Abstract
Denmark and Iceland are countries where the frequency of methicillin-resistant Staphylococcus aureus is very low due to strict infection control and restrictive antibiotic use policies. In contrast, methicillin-resistant S. epidermidis (MRSE) continues to be isolated as a nosocomial pathogen. The molecular typing by pulsed-field gel electrophoresis (PFGE) of 136 MRSE isolates from five hospitals in Denmark and 94 MRSE isolates from one hospital in Iceland collected in 1997 and 1998 defined 40 different patterns. Closely related PFGE types were found in isolates recovered in Iceland, Denmark, Mexico, Uruguay, Greece, and Cape Verde, evidencing for the first time the geographic clonal dissemination of MRSE strains. The large majority (87.4%) of the MRSE isolates studied were multiresistant.
In recent years, Staphylococcus epidermidis, one of the staphylococcal species most frequently isolated from the microflora of humans (20), has emerged as a major pathogen in nosocomial infections. The increasing use of indwelling devices is the primary reason why S. epidermidis is one of the most frequent causes of hospital-acquired bacteremia (18, 23, 32, 41).
Of the S. epidermidis strains circulating in the hospital environment, as many as 70% can be resistant to methicillin (46). Most of these methicillin-resistant S. epidermidis (MRSE) strains are also resistant to other antimicrobial agents (30), and the emergence of S. epidermidis with low-level resistance to glycopeptides (16, 34, 37) is particularly worrisome. Moreover, MRSE strains are considered reservoirs of antimicrobial resistance genes that can be transferred to other staphylococci (3, 45). The acquisition of these elements by other pathogenic species, particularly S. aureus, is a cause of major concern, as it may lead to the redundancy of available therapeutic options.
Infections due to S. epidermidis can be caused by either community- or hospital-acquired isolates (4, 5, 12, 27, 29). Skin of patients and health care workers, medical equipment, clothing of personnel, and environment surfaces can be sources of antibiotic-resistant S. epidermidis strains (2, 19, 39, 40).
Iceland and Denmark are countries with very low frequencies of nosocomial methicillin-resistant S. aureus (MRSA) (22, 44; K. Kristinsson, unpublished data) as a result of the good infection control practices applied in hospitals, strict antimicrobial prescription policies, and low antibiotic consumption (7). The relative geographic isolation of Iceland could be one of the reasons why it has largely avoided the problem of multiresistant bacteria. However, multiresistant pneumococci managed to reach high prevalence levels in a short time (24). In Denmark, the MRSA frequency was reported to be 30% in S. aureus blood isolates in 1967 (14) and 17% in all S. aureus isolates in 1969 (22). The rapid implementation of strict strategies for antimicrobial use containment, particularly the reduction of tetracycline consumption (11), and infection control practices might have been the cause for this decrease in the frequency of MRSA, which gradually declined to less than 0.6% in 1998 (information found at the Statens Serum Institut web site [http://ssi.dk/dk]).
In contrast to the low MRSA frequency currently observed in Iceland and Denmark, MRSE strains continue to be isolated in the hospital environment. The use of molecular epidemiology as a tool for tracking of MRSE clones can help us to define routes of transmission and to control more effectively the spread of resistance among staphylococci, as well as to address problems of nosocomial infections. In this study, we used both molecular typing techniques and phenotypic methods to (i) identify the MRSE clones circulating in one central hospital in Iceland and five hospitals in Greater Copenhagen, Denmark, and (ii) compare the MRSE clones from Iceland and Denmark with representative MRSE strains isolated in hospitals in other countries in Europe, North and South America, and Africa in order to track the geographic spread of MRSE clones.
MATERIALS AND METHODS
Setting.
Collaborating national centers participating in an international surveillance program (RESIST) (33), namely, five hospitals in greater Copenhagen, Denmark (coded AH, B, C, D, and HV), and one hospital in Reykjavik, Iceland, provided the isolates. Hospital designations, codes, and general characteristics are listed in Table 1.
TABLE 1.
Characterization of the hospitals in Denmark and Iceland that provided isolates for this study
| Country | Hospital (code) | Antibiotic consumptiond (1997/1998) | No. of beds in 1997 | No. of hospital admissions (1997/1998) | MRSE frequency in 1998 (%) | MRSA frequency in 1998 (%) | No. of MRSE isolates studiede
|
|
|---|---|---|---|---|---|---|---|---|
| 1997 | 1998 | |||||||
| Denmark | Amager hospitala (AH)b | 444/459 | 377 | 16,600/17,500 | 31.5 | 0 | 9 | 8 |
| Herlev hospital (B)b | 469/483 | 809 | 35,700/36,600 | 40.0 | 0.4 | 15 | 15 | |
| Gentofte hospital (C)b | 376/462 | 930 | 39,600/38,800 | 43.0 | 0.1 | 12 | 14 | |
| Glostrup hospital (D)b | 328/340 | 1,012 | 37,500/37,300 | 32.0 | 0.3 | 3 | 9 | |
| Hvidovre hospital (HV)b | 405/459 | 941 | 43,500/40,900 | 38.5 | 0.6 | 26 | 25 | |
| Total | 65 | 71 | ||||||
| Iceland | Landspitali University hospital (R) | 337c | 911 | 23,739/23,737 | 39 | 0 | 45 | 49 |
Amager hospital is a combination of Sundby hospital and Sankt Elisabeth hospital.
