Abstract
FSH, which binds to specific receptors on granulosa cells in mammals, plays a key role in folliculogenesis. Its biological activity involves stimulation of intercellular communication and upregulation of steroidogenesis, but the entire spectrum of the genes regulated by FSH has yet to be fully characterized.
In order to find new regulated transcripts, however rare, we have used a Suppression Subtractive Hybridization approach (SSH) on pig granulosa cells in primary culture treated or not with FSH. Two SSH libraries were generated and 76 clones were sequenced after selection by differential screening. Sixty four different sequences were identified, including 3 novel sequences. Experiments demonstrated the presence of 25 regulated transcripts.
A gene ontology analysis of these 25 genes revealed (1) catalytic; (2) transport; (3) signal transducer; (4) binding; (5) anti-oxidant and (6) structural activities. These findings may deepen our understanding of FSH's effects. Particularly, they suggest that FSH is involved in the modulation of peroxidase activity and remodelling of chromatin.
Background
The development of ovarian follicles leading to ovulation requires endocrine regulation by the gonadotropins FSH and LH as the main actors. The complex regulatory network also includes steroids and peptides (e.g. growth factors, inhibins) acting via the autocrine and paracrine pathways. Recent studies have highlighted the importance of FSH in ovarian follicle maturation: in FSH-deficient mice the folliculogenesis is blocked prior to antral formation [1,2]. In order to obtain functional gametes, granulosa cell (GC) communication with the oocyte also seems essential [2,3]. GCs constitute an important compartment in the mammalian ovarian follicle, contributing to follicle development. They actively participate in the endocrine function of the ovaries by secreting oestradiol or progesterone under FSH stimulation. Besides their functional importance, GCs have been intensively studied for their convenient isolation. In murine, porcine, and bovine species they constitute a well-standardized model for the in vitro study of GC function, including hormonal regulation.
Even if much data has been accumulated on the action of gonadotropins on GCs, the entire spectrum of genes regulated by FSH is not known. Besides recent advances in the generation of normalized cDNA libraries [4,5] and expression analysis with differential display PCR and microarrays [6,7], SSH approach [8] was more efficient in accessing low-level expressed transcripts. We have therefore used the SSH method to isolate FSH-regulated genes in pig primary GC. These results increase our understanding of the physiological processes involved in the response of GC to FSH. In particular, FSH may play a role in the modulation of peroxidase activities and the remodelling of chromatin.
Methods
Cell cultures
Pig granulosa cells were isolated and cultured as described previously by Hatey et al [9]. Briefly, the granulosa cells were isolated from medium (around 3 mm in diameter) healthy follicles from immature swine ovaries. Cells were plated and grown to confluence in a serum-containing medium, which was replaced after 5 days of culture by a serum-free medium with or without FSH (Gonal-F™ 0.5 UI/ml, Serono laboratory) and incubated for a 48-h period before RNA extraction. FSH stimulation efficiency was tested both by measuring progesterone secretion in the culture medium using HPLC analysis [10] and by controlling the increase in P450scc and IGF1 genes expression using Northern blot analysis.
RNA and polyA extraction
Total RNA extraction was performed according to Chomczynski and Sacchi [11] with minor modifications [12]. Poly(A)-containing RNA was selected with Dynabeads mRNA Purification Kit (Dynal) following the manufacturer's instructions. For quality control, total RNA and mRNA were denatured with formaldehyde and size-fractionated through a 1% agarose gel according to standard methods. The integrity of each RNA sample was checked by ethidium bromide staining of the gel. Before reverse transcription, DNase I treatment of RNA was performed as described [13].
Reverse-Transcription
Total DNase I treated RNA (2 μg) from control and induced cells were used for reverse transcription (RT) using Superscript™ II Reverse Transcriptase (Invitrogen) and oligo-dT15 primers (Roche) according to the manufacturer's recommendations.
Suppression subtractive hybridization (SSH)
SSH was performed with 1 μg of mRNA using the Clontech PCR-Select cDNA Subtraction Kit (Clontech) with minor modifications. The primary PCR amplification was achieved through 30 PCR cycles starting with 1 μl of 6 fold diluted second hybridization reaction. The secondary PCR amplification was achieved through 12 PCR cycles starting with 2 μl of 10 fold diluted primary PCR amplification. The PCR products were cloned using the TOPO™ TA CloningR Kit (Invitrogen). Agarose gel analysis allowed the exclusion of empty clones or those containing more than one product. Forward subtraction H1 (FSH/C) used mRNA of control cells as a driver to select genes induced by FSH and reverse subtraction H2 (C/FSH) used mRNA of FSH-induced cells as a driver to select genes repressed by FSH.
Differential screening
As the final SSH products were enriched, but not strictly composed of differentially expressed cDNAs, a screening procedure was set up to sort out the false positive clones. Inserts were amplified by PCR starting from each colony. Each cDNA was amplified by PCRusing Invitrogen Taq polymerase: initiation with one cycle of 7 min at 94°C and amplification with 20 cycles (94°C for 30 s, 68°C for 30 s and 72°C for 1.5 min). Primers used were nested PCR primer1 (5'-TCGAGCGGCCGCCCGGGCAGGT-3') and nested PCR primer2 (5'-AGGGCGTGGTGCGGAGGGCGGT-3'). PCR products were checked by electrophoresis on agarose gel. PCR products were denaturated in NaOH 0.5 M, EDTA 25 mM and 5% bromophenol blue, vacuum transferred onto two identical Hybond N membranes (Millipore) and UV crossed-linked. Macro-arrays were subsequently hybridized with the different probes (cDNA of control and treated cells). The screening was performed visually.
Probe labelling and hybridization
Probe labelling with dCTP α 32P and hybridization of Northern-blot or macro-arrays were performed as described [9].
Sequencing reaction and analysis
The inserts of regulated clones were amplified from the plasmid using Primer1 (5'-GTAATACGACTCACTAGGGC-3'), and Primer2 (5'-TGTAGCGTGAAGACGACAGAA-3'). PCR products were purified using Wizard® PCR Preps DNA Purification System (Promega). Sequencing was performed using the ABI PRISM™ Big Dye™ Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) and the Perkin-Elmer 377 apparatus (Perkin-Elmer Cetus). Sequences were submitted to EMBL database and accession numbers are given in Table 2. Sequence assembling was done using the Wisconsin Genetic Computer Group software package (Wisconsin Package, version 10.0, Genetic Computer Group). Edited sequence data were analysed with the Advanced BLAST program of the EMBL web site for similarities with known genes or ESTs in the entire EMBL databases.
Table 2.
