted in a perfect match (0.00% distant). The high distance score from sequencing actually implied that the isolates were not M. simiae but something absent from the MicroSeq 500 database. We searched the RIDOM database and found perfect matches for M. lentiflavum. Two different strains are included in the RIDOM database, accounting for the two different distance scores given by the MicroSeq database. Another isolate, identified by HPLC as M. asiaticum, gave a sequence with identical distance scores (1.79%) for each of three different species in the MicroSeq database: M. simiae, M. triplex, and M. interjectum. The Canadian National Reference Centre for Mycobacteriology, as well as the RIDOM database, matched the sequence perfectly with M. kubicae (9), a more recently described slowly growing scotochromogen recovered from sputum.
Identification achieved by sequencing did not agree with any of the five isolates identified by HPLC as M. flavescens. Three of these isolates were sequenced as perfect matches with M. obuense, M. marinum/ulcerans, and M. neoaurum. The fourth species was closely related to M. pulveris (0.59% distant) in both the MicroSeq and RIDOM databases but, from the reference laboratory's evaluation, perfectly matched (0.00% distant) M. elephantis, a unique sequence deposited in GenBank in June of 1999 (25). The last of the five isolates was most closely related to M. moriokaense but, due to the high distance score (2.19%), is probably a unique strain that is not included in the MicroSeq or RIDOM database or that of the reference lab. An ATCC type strain for M. flavescens was identified correctly by 16S rDNA sequencing with the MicroSeq 500 system.
DISCUSSION
The HPLC database of phenotypic characteristics is limited to common species and cannot be upgraded very easily. The MicroSeq database of 16S rDNA sequences is lacking some common species. Sequencing databases, however, are easily upgraded, and individual sequences can be verified with other databases. The confirmation of sequence analyses by another lab can be done electronically without repeating the costly technical procedures to obtain the sequence. Once sequences are confirmed, they can be used for database searches of unknown organisms.
Commercial probe hybridization assays can provide a rapid identification but can test for only one species at a time, and probes are available for only M. tuberculosis complex, M. gordonae, M. kansasii, and M. avium-intracellulare complex. False-negative probe results often occur, requiring repeat analysis for verification. Also, previously published reports describe several subtypes of M. kansasii, some of which do not react with the probe (19, 20). False-positive results have been noted previously with the M. tuberculosis complex DNA probe cross-reacting with isolates of the M. terrae complex (16).
The MicroSeq 500 sequencing system is a commercial bacterial identification assay with a turnaround time of 2 working days and 4 h of technologist time using the ABI 377 Prism. Our findings are consistent with other reports which compared phenotypic identification to molecular sequencing (12, 18, 26). Due to reduced repeat rates, we found the MicroSeq 500 system to be faster than our conventional identification scheme and also more accurate when the RIDOM database was consulted. Sequences of 15 (of 94) isolates clearly demonstrated that misidentifications had been made by our conventional algorithm. For example, HPLC analysis of mycolic acids from 5 of the 15 discrepancies led us to believe that cultures were identified as M. flavescens when sequencing clearly showed that they were not M. flavescens.
Compared to probe hybridization, sequencing technology requires less judgment on the part of the technologists for interpretation. A false-negative probe result may be due to insufficient inoculum, incomplete cell lysis, or variability in the targeted sequence. If this happens using the MicroSeq 500 system, no PCR product is generated and thus no sequence is generated. Therefore, sequencing is not subject to false-negative results.
Another advantage of molecular sequencing for identification of unusual isolates is its more accurate classification of species. Fifteen unusual isolates included in our study were misidentified. Eleven of these isolates resulted in perfect or close matches (<0.80% divergence) within the two libraries employed. Although the remaining isolates did not give us a definitive identification, the sequence and distance score from the closest match in the database give us valuable information: (i) a higher distance score suggests a unique species that is not included in the sequence database, and (ii) the sequence can be matched against other databases such as GenBank, the ribosomal database library (www.cme.msu.edu/RDP), the RIDOM database, or a privately developed database. Interpretation of the distance scores of the sequences may be hampered by a higher percentage of ambiguities to be edited in the forward and reverse sequences. Drancourt et al. (6) recommend 1% ambiguities (∼15 positions) within an approximately 1,500-base sequence. We routinely obtained a 500-base 16S rDNA sequence with less than 2% ambiguities (∼10 positions).