Copenhagen University Hospitals.
Average for the years 1997 to 1999.
The values are defined daily doses per 1,000 patient days.
Totals for 1997 and 1998 combined: Denmark, 136; Iceland, 94.
Clinical strains.
All MRSE isolates were collected at the collaborating national centers and stored in the culture collection of the Molecular Genetics Laboratory of the Instituto de Tecnologia Qu|$$|Aa|fimica e Biológica, Universidade Nova de Lisboa, Oeiras, Portugal.
A representative sample of 230 MRSE isolates was chosen for molecular characterization. These included all of the 136 isolates provided by Denmark and 94 out of 155 isolates from Iceland, chosen on the basis of the greatest diversity possible in terms of clinical origin and patient demographics.
The information provided for each isolate included collection date, service, and source of isolation, as well as patient identification, gender, age, immunodeficiency condition, and period of hospitalization. The isolates were collected from April to October of 1997 and throughout 1998 in the Icelandic hospital and from August to December of 1997 and from June to December of 1998 in the Danish hospitals. The collection studied (n = 230) included strains isolated from single patients, the majority (92%) of whom were inpatients, 79% of whom were hospitalized for more than 48 h. The isolates were reported as having originated from infection (38%) and colonization (54%) sites, mostly from blood (46%) and wounds (30%). There are no data available on the origin of 8% of the remaining isolates.
An additional group of seven MRSE strains previously characterized by PFGE in our laboratory was selected for comparison of PFGE patterns. These strains belong to four MRSE collections from Greece (34 isolates) (I. Spiliopoulou, unpublished data), Cape Verde (48 isolates) (2), Mexico (74 isolates), and Uruguay (10 isolates) (H. de Lencastre, unpublished data). The PFGE patterns of these strains were visually compared with those of the MRSE strains from Iceland and Denmark. The strains that had patterns visually similar or related to those of the strains from Iceland and Denmark were selected and run side by side in PFGE to confirm the putative similarities.
Control strains.
The type strain of S. epidermidis, ATCC 14990T, was used as a reference for internal transcribed spacer identification by PCR. S. aureus ATCC 25923 and Escherichia coli ATCC 25922 were included for quality control of the antimicrobial susceptibility patterns. Partially quinupristin-dalfopristin (Synercid)-resistant S. aureus clinical isolates Syn 1 and Syn 25 (provided by Rhône-Poulenc Rorer S.A.) were included as quality control strains for quinupristin-dalfopristin susceptibility determination (33). S. aureus strain COL was used as the source of the mecA gene.
Species identification.
After bacterial identification at the collaborating national centers, all isolates were tested at the Molecular Genetics Laboratory of the Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa, for mannitol fermentation in solid media (Difco, Detroit, Mich.) and for coagulase production with the BBL Coagulase Plasma Rabbit test (Becton Dickinson Microbiology Systems, Cockeysville, Md.) in accordance with the manufacturer's instructions. Identification to the species level was carried out by internal transcribed spacer PCR as previously described (9).
Antimicrobial susceptibility testing.
Susceptibility to 11 antimicrobial agents was determined by the disk diffusion method in accordance with the National Committee for Clinical Laboratory Standards guidelines (28). The antimicrobial agents tested comprised oxacillin (1 μg), trimethoprim-sulfamethoxazole (25 μg), ciprofloxacin (5 μg), chloramphenicol (30 μg), clindamycin (2 μg), erythromycin (15 μg), gentamicin (10 μg), rifampin (5 μg), tetracycline (30 μg), vancomycin (30 μg), and teicoplanin (30 μg) (Oxoid, Basingstoke, England). Testing for susceptibility to quinupristin-dalfopristin (provided by Rhône-Poulenc Rorer S.A.) was performed as previously described (33). Multiresistance was defined as resistance to three or more antimicrobial classes.
DNA preparation.
Agarose disks containing chromosomal DNA for PFGE were prepared as previously described for S. aureus (8). DNA for PCR was isolated by the guanidine isothiocyanate extraction method as described before (9).
PFGE.
Chromosomal DNAs were digested with the endonuclease SmaI (New England Biolabs, Beverly, Mass.) in accordance with the manufacturer's instructions and separated by PFGE in a contour-clamped homogeneous electric field apparatus (CHEF-DRII; Bio-Rad, Hercules, Calif.) as previously described (8). Lambda ladder DNA (New England Biolabs) was used as molecular weight markers for PFGE. Analysis of SmaI macrorestriction profiles was done by visual inspection, and PFGE patterns were assigned by using the criteria proposed by Tenover et al. (42). Isolates with similar PFGE patterns were included in the same type designated by one or two uppercase letters and were considered to belong to the same clonal type. Isolates with PFGE types differing by up to six bands were assigned to subtypes identified by uppercase letters followed by arabic numbers. The strain chosen as the reference for each type was a representative of the predominant subtype. PFGE type designation was maintained for all of the collections studied.