Identification of cDNA clones. The table gives the identification of the cDNA clones using blast analysis against EMBL/NCBI database. The clones were ranked by HUGO symbol (Human Genome Organisation). H1-clones correspond to forward hybridization (FSH-induced genes) and H2-clones to reverse hybridization (FSH-repressed genes).
| Clone | Matching Sequences | |||||||
| clone | EMBL accession number | insert size (bp) | identification | species | Similarity | accession number | HUGO symbol | |
| % | bp | |||||||
| H2-146 | AJ704910 | 255 | actin, alpha 2, smooth muscle, aorta | HS | 96 | 211 | MN_001613 | ACTA2 |
| H2-96 | AJ704907 | 111 | actin, gamma 1 | HS | 89 | 119 | NM_001614 | ACTG1 |
| H1-2 | AJ704867 | 186 | cytosolic dihydrodiol dehydrogenase 3 | BT | 90 | 115 | D49542 | AKR1C3 |
| H2-222 | AJ704918 | 643 | annexin A5 | HS | 93 | 495 | CR541842 | ANXA5 |
| H1-378 | AJ704899 | 76 | ADP-ribosylation factor 1 | HS | 91 | 74 | AF493881 | ARF1 |
| H1-98 | AJ704876 | 166 | mitochondrial ATPase 6 | SS | 98 | 165 | AF250315 | ATP6 |
| H1-138 | AJ704880 | 354 | calpain I light subunit | SS | 99 | 353 | M11778 | CAPNS1 |
| H1-357 | AJ704896 | 107 | Cctg for chaperonin | HS | 96 | 104 | X74801 | CCT3 |
| H1-124 | AJ704878 | 177 | Pig complement cytolysis inhibitor | SS | 99 | 171 | M84639 | clu |
| H1-61 | AJ704870 | 245 | alpha collagene human type IV(alport syndrome) | SS | 82 | 162 | M31115 | col4A5 |
| H2-108 | AJ704909 | 410 | cytochrome c oxidase II | SS | 99 | 410 | U18827 | COXII |
| H1-253 | AJ704887 | 99 | chondroitin sulfate proteoglycan 6 | HS | 89 | 88 | BC047324 | CSPG6 |
| H1-280 | AJ704890 | 251 | cytochrome P-450 (SCC) | SS | 99 | 249 | X13768 | CYP11A1 |
| H1-365 | AJ704898 | 110 | 7-dehydrocholesterol reductase (DHCR7) | HS | 95 | 111 | AF062481 | DHCR7 |
| H1-140 | AJ704881 | 143 | farnesyl diphosphate synthase | BT | 89 | 75 | AF461050 | FDPS |
| H1-185 | AJ783757 | 366 | glutathione peroxidase 3 (plasma) (GPX3) | HS | 87 | 354 | NM_002084 | GPX3 |
| H2-200 | AJ704914 | 179 | glycogenin | HS | 83 | 179 | NM_004130 | GYG |
| H2-105 | AJ704908 | 228 | Homo sapiens heterochromatin protein 1, binding protein 3 | HS | 92 | 219 | BC045660 | HP1BP3 |
| H1-1 | AJ704866 | 256 | 3 β hydroxysteroidogenase | SS | 99 | 256 | AF232699 | HSD3B1 |
| H2-265 | AJ704919 | 299 | inositol polyphosphate-1-phosphatase | HS | 89 | 229 | NM_002194 | INPP1 |
| H2-175 | AJ704912 | 187 | matrin 3 | HS | 88 | 186 | NM_018834 | MATR3 |
| H1-100 | AJ704877 | 237 | NAC alpha | HS | 92 | 237 | AY034001 | NACA |
| H1-242 | AJ704885 | 481 | complete mitochondrial DNA | SS | 100 | 450 | AJ002189 | NADH1 |
| H2-201 | AJ785755 | 170 | Sus scrofa breed Landrace mitochondrion | SS | 100 | 170 | AF304202 | NADH5 |
| H1-33 | AJ704869 | 141 | peroxiredoxin 2 | BT | 86 | 117 | AF305562 | PDX2 |
| H1-266 | AJ704888 | 135 | non-selenium glutathione phospholipid hydroperoxide peroxidase (phgpx gene) | SS | 100 | 135 | AJ243849 | phgpx |
| H2-29 | AJ704900 | 123 | proteasome (prosome, macropain) 26S subunit, ATPase | HS | 86 | 123 | NM_002806 | PSMC6 |
| H1-358 | AJ704897 | 87 | cDNA DKFZp686F06244 | HS | 97 | 87 | HSM806676 | Q8NEW0 |
| H1-73 | AJ704872 | 120 | ribosomal protein S15A | HS | 93 | 130 | BC001697 | RPS15A |
| H1-129 | AJ704879 | 467 | Sterol regulatory element binding protein cleavage-activating protein (SCAP) | SS | 88 | 486 | AY705447 | SCAP |
| H1-90 | AJ704873 | 199 | high density lipoprotein receptor SR-BI mRNA | SS | 99 | 199 | AF467889 | SCARB1 |
| H1-96 | AJ704875 | 404 | splicing factor 3a, subunit 3 | HS | 93 | 401 | NM_006802 | SF3A3 |
| H1-62 | AJ704871 | 90 | solute carrier family 25 | HS | 85 | 98 | BC008737 | SLC25A6 |
| H2-44 | AJ704902 | 529 | thrombospondin 1 | BT | 90 | 527 | AB005287 | THBS1 |
| H1-94 | AJ704874 | 114 | tenascin | HS | 91 | 114 | D49542.1 | TNC |
| H2-72 | AJ704904 | 133 | ubiquitin associated protein 2 (UBAP2), transcript variant 1 | HS | 91 | 106 | NM_018449.2 | UBAP2 |
| H1-202 | AJ704883 | 178 | ubiquitin-conjugating enzyme E2E 2 | HS | 98 | 178 | BC022332 | UBE2E2 |
| H1-305 | AJ704893 | 178 | angiogenic factor VG5Q, | HS | 85 | 169 | BC032844 | VG5Q |
| H2-37 | AJ704901 | 451 | vimentin | HS | 92 | 438 | NM_003380 | VIM |
| H2-170 | AJ704911 | 191 | breed Landrace mitochondrion | SS | 100 | 190 | AF304202 | unknown |
| H2-202 | AJ704915 | 120 | domestica mitochondrial D-loop, | SS | 100 | 120 | AF276937 | unknown |
| H1-180 | AJ704882 | 178 | 885998 MARC 4PIG | SS | 99 | 178 | CF792994 | unknown |
| H1-246 | AJ704886 | 284 | jns82_C06.f jns 5' | SS | 99 | 284 | CB481752 | unknown |
| H1-276 | AJ704889 | 247 | MI-P-CP0-nvm-c-03-0-UI.s2 MI-P-CP0 | SS | 99 | 236 | BQ603741 | unknown |
| H1-294 | AJ704891 | 131 | 855499 MARC 4PIG 5' | SS | 98 | 118 | CF787582 | unknown |
| H1-345 | AJ704895 | 90 | 820907 MARC 3PIG 3' | SS | 10 | 89 | CF359642 | unknown |
| H2-92 | AJ704905 | 497 | jns96_H04.f jns | SS | 99 | 492 | CB482726 | unknown |
| H2-199 | AJ704913 | 236 | 903820 MARC 4PIG 3' | SS | 99 | 236 | CK450726 | unknown |
| H1-338 | AJ704894 | 97 | , Similar to RIKEN cDNA 2410141M05 gene | HS | 91 | 92 | BC008630 | unknown |
| H2-216 | AJ704917 | 107 | RIKEN full-length enriched library, product: ribosomal protein S17 | MM | 94 | 93 | AK007992 | unknown |
| H2-288 | AJ704920 | 162 | CT02029A1F04 Equine Articular Cartilage | EC | 87 | 114 | CX601260 | unknown |
| H1-14 | AJ704868 | 221 | endogenous retrovirus PERV-MSL | SS | 97 | 221 | AF038600 | |
| H1-302 | AJ704892 | 108 | None | SS | ||||
| H2-95 | AJ704906 | 137 | None | SS | ||||
| H2-206 | AJ704916 | 658 | None | SS | ||||
Comparative RT-PCR analysis
The comparative RT-PCR was performed as described by Tosser-Klopp et al [13]. Briefly, 0.5 μCi of α 32P labelled dCTP (specific activity >3000 Ci.mmole-1, Perkin Elmer) was added to each reverse transcription to monitor the transcription. First strand cDNAs were purified by G50 chromatography and quantified by measuring the incorporated radioactivity. For each cDNA sample (control and FSH-treated cells) the same dilutions were made starting from the same amount of material. The PCR conditions were: denaturation 5 min at 94°C, followed by (22–40) cycles of (30 s, 94°C, 30 s, 56°C, 1.5 min, 72°C) in presence of 1.5 mM MgCl2. The primers are listed in table 4. Ten μl of PCR products were analysed by electrophoresis on 1.5% agarose gel. Each experiment was performed at least three times. An external standard (plant mRNA: I11a accession number: y10291) was added to each RNA sample (200 fg for 2 μg total RNA sample) before reverse transcription and allowed the control of the cDNAs quantifications and dilutions using specific primers (Figure 1).