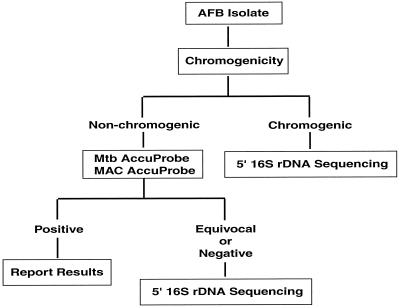
This technology has streamlined our algorithm for identifying mycobacteria. In the past we analyzed cultures for growth rate, chromogenicity, and colony morphology. Then we proceeded to perform AccuProbe testing to rule out M. tuberculosis complex and MAC if the colonies were nonchromogenic and M. gordonae and M. kansasii if the colonies were chromogenic. We subsequently performed HPLC testing to get a species identification that was often questionable or indeterminate, directing us to perform various biochemical reactions in efforts to confirm results. Now we perform only the M. tuberculosis complex and MAC AccuProbe assays. Probe-negative nonchromogenic colonies, rapid growers, and chromogenic slow-growing organisms are sequenced (Fig. 2). Due to a very low repeat rate and less need for biochemical confirmation, sequencing has reduced our turnaround time considerably.
FIG. 2.
Algorithm for identification of Mycobacterium spp. after incorporation of sequencing. AFB, acid-fast bacillus.
Clinical information beneficial to the patient can also be acquired from sequence analysis of 16S rDNA. Since the completion of the present validation, we obtained an identification of M. tuberculosis complex on an isolate with a distance score of 0.39% (MicroSeq database). Since most isolates within M. tuberculosis complex resulted in sequences with no divergence from the type strain, we decided to look into this particular sequence further. We consulted the Canadian National Reference Centre for Mycobacteriology and discovered this isolate to contain mutations within the 16S rRNA gene consistent with streptomycin resistance (17). In retrospect, we realize that a similar isolate was included in our validation data for this study (Table 2).
Dendrograms (or trees) can be constructed based upon the set of distances between all pairs of sequences of the input files which may include unknown sequences, any of the sequences from the MicroSeq database, and sequences from other sources or databases. A scale is included describing a percentage difference that the segment length represents. The spatial relationship of the unknown sequence to known sequences helps to determine if the isolate represents a novel species. Tree-making tools are included in the MicroSeq software. Sequencing 16S rDNA not only is a useful technology for identification and phylogenetic analysis of bacteria but may lead to the discovery of previously uncharacterized species.
When perfect matches are not achieved, the question arises as to when to consider the species identified and when it should be considered only closely related to a species. After the study of large numbers of sequences, cutoff values can be designed specifically for mycobacteria with, of course, exceptions to the rule. Because bacterial genera do not evolve at the same speed, it may be necessary to use different cutoff values depending on the bacterial genus under investigation. Currently, there are no accepted guidelines regarding computer-aided comparison of percent differences or sequence similarity for 16S rDNA-based bacterial identification (6).
As with HPLC, a major drawback of the MicroSeq 500 system is the high cost of the test. Considering the costs of reagents, supplies, technologist time, and repeat rate, the relative costs per test for HPLC and the MicroSeq 500 System are similar (approximately $50 and $54, respectively). While the AccuProbe test costs less (approximately $35 per test), only four species can be identified. We hope to reduce these costs as we apply the sequencing technology for identifying other bacterial genera in the microbiology lab, thus increasing test volume. We are also concerned about the technical demands required by the MicroSeq protocol, specifically the manual extraction of DNA, PCR, and cleanup methods. We are hopeful for less labor-intensive and more automated approaches in the future.
Overall we find sequencing technology to be an excellent tool for species identification of mycobacteria. We reduced the turnaround time from that of HPLC because of shortening the need for repeat analysis and confirmation of questionable results with biochemicals. More importantly, we showed (Table 3) that sequencing provides more accuracy in identifying Mycobacterium species than does HPLC. While the MicroSeq database consists mainly of a single type strain for each species and clearly lacks some important species, we found the RIDOM database to be very complete and a valuable complement to the MicroSeq system. Caution should be exercised when using public databases, which are not monitored, such as GenBank. Sequencing technology is continually giving us new information regarding mycobacterial disease. In our opinion, the amount of information that we achieve through sequence data analysis, both academic and clinical, is an accepted counter for the costs and technical demands of the procedure, which we hope will improve over time.
TABLE 3.