Cluster analysis of electrophoretic band patterns was performed by using the Dice similarity metric and single linkage amalgamation scheme, considering a deviation table defined within the PFGE band weights of lambda ladder DNA (Whole Band Analyzer software, version 3.3; BioImage, Ann Arbor, Mich.). The representativeness of the resulting dendrogram was verified by determining the cophenetic coefficient (36) with the MatLab software, version 6 (R12) (Statistics Toolbox, The Mathworks, Inc., Natick, Mass.).
Detection of the mecA gene.
The presence of the mecA gene was confirmed for all isolates by hybridizing the SmaI digests with a DNA probe internal to the mecA gene of MRSA strain COL.
Statistical analysis of clinical data.
A test of differences between proportions with a degree of confidence of 95% was performed to analyze possible relationships between the different clones defined and the clinical data provided by the hospitals, namely, service, source of isolation, and period of hospitalization, by using the MatLab software, version 6 (R12) (Statistics Toolbox, The Mathworks, Inc.).
RESULTS
PFGE clonal types.
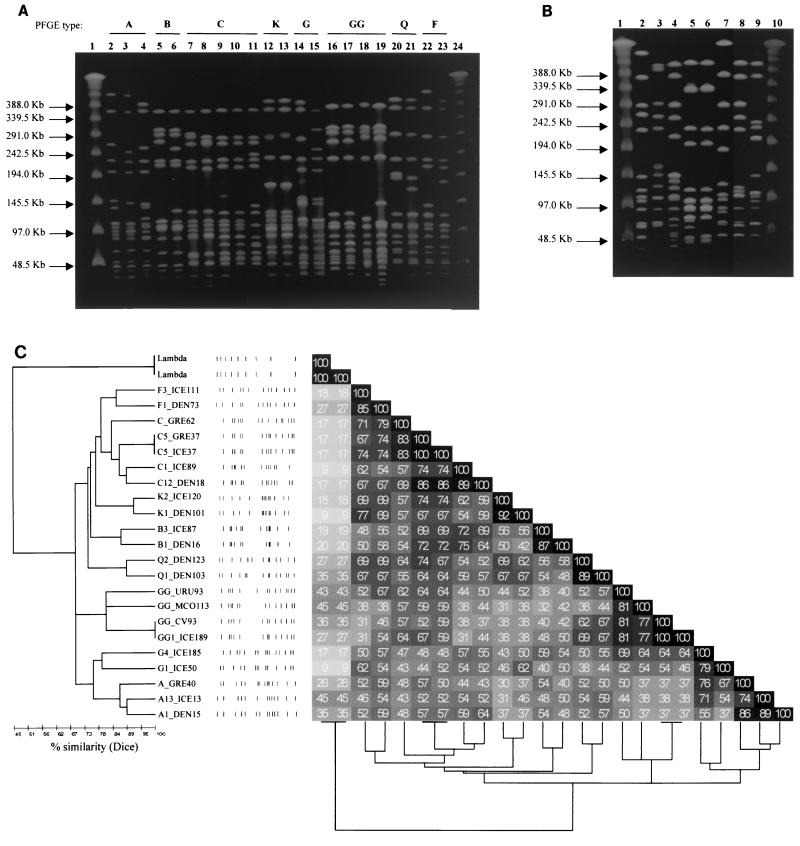
Considering both the Danish and Icelandic collections, 40 different PFGE types were assigned among the 230 MRSE isolates studied, most of which comprised several subtypes (Tables 2 and 3). Twelve PFGE clonal types (A, B, C, D, F, G, H, I, K, Q, N, and V) were common to Denmark and Iceland (Fig. 1A and 2), and some strains from both countries (recovered in the 2-year study period in either the same or different clinical services) even had the same PFGE subtype. Figure 1A presents the PFGE patterns of the most predominant MRSE clones in Denmark and Iceland, as well as the PFGE patterns of strains from other countries; Fig. 1B shows some of the less frequently found clones in the Icelandic and Danish collections.
TABLE 2.