Table 4.
Localisation on pig chromosomes. This table gives the localisation obtained using SCHP and/or ImpRH panel hybrid. The result of SCHP consists in the chromosomal region with statistical scores: error risk and probability. For IMpRH the results were obtained by 2-point analysis. We indicate the number of hybrid clones used (90 or 118 Hybrids), the retention as a % frequency (% ret), the bearing chromosome, the closest marker, the location in Ray, and the load score value. We have indicated the chromosomal localisation in humans when known; the expected localisation in pigs derives from comparative mapping between humans and pigs.
| Clone | Putative identification | Somatic localisation | IMpRH panel used | IMpRH localisation using two point analysis | Human localisation | pig expected localisation | ||||||
| Location | % Error risk | Prob. | % ret | Chr | Name of the most linked marker | Location | LOD | |||||
| H1-2 | AKR1C3 | 90 | 34 | 14 | EST-AR023C05 | 4.74 | 4.81 | 10p 15 p 14 | 10 | |||
| H1-61 | col4A5 | X p 24 | 0.10 | 0.77 | 90 | 50 | X | SW1426 | 7.1 | 14.32 | X q22 | X |
| H1-129 | SCAP | 90 | 25 | 13 | EST-AR062F01 | 11.44 | 19.28 | 3 p 21.31 | 13 | |||
| H1-140 | FDPS | 4 p14-p15 | 0.10 | 1.00 | 90 | 24 | 4 | IMpRH01700 | 26.17 | 15.94 | 1 q 22 | 4 |
| H1-180 | EST | 9 p21-p24 | 5 | 0.63 | 118 | 35 | 9 | SW911 | 3.83 | 10.89 | ||
| H1-246 | EST | 9 p21-p24 | 5 | 0.63 | 118 | 40 | 9 | SW911 | 3.83 | 14.49 | ||
| H1-276 | EST | 10 p1-p16 | 0.10 | 0.80 | 118 | 29 | 2 | PTH3 | 10.97 | 5.05 | ||
| H2-44 | THBS1 | 1 p24-p25 | 1 | 0.81 | 118 | 29 | 1 | 348A10F | 27.08 | 5.95 | 15 q 15 | 1 |
| H2-92 | EST | 14? | 0.50 | 1.00 | 90 | 31 | 14 | RhC21D6F | 19.69 | 15.71 | ||
| H2-95 | EST | 4 p14-p15 | 1 | 0.76 | 118 | 35 | 4 | S0107 | 18.07 | 12.08 | ||
Figure 1.
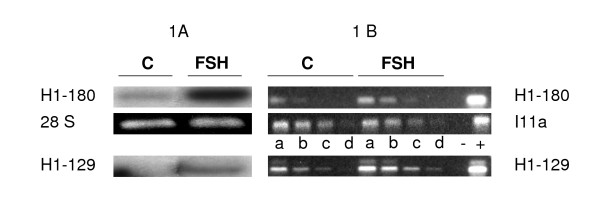
validation of comparative RT-PCR. Comparison ofNorthern and RT-PCR analysis with H1-180 and H1-129. A shows Northern analysis of total RNA (5 μg) from control (C) or FSH-treated GCs (FSH). Hybridization was performed with H1-180 and H1-129 cDNA probes coming from forward hybridization (H1 = FSH-induced genes). The amount and integrity of RNA in each lane was checked with ethidium bromide staining of the gels before transfer (28 S lane). B shows the results of PCR amplification of H1-180 and H1-129 cDNAs using specific primers. Total RNA was extracted from control (C) or FSH-treated (FSH) cells and was reverse-transcribed. For PCR, four different amounts of each cDNA were used: a: 1 ng, b: 250 pg, c: 50 pg, d: 10 pg and 2 controls (-: water, +: corresponding insert). PCR amplification of I11a (plant external control added to each RNA before RT) was performed on the same samples (control and FSH-treated cDNAs) to check efficiency of the RT and PCR processes.
Regional assignment
Genomic localisation of the PCR products was done using the pig somatic cell hybrid panel (SCHP) as described previously by Tosser-Klopp et al. [13]. Each hybrid was scored for the presence of a pig specific DNA fragment and the assignments were performed using online software [14].
We also used the IMpRH panel 7000 rads [15] as described by Lahbib-Mansais et al.[16]. The results of radiation hybrid PCR products were analysed using the IMpRH mapping tool [17,18].
Functional Gene Ontology annotation
The functional annotation analysis was performed on the 25 genes found to be regulated by Northern or comparative RT-PCR.
Since our sequences are rather short, and mainly belong to 3'UTR portions of genes, available public tools to assign GO molecular function to nucleic acid sequences lack sensitivity. So we set up the following workflow:
- Each sequence was compared to EMBL database via a NCBI blastn with an e-value limit set to 1e-3
- Blast results were parsed and filtered to keep hits having a) at least a coverage of 50% of the query sequence, b) 70% of identities, c) its coordinates inside a gene/mRNA/CDS location on the subject sequence.
- For each query sequence, candidate hits were sorted by a) their closeness to the pig genome, and b) their coverage and percent of identity.
- Annotations present in the EMBL record of the hit were parsed to look for cross references to proteins from UniProt/SwissProt databases
- Finally, using cross reference tables provided by GOA http://www.ebi.ac.uk/GOA/, molecular function GO was inferred from these best hits and associated to the sequences.