Sequencing results discordant with conventional identification
| ARUP sample no. | Conventional identification (HPLC) | Sequencing identification by:
|
|||||
|---|---|---|---|---|---|---|---|
| MicroSeq 500 and RIDOM databases
|
Reference lab
|
||||||
| Identification | Distance score (%) | Identification | Distance score (%) | ||||
| 02709 | M. szulgai | M. gordonae | 1.37 | M. gordonae | 1.30 | ||
| 31226 | M. szulgai | M. gordonae | 1.17 | M. gordonae | 1.30 | ||
| 04382 | M. flavescens | M. obuense | 0.00 | M. obuense | 0.00 | ||
| 30118 | M. flavescens | M. marinum/M. ulceransa | 0.00 | M. marinum/M. ulceransa | 0.00 | ||
| 30783 | M. flavescens | M. moriokaense | 2.19 | M. goodii | 3.8 | ||
| 31010 | M. flavescens | M. pulveris | 0.59 | M. elephantis | 0.00 | ||
| 30397 | M. flavescens | M. neoaurum | 0.00 | M. neoaurum | 0.00 | ||
| 30089 | M. asiaticum | M. kubicae | 0.00 | M. kubicae | 0.00 | ||
| 30283 | M. mucogenicum | M. sphagni/M. aichienseb | 1.40 | M. sphagni | 1.5 | ||
| 31037 | M. xenopi | M. interjectum | 0.20 | M. interjectum | 0.00 | ||
| 30318 | M. simiae | M. lentiflavum | 0.00 | M. lentiflavum | 0.00 | ||
| 30770 | M. simiae | M. lentiflavum | 0.00 | M. lentiflavum | 0.00 | ||
| 30292 | M. simiae | M. lentiflavum | 0.00 | M. lentiflavum | 0.00 | ||
| 06201 | M. simiae | M. lentiflavum | 0.00 | M. lentiflavum | 0.00 | ||
| 31049 | M. simiae | M. lentiflavum | 0.00 | M. lentiflavum | 0.00 | ||
Strains indistinguishable by rDNA sequencing.
Strains with different rDNA sequences but the same distance scores.
Acknowledgments
The work was supported by the ARUP Institute for Clinical and Experimental Pathology.
We are very grateful to Sam Page for his computer expertise and assistance.
REFERENCES
- 1.American Thoracic Society. 1997. Diagnosis and treatment of disease caused by nontuberculous mycobacteria. Am. J. Respir. Crit. Care Med. 156:S1-S25. [DOI] [PubMed] [Google Scholar]
- 2.Beggs, M. L., R. Stevanova, and K. D. Eisenack. 2000. Species identification of Mycobacterium avium complex isolates by a variety of molecular techniques. J. Clin. Microbiol. 38:508-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwood, K. S., S. He, J. Gunton, C. Y. Turenne, J. Wolfe, and A. M. Kabani. 2000. Evaluation of recA sequences for identification of Mycobacterium species. J. Clin. Microbiol. 38:2846-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler, W. R., K. C. Jost, Jr., and J. O. Kilburn. 1991. Identification of mycobacteria by high-performance liquid chromatography. J. Clin. Microbiol. 29:2468-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler, W. R., and J. O. Kilburn. 1990. High-performance liquid chromatography patterns of mycolic acids as criteria for identification of Mycobacterium chelonae, Mycobacterium fortuitum, and Mycobacterium smegmatis. J. Clin. Microbiol. 28:2094-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drancourt, M., C. Bollet, A. Carlioz, R. Martelin, J.-P. Gayral, and D. Raoult. 2000. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol. 38:3623-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El Amin, N. M., H.-S. Hanson, B. Pettersson, B. Petrini, and L. V. von Stedingk. 2000. Identification of non-tuberculous mycobacteria: 16S rRNA gene sequence analysis vs. conventional methods. Scan. J. Infect. Dis. 32:47-50. [DOI] [PubMed] [Google Scholar]
- 8.Falkinham, J. O., III. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9:177-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Floyd, M. M., W. M. Gross, D. A. Bonato, V. A. Silcox, R. W. Smithwick, B. Metchock, J. T. Crawford, and W. R. Butler. 2000. Mycobacterium kubicae sp. nov., a slowly growing, scotochromogenic mycobacterium. Int. J. Syst. Evol. Microbiol. 50:1811-1816. [DOI] [PubMed] [Google Scholar]
- 10.Goto, M., S. Ika, K. Okuzumi, S. Kimura, and K. Shimada. 1991. Evaluation of acridinium-ester-labeled DNA probes for identification of Mycobacterium tuberculosis and Mycobacterium avium-Mycobacterium intracellulare complex in culture. J. Clin. Microbiol. 29:2473-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harmsen, D., J. Rothganger, C. Singer, J. Albert, and M. Frosch. 1999. Intuitive hypertext-based molecular identification of micro-organisms. Lancet 353:291.. [DOI] [PubMed] [Google Scholar]