Summary of results obtained for MRSE isolates from Denmark
| PFGE type; no. (%) of isolatesa | PFGE subtype (no. of isolates) | Antibiotype (>50%)b,c | Clinical data
|
||
|---|---|---|---|---|---|
| Hospital(s)d | Source(s)e | Infection or colonizatione | |||
| A; 25 (18.4) | A1 (8) | CIP, GEN, OXA, SXT | AH, B, C, HV | Bl, W, ot | I, C |
| A2 (5) | |||||
| A3 (3) | |||||
| A4 (2) | |||||
| A5 (2) | |||||
| A6 (1) | |||||
| A7 (1) | |||||
| A8 (1) | |||||
| A9 (1) | |||||
| A10 (1) | |||||
| B; 16 (11.8) | B1 (15) | CIP, GEN, OXA, SXT | AH, B, C, D, HV | Bl, A, W, ot | I, C |
| B2 (1) | |||||
| C; 16 (11.8) | C1 (1) | ERY, GEN, OXA, SXT | AH, B, C, D, HV | Bl, A, W, ot | I, C |
| C2 (5) | |||||
| C3 (3) | |||||
| C4 (2) | |||||
| C5 (1) | |||||
| C6 (1) | |||||
| C7 (1) | |||||
| C12 (2) | |||||
| K; 15 (11.0) | K1 (12) | ERY, GEN, OXA | AH, B, C, D, HV | BI, R, W, A, ot | I, C |
| K2 (3) | |||||
| D; 10 (7.4) | D1 (6) | CHL, ERY, GEN, OXA, SXT | C, HV | Bl, R, ot | I, C |
| D2 (3) | |||||
| D3 (1) | |||||
| F; 9 (6.6) | F1 (3) | GEN, OXA | AH, C, D, HV | Bl, ot | I, C |
| F2 (1) | |||||
| F3 (1) | |||||
| F4 (1) | |||||
| F5 (1) | |||||
| F6 (1) | |||||
| F7 (1) | |||||
| G; 5 (3.7) | G1 (2) | CIP, CLI, ERY, GEN, OXA, SXT | B, C, HV | Bl, R, W, ot | I, C |
| G2 (1) | |||||
| G3 (2) | |||||
| H; 5 (3.7) | H1 (3) | GEN, RIF, OXA, SXT | AH, C, HV | Bl, R, ot | C |
| H2 (1) | |||||
| H4 (1) | |||||
| I; 4 (2.9) | I1 (1) | ERY, GEN, OXA, TET | C, HV | Bl, W | C |
| I2 (2) | |||||
| I3 (1) | |||||
| J; 4 (2.9) | J1 (3) | ERY, OXA | C, D, HV | Bl | C |
| J2 (1) | |||||
| E; 3 (2.2) | E1 (2) | CHL, GEN, OXA, RIF, TET | AH, B, C | Bl | I, C |
| E2 (1) | |||||
| L; 3 (2.2) | L1 (3) | GEN, OXA | C, HV | Bl, W | C |
| Q; 3 (2.2) | Q1 (2) | ERY, GEN, OXA, SXT | B, C, HV | Bl, ot | I, C |
| Q2 (1) | |||||
| M; 2 (1.5) | M1 (2) | CIP, CLI, ERY, GEN, OXA, RIF, SXT | C, HV | Bl, A | C |
| N; 2 (1.5) | N1 (1) | CLI, ERY, GEN, OXA, SXT | B, HV | Bl, ot | I, C |
| N2 (1) | |||||
| O; 2 (1.5) | O1 (1) | CHL, ERY, GEN, OXA, SXT | B, D | Bl | C |
| O2 (1) | |||||
| P; 2 (1.5) | P1 (1) | ERY, GEN, OXA | HV | Bl | C |
| P2 (1) | |||||
| R; 2 (1.5) | R1 (2) | CIP, CLI, ERY, GEN, OXA, SXT, TET | HV | W | I, C |
| V; 1 (0.7) | V1 (1) | CIP, ERY, GEN, OXA, SXT, TET | AH, HV | Bl | C |
| T; 1 (0.7) | T1 (1) | CHL, ERY, OXA, TET | HV | Bl | C |
| U; 1 (0.7) | U1 (1) | OXA | AH | Bl | |
| W; 1 (0.7) | W1 (1) | OXA | B | Bl | C |
| X; 1 (0.7) | X1 (1) | ERY, GEN, OXA, SXT | C | ot | |
| Z; 1 (0.7) | Z1 (1) | OXA, SXT, TET | AH | Bl | C |
| AA; 1 (0.7) | AA1 (1) | ERY, OXA | HV | Bl | C |
| CC; 1 (0.7) | CC1 (1) | ERY, OXA, TEC | HV | Bl | C |
Numbers in parentheses correspond to the frequency of each PFGE type in relation to the total number of MRSE isolates in Denmark.
Antibiogram for more than 50% of isolates with this PFGE type.
CHL, chloramphenicol; CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; GEN, gentamicin; OXA, oxacillin; RIF, rifampin; TEC, teicoplanin; TET, tetracycline; SXT, trimethoprim-sulfamethoxazole.
Hospital codes: AH, Amager hospital (Sundby hospital); B, Herlev hospital; C, Gentofte hospital; D, Glostrup hospital; E, Amager hospital (Sankt Elisabeth hospital); HV, Hvidovre hospital.
A, abscess; Bl, blood; C, colonization; I, infection; ot, other sources; R, respiratory tract; W, wound.
TABLE 3.