Results
Suppression subtractive hybridization
The SSH approach was performed on primary granulosa cells from cultures treated or not treated by FSH. mRNA from control granulosa cells was subtracted from the mRNA from FSH treated granulosa cells and vice versa. We isolated 226 clones from the forward hybridization H1 (FSH/C, FSH-induced genes) and 275 from the reverse subtraction H2 (C/FSH, FSH-repressed genes). In order to eliminate false positive clones, differential screening was used. The clones were spotted on two sets of macro-arrays and differential screening was performed using cDNA from the control and the treated cells as probes. We identified 28 (12.4%) induced cDNA clones and 19 (6.9%) repressed cDNA clones (Table 1). We also randomly selected 29 cDNA clones in both forward and reverse subtraction which provided no differential signals or no signal at all.
Table 1.
Differential screening by macro-arrays. This table summarises the results of differential screening by SSH and the results of sequence analysis: Forward subtraction corresponds to H1 SSH and up-regulated genes by FSH (FSH/C). Reverse one corresponds to H2 SSH and down-regulated genes by FSH (C/FSH). Random selection corresponds to clones that provided no differential signals or no signal at all in both forward and reverse subtraction.
| Forward (FSH/C) | reverse (C/FSH) | random selection | total | |
| number of clones | 226 | 275 | 501 | |
| number of selected clones after screening | 28 (12.4%) | 19 (6.9%) | 29 | 76 |
| different clones | 20 | 15 | 29 | 64 |
| different genes | 18 | 12 | 27 | 55 |
Sequence analysis
These 76 selected clones (28+19+29) were sequenced and compared against public sequence databases. We identified 64 different sequences of which 3 were novel and 14 corresponded to different regions of the same 5 genes. We finally dealt with 55 genes. The results are summarized in Table 1. The complete list of genes is given in Table 2.
Regulation study
Northern and comparative RT-PCR were used to analyse the differential expression of the selected genes. Out of the 55 genes, 18 could not be analysed according to their short length.
The results of the 37 genes left are presented in Table 3. Some genes could not be analysed by Northern (no signal or non interpretable results) and were thus analysed by RT-PCR. Also, 9 genes which gave a good signal by Northern analysis were also analysed by RT-PCR to compare the results (Fig 1).
Table 3.
RNA expression. Results of RNA expression for the 37 genes. Nine genes are analysed by both Northern and comparative RT-PCR. For comparative RT-PCR the regulation was determined using cDNA dilutions from control cells and FSH treated cells as in Figure 1 and 2. In the Table, C = FSH corresponds to non regulated genes by FSH, FSH>C corresponds to up-regulated genes and C>FSH corresponds to down-regulated genes. NI corresponds to Non Interpretable, 0 corresponds to no signal. * these genes gave no signals during macro-array screening and thus were not controlled by Northern.
| clones | putative identification | Northern | RT-PCR result | primers | Size of PCR product (pb) |
| H2-146 | ACTA2 | C>FSH | |||
| H2-96 | ACTG1 | C>FSH | |||
| H1-2 | AKR1C3 | FSH>C | U: GATCATCTCCAGCTGCTTGT L: AGGAGCTTCTGCCTAAGGAT |
123 | |
| H2-222 | ANXA5 | C>FSH | |||
| H1-98 | ATP6 | NI | C = FSH | U: TACGGCTAGGGCTACTGG L: TTCCGCCATAAAACCAAAAC |
126 |
| H1-138 | CAPNS1 | C = FSH | C = FSH | U: ACGGGGTGAGTCTAATGC L: CCAGTGAAGTGCCCAGAA |
348 |
| H1-357 | CCT3 | FSH>C | |||
| H1-124 | CLU | C = FSH | C = FSH | U: ACCCCAGCGTG CCTCT L: TTCCTCCGCCACAGTCTC |
122 |
| H1-61 | Col4A5 | FSH>C | FSH>C | U: CATAAAAAGCACAGGGAAAAG L: TTTCTCTTCTGCTGTATTCG |
172 |
| H2-108 | COX II | FSH>C | |||
| H1-280 | CYP11A1 | NI | FSH>C | U: CAAGTCATTCACGAGGTATC L: AGGAGATGCTGCGGGAG |
161 |
| H1-185 | GPX3 | FSH>C | |||
| H2-105 | HP1BP3 | * | FSH>C | U: CTCAAGTCCATTTCCCAGC L: TCTGGCTCCTTGCGTGCT |
114 |
| H1-1 | HSD3B1 | FSH>C | |||
| H2-265 | INPP1 | C>FSH | |||
| H2-175 | MATR3 | * | C>FSH | U: GTTTTTGTTTTATCAGAATGG L: CTTATCTTTCTTAGTGGTGC |
91 |
| H1-100 | NACA | C = FSH | C = FSH | U: AAG AGG AGA GTG AAG AGG A L: TTGAAACACCAAAAAAAGGGT |
197 |
| H1-242 | NADH1 | 0 | C = FSH | U: TTGGTGAATAGTTTTAGGGC L: CGAAAGGACAAGAGAAATGG |
360 |
| H1-33 | PDX2 | NI | C = FSH | U: TGACCCAGGAAAGCCAGA L: CTATGTCGCTCCAGGAAA |
129 |
| H1-14 | porcine endogenous retrovirus perv-MSL | 0 | FSH>C | U: AAAACAGCAAAAAACCTAACC L: GCCCGTCTAACAAGAAAGC |
180 |
| H1-73 | RP515A | C = FSH | |||
| H1-129 | SCAP | FSH>C | FSH>C | U: CCAGCCCAGATCCAGTG L: TAAACTACGGGGACCTGT |
250 |
| H1-90 | SCARB1 | NI | FSH>C | U: CAAGAAGCAAGACTGTAGG L: TGGCAACGGGAGGTGAG |
160 |
| H1-62 | SLC25A6 | NI | C = FSH | U: GTTCTCTTTTGCACAGCCG L: TTTTTTTGTGTCCTGATTTTATT |
86 |
| H2-44 | THBS1 | C>FSH | U: CTTTTCGTCTCCCTGGAAAT L: TGAAACTGATGGGCAAATCT |
529 | |
| H1-94 | TNC | 0 | C>FSH | U: GCTCCGTGGTGGACCTG L: CCCTCCACCTTCAGCTTG |
104 |
| H1-305 | VG5Q | 0 | C = FSH | U: CCAGCATAAGCATCATTTTC L: AACAGAGAACCACCCTCCT |
174 |
| H2-37 | VIM | 0 | C = FSH | U: GCTCAAGGGGACCAACGA L: TGGAAGAGGCAGAGAAATC |
253 |
| H2-95 | New | 0 | C>FSH | U: AGTCACCAATCTTATCTCCA L: ATGAAACAATAGTCCAGGAAG |
119 |
| H1-202 | unknown | C = FSH | C = FSH | U: ATCCTCAGTCAAAAGTGGCA L: CAATCAGAATCACTGTGCGT |
154 |
| H2-288 | unknown | * | C = FSH | U: AAGGTGTGTTGATGTATTTTA L: TACAGGAAGGGAAGCATC |
77 |
| H1-294 | unknown | FSH>C | FSH>C | U: CTTAGTTTCAGACTGGAGTT L: TTTAGCGGCTTGTTCACTCA |
104 |
| H2-92 | unknown | * | FSH>C | U: ATGCCAACATCATCACCTCT L: CCCTCTAAACTGGGATCCAT |
351 |
| H2-170 | unknown | FSH>C | FSH>C | U: AGACTTGTAGGTAGAGGTGA L: TAGGGTTTTTGGGGTATTTTT |
99 |
| H1-180 | unknown | FSH>C | FSH>C | U: AAA TTG TAG GTA TAT GTG TCA L: TAGAGAATAAAATGGTCAGTAA |
123 |
| H1-246 | unknown | FSH>C | U: CAATTCACCATCATCCACAA L: CCCAGCATTTCATACTGACC |
284 | |
| H1-276 | unknown | FSH>C | U: ACGATTCCTCTACTGAAAGCTG L: TGTTTTTGGCAAACAGCAG |
247 |
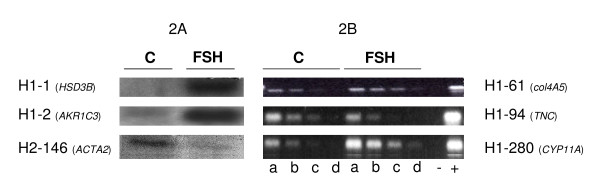
Finally, out of the 37 genes, 25 were FSH-regulated (Table 3). In agreement with literature, HSD3B1 and CYP11A expression was induced by FSH in granulosa cells and ACTA2 expression was repressed by FSH (Fig 2).