- 12.Holberg-Petersen, M., M. Steinbakk, K. J. Figenschau, E. Jantzen, J. Eng, and K. K. Melby. 1999. Identification of clinical isolates of Mycobacterium spp. by sequence analysis of the 16S ribosomal RNA gene. APMIS 107:231-239. [PubMed] [Google Scholar]
- 13.Kirschner, P., and E. C. Böttger. 1992. Microheterogeneity within rRNA of Mycobacterium gordonae. J. Clin. Microbiol. 30:1049-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirschner, P., B. Springer, U. Vogel, A. Meier, A. Wrede, M. Kiekenbeck, F. C. Bange, and E. C. Böttger. 1993. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J. Clin. Microbiol. 31:2882-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirschner, P., M. Kiekenbeck, D. Meissner, J. Wolters, and E. C. Böttger. 1992. Genetic heterogeneity within Mycobacterium fortuitum complex species: genotypic criteria for identification. J. Clin. Microbiol. 30:2772-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin, C., V. Vincent Levy-Frebault, B. Cattier, A. Legras, and A. Goudeau. 1993. False positive result of Mycobacterium tuberculosis complex DNA probe hybridization with a Mycobacterium terrae isolate. Eur. J. Clin. Microbiol. Infect. Dis. 12:309-310. [DOI] [PubMed] [Google Scholar]
- 17.Meier, A., P. Kirshchner, F. C. Bange, U. Vogel, and E. C. Böttger. 1994. Genetic alterations in streptomycin-resistant Mycobacterium tuberculosis: mapping of mutations conferring resistance. Antimicrob. Agents Chemother. 38:228-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel, J. B., D. G. B. Leonard, X. Pan, J. M. Musser, R. E. Berman, and I. Nachamkin. 2000. Sequence-based identification of Mycobacterium species using the MicroSeq 500 16S rDNA bacterial identification system. J. Clin. Microbiol. 38:246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Picardeau, M., G. Prod'hom, L. Raskine, M. P. LePennec, and V. Vincent. 1997. Genotypic characterization of five subspecies of Mycobacterium kansasii. J. Clin. Microbiol. 35:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richter, E., S. Niemann, S. Rüsch-Gerdes, and S. Hoffner. 1999. Identification of Mycobacterium kansasii by using a DNA probe (AccuProbe) and molecular techniques. J. Clin. Microbiol. 37:964-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ringuet, H., C. Akoua-Koffi, S. Honore, A. Varnerot, V. Vincent, P. Berche, J. L. Gaillard, and C. Pierre-Audigier. 1999. hsp65 sequencing for identification of rapidly growing mycobacteria. J. Clin. Microbiol. 37:852-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogall, T., T. Flohr, and E. C. Böttger. 1990. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J. Gen. Microbiol. 136:1915-1920. [DOI] [PubMed] [Google Scholar]
- 23.Shinnick, T. M., and R. C. Good. 1994. Mycobacterial taxonomy. Eur. J. Clin. Microbiol. Infect. Dis. 13:884-901. [DOI] [PubMed] [Google Scholar]
- 24.Shinnick, T. M., and R. C. Good. 1995. Diagnostic mycobacteriology laboratory practices. Clin. Infect. Dis. 21:291-292. [DOI] [PubMed] [Google Scholar]
- 25.Shojaei, H., J. G. Magee, R. Freeman, M. Yates, N. U. Horadagoda, and M. Goodfellow. 2000. Mycobacterium elephantis sp. nov., a rapidly growing non-chromogenic mycobacterium isolated from an elephant. Int. J. Syst. E vol. Microbiol. 50:1817-1820. [DOI] [PubMed] [Google Scholar]
- 26.Springer, B., L. Stockman, K. Teschner, G. D. Roberts, and E. C. Böttger. 1996. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J. Clin. Microbiol. 34:296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thibert, L., and S. LaPierre. 1993. Routine application of high-performance liquid chromatography for identification of mycobacteria. J. Clin. Microbiol. 31:1759-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wayne, L. G. 1985. The “atypical” mycobacteria: recognition and disease association. Crit. Rev. Microbiol. 12:185-222. [DOI] [PubMed] [Google Scholar]
- 29.Witebsky, F. G., and P. S. Conville. 1993. The laboratory diagnosis of mycobacterial diseases. Infect. Dis. Clin. N. Am. 7:359-377. [PubMed] [Google Scholar]