Summary of results obtained for MRSE isolates from Iceland
| PFGE type; no. (%) of isolatesa | PFGE subtype (no. of isolates) | Antibiotype (>50%)b,c | Clinical datad
|
||
|---|---|---|---|---|---|
| Source(s) | Infection or colonization | ||||
| C; 23 (24.5) | C1 (11) | CIP, GEN, OXA, SXT, TET | Bl, W | I, C | |
| C2 (4) | |||||
| C5 (3) | |||||
| C8 (1) | |||||
| C9 (1) | |||||
| C10 (2) | |||||
| C11 (1) | |||||
| B; 12 (12.8) | B3 (9) | GEN, OXA, SXT | Bl, W | I, C | |
| B4 (3) | |||||
| A; 9 (9.6) | A11 (2) | CHL, GEN, OXA, SXT | U, Bl, W, R | I, C | |
| A12 (2) | |||||
| A13 (1) | |||||
| A14 (1) | |||||
| A15 (1) | |||||
| A16 (1) | |||||
| A17 (1) | |||||
| G; 8 (8.5) | G1 (2) | CLI, ERY, GEN, OXA, RIF, SXT | I, C | ||
| G3 (2) | |||||
| G4 (1) | |||||
| G5 (2) | |||||
| G6 (1) | |||||
| D; 7 (7.4) | D1 (3) | CHL, ERY, GEN, OXA, SXT | U, W, Bl | I, C | |
| D4 (1) | |||||
| D5 (1) | |||||
| D6 (1) | |||||
| D7 (1) | |||||
| EE; 6 (6.4) | EE1 (6) | CLI, ERY, GEN, OXA, RIF, SXT | Bl, W | I, C | |
| FF; 4 (4.3) | FF1 (1) | OXA, SXT | W | I, C | |
| FF2 (1) | |||||
| FF3 (1) | |||||
| FF4 (1) | |||||
| I; 3 (3.2) | I1 (3) | CIP, OXA | Bl, W | C | |
| BB; 2 (2.1) | BB1 (2) | OXA, SXT, TEC, TET | Bl, W | C | |
| GG; 2 (2.1) | GG1 (2) | CIP, CLI, ERY, GEN, OXA, RIF, SXT | U, W | I, C | |
| HH; 2 (2.1) | HH1 (1) | CHL, CLI, ERY, GEN, OXA, SXT, TEC, TET | W | C | |
| HH2 (1) | |||||
| II; 2 (2.1) | II1 (1) | CLI, ERY, OXA, SXT, TET | U, W | I | |
| II2 (1) | |||||
| F; 1 (1.1) | F3 (1) | OXA, SXT | W | C | |
| H; 1 (1.1) | H3 (1) | CLI, ERY, OXA, SXT | W | C | |
| K; 1 (1.1) | K2 (1) | CHL, CLI, ERY, GEN, OXA | W | I | |
| N; 1 (1.1) | N3 (1) | CLI, ERY, OXA | R | C | |
| Q; 1 (1.1) | Q1 (1) | GEN, OXA, RIF, SXT | B | I | |
| V; 1 (1.1) | V2 (1) | ERY, GEN, OXA, SXT | B | C | |
| S; 1 (1.1) | S1 (1) | CIP, ERY, GEN, OXA, SXT, TEC, TET | U, W | I, C | |
| DD; 1 (1.1) | DD1 (1) | CLI, ERY, GEN, OXA, RIF, SXT | W | I | |
| KK; 1 (1.1) | KK1 (1) | CHL, CIP, CLI, ERY, GEN, OXA, SXT | Bl | I | |
| LL; 1 (1.1) | LL1 (1) | OXA | Bl | C | |
| MM; 1 (1.1) | MM1 (1) | OXA | W | C | |
| OO; 1 (1.1) | OO1 (1) | ERY, GEN, OXA | Bl | C | |
| PP; 1 (1.1) | PP1 (1) | CIP, CLI, ERY, GEN, OXA, SXT, TET | U | I | |
| QQ; 1 (1.1) | QQ1 (1) | CIP, CLI, ERY, GEN, OXA, SXT | U | I | |
Numbers in parentheses correspond to the frequency of each PFGE type in relation to the total number of MRSE isolates in Iceland.
Antibiogram for more than 50% of isolates with this PFGE type.
CHL, chloramphenicol; CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; GEN, gentamicin; OXA, oxacillin; RIF, rifampin; TEC, teicoplanin; TET, tetracycline; SXT, trimethoprim-sulfamethoxazole.
Bl, blood; C, colonization; I, infection; ot, other sources; R, respiratory tract; W, wound; U, urine.
FIG. 1.
SmaI PFGE patterns of MRSE clones identified in this work. (A) SmaI PFGE patterns of the major MRSE clones found in Iceland and Denmark and comparison with PFGE patterns of MRSE strains isolated in Mexico, Uruguay, Greece, and Cape Verde. Lanes (PFGE subtypes are in parentheses): 1 and 24, lambda ladder molecular size markers; 2, DEN15 (A1); 3, ICE13 (A13); 4, GRE40 (A); 5, DEN16 (B1); 6, ICE87 (B3); 7, DEN18 (C12); 8, ICE89 (C1); 9, ICE37 (C5); 10, GRE37 (C); 11, GRE62 (C); 12, DEN101 (K1); 13, ICE120 (K2); 14, ICE50 (G1); 15, ICE185 (G4); 16, ICE189 (GG1); 17, CV93 (GG); 18, MCO113 (GG); 19, URU93 (GG); 20, DEN103 (Q1); 21, DEN123 (Q2); 22, DEN73 (F1); 23, ICE111 (F3). Each isolate designation indicates the country of origin as follows: CV, Cape Verde; DEN, Denmark; GRE, Greece; ICE, Iceland; MCO, Mexico; URU, Uruguay. (B) SmaI PFGE patterns of less frequent MRSE clones found in Iceland and Denmark. Lanes: 1 and 10, lambda ladder molecular size markers; 2, DEN105 (Z1); 3, ICE18 (HH1); 4, ICE96 (II1); 5, DEN94 (P1); 6, DEN176 (P2); 7, ICE9 (QQ1); 8, ICE24 (HH2); 9, ICE26 (V2). (C) Dendrogram representing the clustering of the strains described in panel A. PFGE subtype and strain designation are specified. Each strain designation indicates the country of origin as follows: CV, Cape Verde; DEN, Denmark; GRE, Greece; ICE, Iceland; MCO, Mexico; URU, Uruguay. See Materials and Methods for details of the similarity measurement and the amalgamation scheme.