Figure 2.
Regulation studies. Regulation by FSH of 6 genesusing Northern or comparative RT-PCR. H1 corresponds to forward hybridization (FSH-induced genes) and H2 to reverse hybridization (FSH-repressed genes). A: Northern analysis. B: comparative RT-PCR. Same conditions as in Figure 1.
FSH regulation of the expression of some other genes (AKR1C3, col4A5, TNC) is illustrated in Fig 2.
Chromosomal localisation
Among the same 37 genes, 11 had already been located on pig chromosomes. Using the SCHP and the IMpRH hybrid panel, and the same primer pairs as for regulation studies, we tried to localise the 26 genes left. We successfully localised 10 genes, for the other 16 genes, the primers used amplified rodent DNA and stopped the analysis of the results (Table 4).
Functional gene ontology analysis
The 25 regulated genes were analysed using gene ontology annotation in order to document the processes involved in FSH effect on granulosa cells from medium sized follicles.
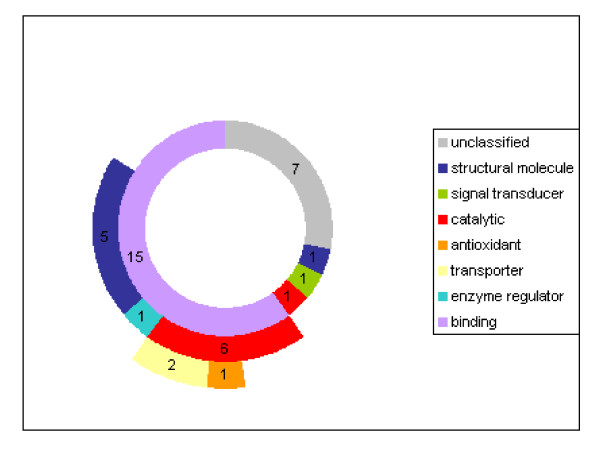
For each sequence, we tried to assign molecular function GO identifiers using the methodology explained previously. We limited the analysis to the first levels of the molecular function ontology tree, just counting genes present in each node. The donut chart of figure 3 shows the distribution of the regulated genes according to their molecular function (see Additional file: supplementaldata-fig 3 for the data used to draw the chart). As genes can be assigned to more than one molecular function, some categories are drawn as outer partial rings in the donut chart (see also supplemental data). Numbers in this chart represent the number of genes observed for each category.
Figure 3.
Functional annotation: Molecular function. The donut chart shows the distribution of the 25 regulated genes according to their molecular function (inner circle). Genes that can be assigned to more than one molecular function are indicated by outer partial rings in the donut chart. Numbers in this chart represent the number of sequences for each category.
Discussion
Our objective has been to identify the genes regulated in granulosa cells in response to FSH stimulation. The identification of such genes will give valuable information about the molecular processes associated with follicular growth. An increasing number of studies are being undertaken on candidate genes that regulate folliculogenesis, e.g. IGF [18], EGF [19], TGF-β [20] and other members of the transforming growth factor superfamily, like BMPs [21,22] and GDF-9 [23,24]. In parallel with such individual studies, we now have access to technologies that allow us to study a large number of genes simultaneously. We applied the SSH technique coupled with a differential screening procedure, to GCs treated with FSH compared with untreated controls. Among 501 clones initially obtained, 76 were sequenced and were found to correspond to 55 different genes (Table 2). Sequence analysis revealed redundancy from 2 different mechanisms: 1) gene redundancy, and 2) clone redundancy. This phenomenon is observed for highly expressed genes like- HSD3B1 and alpha-actin. High level of expression can also explain why two genes (alpha-actin and complement cytosolic inhibitor) are present in both the forward and reverse libraries (data not shown): the subtraction process may not be efficient for them.
Regulation studies
Thirty seven genes were analysed for regulation. Ten genes did not give a signal with Northern blot or macroarrays (table 3) and were successfully analysed by RT-PCR, underlining the low level of expression of some genes identified through SSH: they probably correspond to rare transcripts.
The Cox2 gene and H2-170 gave discordant results when using subtraction or Northern analysis, possibly due to the high abundance of their mRNAs [25,26]. However, Northern experiments demonstrated clearly their high expression and regulation in granulosa cells.
We found some genes already known to be regulated by FSH in pigs e.g. HSD3B and CYP11A which are upregulated by FSH (Figure 2). We corroborated in pigs that alpha and gamma actin were downregulated by FSH, whereas vimentin was not affected, as observed in rats [27]. These genes validate the biological model and the analytical methods, as well as reinforcing previous studies [28].
Localisation of cDNA and comparative mapping
This paper describes 10 localisations of genes or ESTs on the pig genome using the somatic or IMpRH hybrid panel. Three genes and 4 ESTs were assigned with a LOD score of >10. Localisations of these genes were in accordance with the data from the human genome. In spite of a relatively low LOD score of 5.95 (significance limit 6), THBS1 was assigned on chromosome 1 in agreement with the expected localisation, using a comparative map between humans and pigs [29,30].
The aim of these localisations was to identify candidate genes for reproductive Quantitative Trait Loci (QTL). This could be the case for col4A5 which is located on chromosome X at the same place as a plasma FSH concentration QTL [31]. Col4A5 was up regulated by FSH and is a positional candidate for this QTL. This result obviously deserves further investigation.