FIG. 2.
MRSE clones identified among Danish and Icelandic isolates. The PFGE type is specified for the clones found in both countries. The percentages are relative to the total number of isolates characterized (n = 230).
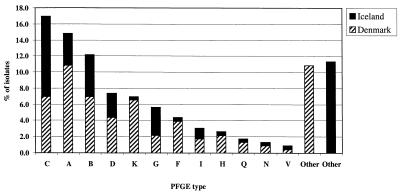
The 136 MRSE isolates collected in Denmark comprised a total of 26 PFGE types, although the majority of the isolates (53.0%) were included in only four major types (A, B, C, and K). Overall, the MRSE clone defined by PFGE type A was found to be the predominant one (18.4% of the total Danish MRSE isolates), followed by PFGE types B and C (11.8% each) and type K (11.0%) (Table 2). In hospitals AH, B, and C, these four clones were the predominant ones, while in hospital D, the majority of the isolates belonged to PFGE types B and K and in hospital HV, the predominant clone was PFGE type D.
The 94 MRSE isolates collected in Iceland were distributed among 26 clonal types, although the majority (55.4%) of the isolates could be included in only four PFGE types (A, B, C, and G). The predominant clone was PFGE type C (24.5%), followed by B (12.8%), A (9.6%), and G (8.5%) (Table 3).
The cluster analysis of electrophoretic band patterns identified among isolates from Denmark and Iceland and among strains from other countries (Fig. 1C) reflects the correctness of our visual PFGE pattern analysis if we consider a similarity distance of 78%. In fact, considering this value of similarity, the patterns shown in Fig. 1A are clustered in eight different groups, which corroborates our PFGE type assignment. The cophenetic value (36; see also Materials and Methods) of 0.84 indicates a good correlation between the dissimilarity matrix and the dendrogram, validating the representation.
Geographic dissemination of MRSE clones.
PFGE patterns of MRSE strains from Iceland and Denmark were compared with those of MRSE isolates from countries in North America (Mexico), South America (Uruguay), Europe (Greece), and Africa (Cape Verde). Besides the similarities already described between Danish and Icelandic MRSE isolates, in this additional international sample, we also found several strains with identical or closely related PFGE types despite their distinct geographic origins. Two isolates, one from Greece and the other from Iceland, had identical PFGE subtypes, here designated C5, while three isolates (one from Cape Verde, another from Uruguay, and a third one from Mexico) showed patterns that could be assigned to subtypes of pattern GG (Fig. 1A). Three additional MRSE isolates from Greece had PFGE profiles related to PFGE types A and C identified in Iceland and Denmark (Fig. 1A). PFGE patterns GG, A, and C were the most or second most frequently found in the MRSE collection of each country: PFGE profiles related to PFGE pattern GG were found in 23% (11 of 48) of the isolates from Cape Verde, 12% (9 of 74) of those from Mexico, and 20% (2 of 10) of those from Uruguay. Moreover, 18% (6 of 34) and 6% (2 of 34) of the isolates from Greece had patterns similar or related to PFGE types A and C, respectively.
Antimicrobial susceptibility patterns.
Each PFGE type could be assigned to a characteristic multiresistance pattern. Generally, isolates with the same PFGE type but from different countries differed in resistance to one or two antimicrobials (Tables 2 and 3).
In Denmark, the majority (>50%) of the isolates belonging to the predominant clones (A, B, C, and K) were resistant to gentamicin. All isolates characterized by PFGE types A and B were also resistant to trimethoprim-sulfamethoxazole, and the majority of them showed resistance to ciprofloxacin. More than half of the isolates belonging to PFGE type C were resistant to trimethoprim-sulfamethoxazole and erythromycin, whereas most isolates of type K only showed additional resistance to erythromycin. Variations within the antibiotype associated with a particular PFGE subtype profile were found. We also detected cases in which a given PFGE subtype was associated with a particular antibiotype (data not shown).
In Iceland, most (>50%) of the MRSE isolates belonging to predominant PFGE types A, B, C, and G showed resistance to trimethoprim-sulfamethoxazole and gentamicin. Isolates belonging to type A were characterized by additional resistance to chloramphenicol; the majority of the isolates of type C were resistant to ciprofloxacin and tetracycline, while most isolates of type G showed resistance to clindamycin, erythromycin, and rifampin. As described for Denmark, MRSE isolates from Iceland belonging to particular PFGE subtypes showed variation in the antimicrobial resistance pattern characteristic of that type. We also detected isolates with the same PFGE subtype that differed in the antibiotype (data not shown).
Most of the less representative clones also showed a pattern of multiresistance, except for those defined by PFGE types U and W from Denmark and LL and MM from Iceland, which showed resistance to β-lactam antibiotics only, or clones F, I, and FF from Iceland and clones F, J, L, and AA from Denmark, which showed resistance to β-lactams and to an additional antimicrobial (gentamicin, ciprofloxacin, trimethoprim-sulfamethoxazole, or erythromycin) (Tables 2 and 3).
Statistical analysis of clinical data.
Associations between the PFGE patterns of particular MRSE clones and clinical data were examined with a test for differences between proportions (see Materials and Methods) in order to test for the clinical significance of the isolates. No significant correlation within a 95% confidence level was found, except for clone K, which was associated with an infection origin (P < 0.05).
DISCUSSION
In this study, we documented the existence of MRSE strains originating from various countries that had identical genotypic structures, as defined by PFGE. In particular, we observed that (i) 12 common MRSE PFGE types, including the 5 predominant ones (A, B, C, G, and K), were found among isolates from Iceland and Denmark and (ii) 3 MRSE PFGE types identified among isolates from Iceland and Denmark were also found in isolates collected during equivalent time periods at health care institutions in Greece, Mexico, Uruguay, and Cape Verde and corresponded to the most or second most representative clones in the collections studied.
The existence of common PFGE types among MRSE isolates from distant origins can result either from geographic dissemination of these MRSE strains among the various countries or from independent and convergent evolution of distinct strains within these different locations. The first hypothesis seems to be more plausible than the second, based on the following observations. S. epidermidis is endogenous to the human skin flora and is therefore easily transmissible. In fact, the dissemination of the resistant counterparts (MRSE) was already described in the hospital environment (12, 43), as well as in a community (35). Recent studies have also documented MRSE carriage by students (35) and young soldiers (T. L. Sørensen, personal communication; I. Santos-Sanches, unpublished data). The colonization of humans by MRSE and the increasing mobility of individuals suggest that the community can act as a vehicle for MRSE dissemination between distant locations. The geographic dissemination of MRSA clones is well established (1), and the same might occur with MRSE strains. Finally, the existence of 12 different clonal types common to Iceland and Denmark also supports the dissemination hypothesis, since the independent and convergent evolution of 12 distinct S. epidermidis strains from different locations seems highly improbable.
In the present work, we observed that isolates with PFGE types and subtypes in common were collected in 2 consecutive years from the same hospital service, suggesting that these MRSE strains are endemic to the hospital environment. The persistence and dissemination of MRSE clones within Danish and Icelandic hospitals have two possible explanations: (i) the MRSE clones originated in the community that attends the various hospitals, or (ii) the MRSE clones are endemic to the hospitals studied and replace the normal susceptible flora of patients in the hospital environment. As mentioned above, MRSE strains were already isolated from healthy carriers. Our own data showed that strains from outpatients (16 isolates) or from inpatients hospitalized for less than 48 h (35 isolates) belonged to the most representative PFGE types (A, B, C, G, and K) and to other less representative types, suggesting that the same MRSE clones can be identified in nosocomial and community settings. Data from one Danish hospital indicate that, among blood cultures positive for coagulase-negative staphylococci (CNS), the frequency of MRSE isolated during the first 1 or 2 days of hospitalization is 15.7%, whereas for a longer hospitalization, the frequency rises to 30.4% (H. Westh, unpublished data). The MRSE rates among CNS sampled in Iceland between 1997 and 2000 were 50, 39, 39, and 33% for hospital isolates and 21, 16, 18, and 20% for outpatient isolates (K. Kristinsson, unpublished). These observations suggest colonization by MRSE during hospital admission in Danish and Icelandic hospitals. In conclusion, both community and nosocomial origins for MRSE are likely, although resistant strains must have been selected primarily in the hospital environment.
In this study, we documented the existence of five predominant clones among 230 MRSE isolates from Denmark and Iceland defined by PFGE. Previous studies on the PFGE genotyping of MRSE isolates from particular hospital units demonstrated the prevalence of a few MRSE clones (35, 43). The detection of few major MRSE clones in our global sample was surprising in view of the high heterogeneity of the isolates in terms of the clinical specimens, patient demographics, and geographic origins. These five predominant MRSE clones were spread among different services of a single hospital, among different hospitals, and even among different countries.
The higher dissemination capacity of these five MRSE clones can be explained by a selective pressure caused by antimicrobial and disinfectant use. The prevalence of these MRSE clones could also be explained by particular colonization advantages or by specific virulence characteristics, such as slime production and synthesis of extracellular proteins or factors involved in interactions with host defense mechanisms (20). The genomic plasticity of the S. epidermidis species (15, 17, 25, 35) may allow these strains to adapt rapidly to environmental changes. The fact that some of the major MRSE clones found in Iceland and Denmark comprised a high number of PFGE subtypes may be the result of such a putative fitness process. Galdbart et al. (15) described patients infected by a single S. epidermidis clone, which subsequently underwent genomic rearrangements, yielding derivatives with divergent phenotypes and macrorestriction patterns. Another event that may reflect the dynamics of the S. epidermidis genome is the in vivo transfer of mecA from S. epidermidis to S. aureus clinical isolates (45). Interestingly, despite the high MRSE prevalence in Danish and Icelandic hospitals, the MRSA prevalence is quite low (Table 1), indicating that transfer of mecA from MRSE to MRSA must be a rare event.