Functional gene ontology analysis
In order to understand the pathways involved in the GC response to FSH, GO was used to cluster regulated genes according to their molecular function. Most of the tools used for automatic GO annotation of EST sequences, such as GOst [32] or OntoBlast [33], are mainly based on homology searches against well annotated protein databases. In our case, the sequence information obtained from the SSH clones is relatively short, and is biased towards 3' UTR; thus these tools are not convenient. Some other tools, such as Goblet [34], GoFigure [35] or Blast2GO [36] allow homology searches against nucleic databases, but once again with low sensitivity for short sequences. We thus used an in-house procedure to browse the GO classification and extract valuable information. The resulting classification (Fig 3 and Additional file) brings to light mainly 5 functional activities: "catalytic" and "signal transducer" activities directly linked with the steroid activity then "binding", "antioxidant" and "structural molecule" activities predominantly linked with differentiation pathways. On the other hand, our data demonstrate for the first time the involvement and/or the regulation of different genes in:
Catalytic activities
COX-2 was upregulated by FSH in our study and has been implicated as an important factor in female fertility [37]. In response to FSH, COX-2 induction could then stimulate progesterone production via the PGE2/EP2 pathway and play a role in ovulation by supporting cumulus expansion [38,39]. Indeed, mice deficient for COX-2 failed to ovulate, showing that COX-2 is necessary for ovulation [40].
Binding activity
Our data underline the FSH regulation of 4 genes involved in 1) DNA binding (HP1-BP74) 2) nucleic acid binding (CCT3), 3) lipid binding (annexin V), and 4) metal ion binding (matrin).
1) The upregulation of the HP1-BP74 gene (histone h1/h5 family), has never been shown before to be involved in granulosa cells development. Histones are highly conserved proteins involved in the package of chromosomes by interacting with DNA. Particularly, the HP1 family plays an important role in chromosomal biology and gene silencing. The HP1-BP74 gene was also shown to be involved in neuronal functional maturation [41]. This finding suggests an important role of H2-105 in GC differerentiation.
2) This study shows also the FSH up regulation of the CCT3 gene (chaperonin containing TCP1) known to mediate the folding of alpha- and beta-tubulin. FSH could thus intensify the remodelling of the microtubule cytoskeleton. Such a modification has been shown to be related to the maintenance and remodelling of heterochromatin during mammalian spermiogenesis [42].
3) We demonstrated the downregulation of annexin V: its binding to the cell membrane corresponds to the earlier events of apoptosis and is used to detect healthy live cells (annexin V negative) [43,44]. It is also a protein kinase C inhibitor which plays a potential role in cellular signal transduction [45]. This downregulation by FSH in GC may prevent apoptosis.
4) Finally, we found the downregulation of matrin by FSH. This protein that is localized in the nuclear matrix may have a role in RNA transcription thanks to its acidic region [46] and could be phosphorylated by nuclear PKC epsilon [47].
Signal transducer activity
Among the genes involved in signal transducer activity, our study showed the upregulation by FSH of the Scavenger Receptor Class B Type I (SCARB1). This receptor is involved in both cholesterol delivery for steroid hormone production and in the recognition of apoptotic granulosa cells. We underline here the relationship with steroid hormone production, according to the downregulation of annexin V by FSH that suggests a differentiation process rather than an atretic one [48].
Anti-oxidant activities
During follicle growth, swine granulosa cells are physiologically exposed to a progressive oxygen shortage. In vitro reactive oxygen species (ROS), such as hydrogen peroxide (H2O2) and lipid hydroperoxide (LOOHs) provided by oxido-reduction reactions, can either negatively or positively affect the differentiation of the gametes notably during spermatogenesis [49,50]. Among the anti-oxidant activities, we noticed the upregulation by FSH of genes like glutathione peroxydase 3 (GPX3). GPX3 seems to regulate free hydoxyperoxides. The upregulation of this peroxidase activity by FSH could prevent atresia by protecting the membrane lipids and allowing differentiation of the follicle.
Structural molecule activity
We identified several interesting regulated genes in the structural molecule activity because they intervene in the extracellular matrix and cytoskeleton. Adhesion proteins, such as type IV collagen, increase the connections between cells and have also been shown to increase FSH receptors and progesterone production of GCs from immature porcine ovarian follicles [51]. The stimulation by FSH of type IV collagen gene expression could thus reinforce the effect of FSH and play a role in the local control of ovarian follicular dynamics [52].
Conclusion
These results demonstrate the validity of both our cellular model and the SSH approach in identifying genes involved in response to FSH. In this way, and in addition to the regulation of the steroidogenesis and morphological changes already described, our data suggests that there is a role of FSH in the chromatin remodelling and protection against peroxides leading the follicle into a differentiation process rather than into atresia. Interestingly, we have been able to demonstrate the involvement and/or regulation of new genes such as HP1-BP74, cox-2, CCT3, SCARB1, GPX3, and also of unidentified genes. These new or unidentified genes will require further studies. Particularly, expression studies associated with histological techniques (in situ hybridization, immunohistochemistry) will allow a better understanding of the involvement of these genes.
SSH has demonstrated its efficiency in our hands. We will now use it to further improve our knowledge of folliculogenesis in pigs by analysing fresh GC from ovarian follicles at different developmental stages.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
AB conceived the design of the study, carried out the cell culture, mRNA extraction, regional assignment and gene ontology annotation, participated in the regulation study and drafted the manuscript. POF carried out the SSH, screening procedure, sequencing and regulation study. PD performed gene ontology annotation. GTK participated in its design and helped to write the manuscript. FH conceived of the study, supervised the experiment and helped to write the manuscript.
Supplementary Material
Functional annotation data. This table gives the results of the analysis for the first levels of the molecular function ontology tree. The regulated genes are listed according to their molecular function(s). These data are used to draw the donut chart (fig 3)
Acknowledgments
Acknowledgements
We thank Francis Benne for technical assistance, Janine Rallières and Florence Mompart for regional mapping and Philippe Monget for helping to produce the manuscript.
Contributor Information
A Bonnet, Email: abonnet@toulouse.inra.fr.
PO Frappart, Email: Pierre-Olivier.Frappart@STJUDE.ORG.
P Dehais, Email: dehais@toulouse.inra.fr.
G Tosser-Klopp, Email: tosser@toulouse.inra.fr.
F Hatey, Email: hatey@toulouse.inra.fr.
References
- Otsuka F, Moore RK, Wang X, Sharma S, Miyoshi T, Shimasaki S. Essential role of the oocyte in estrogen amplification of follicle-stimulating hormone signaling in granulosa cells. Endocrinology. 2005;146:3362–3367. doi: 10.1210/en.2005-0349. [DOI] [PubMed] [Google Scholar]
- Kumar TR, Wang Y, Lu N, Matzuk MM. Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nat Genet. 1997;15:201–204. doi: 10.1038/ng0297-201. [DOI] [PubMed] [Google Scholar]
- Plancha CE, Sanfins A, Rodrigues P, Albertini D. Cell polarity during folliculogenesis and oogenesis. Reprod Biomed Online. 2005;10:478–484. doi: 10.1016/s1472-6483(10)60824-3. [DOI] [PubMed] [Google Scholar]
- Tosser-Klopp G, Benne F, Bonnet A, Mulsant P, Gasser F, Hatey F. A first catalog of genes involved in pig ovarian follicular differentiation. Mamm Genome. 1997;8:250–254. doi: 10.1007/s003359900403. [DOI] [PubMed] [Google Scholar]
- Caetano AR, Johnson RK, Pomp D. Generation and sequence characterization of a normalized cDNA library from swine ovarian follicles. Mamm Genome. 2003;14:65–70. doi: 10.1007/s00335-002-2220-5. [DOI] [PubMed] [Google Scholar]
- Caetano AR, Johnson RK, Ford JJ, Pomp D. Microarray profiling for differential gene expression in ovaries and ovarian follicles of pigs selected for increased ovulation rate. Genetics. 2004;168:1529–1537. doi: 10.1534/genetics.104.029595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladney CD, Bertani GR, Johnson RK, Pomp D. Evaluation of gene expression in pigs selected for enhanced reproduction using differential display PCR and human microarrays: I. Ovarian follicles. J Anim Sci. 2004;82:17–31. doi: 10.2527/2004.82117x. [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci U S A. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatey F, Langlois I, Mulsant P, Bonnet A, Benne F, Gasser F. Gonadotropins induce accumulation of insulin-like growth factor I mRNA in pig granulosa cells in vitro. Mol Cell Endocrinol. 1992;86:205–211. doi: 10.1016/0303-7207(92)90145-V. [DOI] [PubMed] [Google Scholar]
- Hatey F, Gasparoux JP, Mulsant P, Bonnet A, Gasser F. P450scc regulation in pig granulosa cells: Investigation into the mechanism of induction. J Steroid Biochem Mol Biol. 1992;43:869–874. doi: 10.1016/0960-0760(92)90314-9. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- Hatey F, Mulsant P, Bonnet A, Benne F, Gasser F. Protein kinase C inhibition of in vitro FSH-induced differentiation in pig granulosa cells. Mol Cell Endocrinol. 1995;107:9–16. doi: 10.1016/0303-7207(94)03420-X. [DOI] [PubMed] [Google Scholar]
- Tosser-Klopp G, Bonnet A, Yerle M, Hatey F. Functional study and regional mapping of 44 hormono-regulated genes isolated from a porcine granulosa cell library. Genet Sel Evol. 2001;33:69–87. doi: 10.1051/gse:2001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHP. p. [http://www.toulouse.inra.fr/lgc/pig/pcr/pcr.htm].
- Yerle M, Pinton P, Robic A, Alfonso A, Palvadeau Y, Delcros C, Hawken R, Alexander L, Beattie C, Schook L, Milan D, Gellin J. Construction of a whole-genome radiation hybrid panel for high-resolution gene mapping in pigs. Cytogenet Cell Genet. 1998;82:182–188. doi: 10.1159/000015095. [DOI] [PubMed] [Google Scholar]
- Lahbib-Mansais Y, Tosser-Klopp G, Leroux S, Cabau C, Karsenty E, Milan D, Barillot E, Yerle M, Hatey F, Gellin J. Contribution to high-resolution mapping in pigs with 101 type I markers and progress in comparative map between humans and pigs. Mamm Genome. 2003;14:275–288. doi: 10.1007/s00335-002-2236-x. [DOI] [PubMed] [Google Scholar]
- IMpRH mapping tool. p. [http://imprh.toulouse.inra.fr].
- Minegishi T, Hirakawa T, Abe K, Kishi H, Miyamoto K. Effect of insulin-like growth factor-1 and 2,3,7,8-tetrachlorodibenzo-p-dioxin on the expression of luteinizing hormone receptors in cultured granulosa cells. Environ Sci. 2004;11:57–71. [PubMed] [Google Scholar]
- Mao J, Smith MF, Rucker EB, Wu GM, McCauley TC, Cantley TC, Prather RS, Didion BA, Day BN. Effect of epidermal growth factor and insulin-like growth factor I on porcine preantral follicular growth, antrum formation, and stimulation of granulosal cell proliferation and suppression of apoptosis in vitro. J Anim Sci. 2004;82:1967–1975. doi: 10.2527/2004.8271967x. [DOI] [PubMed] [Google Scholar]
- May JV, Stephenson LA, Turzcynski CJ, Fong HW, Mau YH, Davis JS. Transforming growth factor beta expression in the porcine ovary: evidence that theca cells are the major secretory source during antral follicle development. Biol Reprod. 1996;54:485–496. doi: 10.1095/biolreprod54.2.485. [DOI] [PubMed] [Google Scholar]
- Mulsant P, Lecerf F, Fabre S, Schibler L, Monget P, Lanneluc I, Pisselet C, Riquet J, Monniaux D, Callebaut I, Cribiu E, Thimonier J, Teyssier J, Bodin L, Cognie Y, Chitour N, Elsen JM. Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Merino ewes. Proc Natl Acad Sci U S A. 2001;98:5104–5109. doi: 10.1073/pnas.091577598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monget P, Fabre S, Mulsant P, Lecerf F, Elsen JM, Mazerbourg S, Pisselet C, Monniaux D. Regulation of ovarian folliculogenesis by IGF and BMP system in domestic animals. Domest Anim Endocrinol. 2002;23:139–154. doi: 10.1016/S0739-7240(02)00152-2. [DOI] [PubMed] [Google Scholar]
- McNatty KP, Juengel JL, Reader KL, Lun S, Myllymaa S, Lawrence SB, Western A, Meerasahib MF, Mottershead DG, Groome NP, Ritvos O, Laitinen MP. Bone morphogenetic protein 15 and growth differentiation factor 9 co-operate to regulate granulosa cell function in ruminants. Reproduction. 2005;129:481–487. doi: 10.1530/rep.1.00517. [DOI] [PubMed] [Google Scholar]
- Vitt UA, Hayashi M, Klein C, Hsueh AJ. Growth differentiation factor-9 stimulates proliferation but suppresses the follicle-stimulating hormone-induced differentiation of cultured granulosa cells from small antral and preovulatory rat follicles. Biol Reprod. 2000;62:370–377. doi: 10.1095/biolreprod62.2.370. [DOI] [PubMed] [Google Scholar]
- Rockett JC, Swales KE, Esdaile DJ, Gibson GG. Use of suppression-PCR subtractive hybridisation to identify genes that demonstrate altered expression in male rat and guinea pig livers following exposure to Wy-14,643, a peroxisome proliferator and non-genotoxic hepatocarcinogen. Toxicology. 2000;144:13–29. doi: 10.1016/S0300-483X(99)00214-0. [DOI] [PubMed] [Google Scholar]
- Li WB, Gruber CE, Lin JJ, Lim R, D'Alessio JM, Jessee JA. The isolation of differentially expressed genes in fibroblast growth factor stimulated BC3H1 cells by subtractive hybridization. Biotechniques. 1994;16:722–729. [PubMed] [Google Scholar]
- Ben-Ze'ev A, Amsterdam A. In vitro regulation of granulosa cell differentiation. Involvement of cytoskeletal protein expression. J Biol Chem. 1987;262:5366–5376. [PubMed] [Google Scholar]
- Clouscard-Martinato C, Mulsant P, Robic A, Bonnet A, Gasser F, Hatey F. Characterization of FSH-regulated genes isolated by mRNA differential display from pig ovarian granulosa cells. Anim Genet. 1998;29:98–106. doi: 10.1046/j.1365-2052.1998.00279.x. [DOI] [PubMed] [Google Scholar]
- Goureau A, Yerle M, Schmitz A, Riquet J, Milan D, Pinton P, Frelat G, Gellin J. Human and porcine correspondence of chromosome segments using bidirectional chromosome painting. Genomics. 1996;36:252–262. doi: 10.1006/geno.1996.0460. [DOI] [PubMed] [Google Scholar]
- Lahbib-Mansais Y, Dalias G, Milan D, Yerle M, Robic A, Gyapay G, Gellin J. A successful strategy for comparative mapping with human ESTs: 65 new regional assignments in the pig. Mamm Genome. 1999;10:145–153. doi: 10.1007/s003359900960. [DOI] [PubMed] [Google Scholar]
- Rohrer GA, Wise TH, Lunstra DD, Ford JJ. Identification of genomic regions controlling plasma FSH concentrations in Meishan-White Composite boars. Physiol Genomics. 2001;6:145–151. doi: 10.1152/physiolgenomics.2001.6.3.145. [DOI] [PubMed] [Google Scholar]
- Gost. pp. [http://www.godatabase.org/cgi–bin/gost/gost.cgi].
- Zehetner G. OntoBlast function: From sequence similarities directly to potential functional annotations by ontology terms. Nucleic Acids Res. 2003;31:3799–3803. doi: 10.1093/nar/gkg555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth D, Lehrach H, Hennig S. GOblet: a platform for Gene Ontology annotation of anonymous sequence data. Nucleic Acids Res. 2004;32:W313–7. doi: 10.1093/nar/gkh406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Situ G, Decker K, Schmidt CJ. GoFigure: automated Gene Ontology annotation. Bioinformatics. 2003;19:2484–2485. doi: 10.1093/bioinformatics/btg338. [DOI] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Wang H, Ma WG, Tejada L, Zhang H, Morrow JD, Das SK, Dey SK. Rescue of female infertility from the loss of cyclooxygenase-2 by compensatory up-regulation of cyclooxygenase-1 is a function of genetic makeup. J Biol Chem. 2004;279:10649–10658. doi: 10.1074/jbc.M312203200. [DOI] [PubMed] [Google Scholar]
- Wu YL, Wiltbank MC. Transcriptional regulation of the cyclooxygenase-2 gene changes from protein kinase (PK) A- to PKC-dependence after luteinization of granulosa cells. Biol Reprod. 2002;66:1505–1514. doi: 10.1095/biolreprod66.5.1505. [DOI] [PubMed] [Google Scholar]
- Elvin JA, Yan C, Matzuk MM. Growth differentiation factor-9 stimulates progesterone synthesis in granulosa cells via a prostaglandin E2/EP2 receptor pathway. Proc Natl Acad Sci U S A. 2000;97:10288–10293. doi: 10.1073/pnas.180295197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BJ, Lennard DE, Lee CA, Tiano HF, Morham SG, Wetsel WC, Langenbach R. Anovulation in cyclooxygenase-2-deficient mice is restored by prostaglandin E2 and interleukin-1beta. Endocrinology. 1999;140:2685–2695. doi: 10.1210/en.140.6.2685. [DOI] [PubMed] [Google Scholar]
- Li Q, Li Z, Sun CX, Yu AC. Identification of transcripts expressed under functional differentiation in primary culture of cerebral cortical neurons. Neurochem Res. 2002;27:147–154. doi: 10.1023/A:1014871109943. [DOI] [PubMed] [Google Scholar]
- Soues S, Kann ML, Fouquet JP, Melki R. The cytosolic chaperonin CCT associates to cytoplasmic microtubular structures during mammalian spermiogenesis and to heterochromatin in germline and somatic cells. Exp Cell Res. 2003;288:363–373. doi: 10.1016/S0014-4827(03)00248-9. [DOI] [PubMed] [Google Scholar]
- Prange-Kiel J, Kreutzkamm C, Wehrenberg U, Rune GM. Role of tumor necrosis factor in preovulatory follicles of swine. Biol Reprod. 2001;65:928–935. doi: 10.1095/biolreprod65.3.928. [DOI] [PubMed] [Google Scholar]
- Rosales-Torres AM, Avalos-Rodriguez A, Vergara-Onofre M, Hernandez-Perez O, Ballesteros LM, Garcia-Macedo R, Ortiz-Navarrete V, Rosado A. Multiparametric study of atresia in ewe antral follicles: histology, flow cytometry, internucleosomal DNA fragmentation, and lysosomal enzyme activities in granulosa cells and follicular fluid. Mol Reprod Dev. 2000;55:270–281. doi: 10.1002/(SICI)1098-2795(200003)55:3<270::AID-MRD5>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Dubois T, Mira JP, Feliers D, Solito E, Russo-Marie F, Oudinet JP. Annexin V inhibits protein kinase C activity via a mechanism of phospholipid sequestration. Biochem J. 1998;330 ( Pt 3):1277–1282. doi: 10.1042/bj3301277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P, Dey R, Berezney R. Molecular cloning of matrin 3. A 125-kilodalton protein of the nuclear matrix contains an extensive acidic domain. J Biol Chem. 1991;266:9893–9899. [PubMed] [Google Scholar]
- Xu TR, Rumsby MG. Phorbol ester-induced translocation of PKC epsilon to the nucleus in fibroblasts: identification of nuclear PKC epsilon-associating proteins. FEBS Lett. 2004;570:20–24. doi: 10.1016/j.febslet.2004.05.080. [DOI] [PubMed] [Google Scholar]
- Svensson PA, Johnson MS, Ling C, Carlsson LM, Billig H, Carlsson B. Scavenger receptor class B type I in the rat ovary: possible role in high density lipoprotein cholesterol uptake and in the recognition of apoptotic granulosa cells. Endocrinology. 1999;140:2494–2500. doi: 10.1210/en.140.6.2494. [DOI] [PubMed] [Google Scholar]
- Aitken RJ, Clarkson JS, Fishel S. Generation of reactive oxygen species, lipid peroxidation, and human sperm function. Biol Reprod. 1989;41:183–197. doi: 10.1095/biolreprod41.1.183. [DOI] [PubMed] [Google Scholar]
- Foresta C, Flohe L, Garolla A, Roveri A, Ursini F, Maiorino M. Male fertility is linked to the selenoprotein phospholipid hydroperoxide glutathione peroxidase. Biol Reprod. 2002;67:967–971. doi: 10.1095/biolreprod.102.003822. [DOI] [PubMed] [Google Scholar]
- Sites CK, Kessel B, LaBarbera AR. Adhesion proteins increase cellular attachment, follicle-stimulating hormone receptors, and progesterone production in cultured porcine granulosa cells. Proc Soc Exp Biol Med. 1996;212:78–83. doi: 10.3181/00379727-212-43994. [DOI] [PubMed] [Google Scholar]
- Bortolussi M, Zanchetta R, Doliana R, Castellani I, Bressan GM, Lauria A. Changes in the organization of the extracellular matrix in ovarian follicles during the preovulatory phase and atresia. An immunofluorescence study. Basic Appl Histochem. 1989;33:31–38. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Functional annotation data. This table gives the results of the analysis for the first levels of the molecular function ontology tree. The regulated genes are listed according to their molecular function(s). These data are used to draw the donut chart (fig 3)