The fact that no association was found between the disseminated clones and particular clinical sources (except for clone K) suggests that their spread in the nosocomial environment is not correlated with a specific transmission route or pathology. The association of clone K with infection sites, together with the fact that this clone has a low number of PFGE subtypes, indicates that this lineage may represent a more recent or stable epidemic clone.
PFGE met our requirements for the molecular characterization of MRSE strains, identifying the major clones, as well as the diversity among them. Nevertheless, due to the high variation rate shown by the S. epidermidis species, this technique will be more useful for determining the MRSE sources and transmission routes in short periods and in specific wards or patients (12, 43) than for studying large collections over time. The dendrogram presented in Fig. 1C reflects our assignment for PFGE typing if a value of 78% similarity is considered, illustrating the variability observed in the S. epidermidis species. In fact, if we had considered a higher percentage of similarity, the clusters would have split, except for those defined by the more related strains. For these reasons, we anticipate that for the study of MRSE strains over time and from different geographic regions, other, less discriminatory typing methods, based on more conserved regions of the genome, such as multilocus sequence typing (13), when established for S. epidermidis or ribotyping (38), may prove to be more adequate. Such studies on MRSE would give us insight into the evolution mechanisms of distinct strains dispersed among many geographical sites (10).
The MRSE frequencies described for other countries in Europe (26, 33) and for the United States, Canada, and Latin America (31) are higher than those described for Iceland and Denmark. Moreover, the overall frequency of antimicrobial resistance shown by the majority of the MRSE strains studied in this work is low compared to that of strains from other countries (33), probably as a result of the antimicrobial use practices and strict infection control measures applied in both Iceland and Denmark. Besides being resistant to the β-lactams, the majority of the MRSE strains studied were also resistant to trimethoprim-sulfamethoxazole and gentamicin. Several clones were found in which all of the isolates were resistant to these three types of antimicrobial agents. A nationwide study performed during 1991 and 1992 that included five Danish counties revealed that S. epidermidis resistant to gentamicin had a prevalence of 33% (21) and a survey performed 1 year later found that 57% of CNS blood isolates were resistant to aminoglycosides (6), showing that the resistance to this class of antimicrobial agents has been rising in the last decade. In a 1997 to 1998 multicenter study in several European countries, resistance of MRSE to trimethoprim-sulfamethoxazole was reported to vary between 46 and 80% (33).
The poor ability of antibiotypes to distinguish clonal types was already described for MRSE (43), as well as for a number of different organisms (38). In our study, we observed that the majority (>50%) of the isolates with a particular PFGE pattern were associated with a particular antibiotype, although we found isolates with the same PFGE subtype with different antibiotypes and vice versa.
The establishment of MRSE as resident flora in the hospital environment is worrisome since these strains, predominantly involved in nosocomial infections, can accumulate resistance determinants to practically all classes of antimicrobials, and these are potentially transferable to S. aureus and other microorganisms. Particularly important is the evidence resulting from this study that some multiresistant MRSE clones may be disseminated geographically. These data suggest that it may become necessary to establish infection control procedures directed toward MRSE that prevent colonization of patients with hospital flora, especially patients at a higher risk of nosocomial infection, such as those receiving foreign bodies, neonates, and immunocompromised patients.
Acknowledgments
This work was partially supported by Projects 2/2.2/SAU/1295/95, POCTI/1999/ESP/34872, and POCTI/1999/CVT/34842 from Fundação para a Ciência e Tecnologia, Portugal, and by a grant from Fundação Calouste Gulbenkian, awarded to H. de Lencastre. The 1997 and 1998 S. epidermidis strains from Denmark and Iceland were obtained with a grant from Rhône-Poulenc Rorer, S. A., awarded to Alexander Tomasz and Hermínia de Lencastre. M. Miragaia was supported by a grant from Fundação Calouste Gulbenkian. I. Couto, J. Carriço, and S. Pereira were supported, respectively, by grants SFRH/BPD/3645/2000 and SFRH/BD/3123/2000 and grant BIC 023/BIC/2000 from Fundação para a Ciência e Tecnologia, Portugal.
We thank I. Spiliopolou, M. Aires de Sousa, R. Palacio, L. Dell'Acqua, and M. E. Velazquez-Meza, who performed the original PFGE analysis of the isolates from Greece, Cape Verde, Uruguay, and Mexico.
REFERENCES
- 1.Aires De Sousa, M., M. Miragaia, I. Santos-Sanches, S. Ávila, I. Adamson, S. T. Casagrande, M. C. Brandileone, R. Palacio, L. Dell'Acqua, M. Hortal, T. Camou, A. Rossi, M. E. Velazquez-Meza, G. Echaniz-Aviles, F. Solorzano-Santos, I. Heitmann, and H. de Lencastre. 2001. Three-year assessment of methicillin-resistant Staphylococcus aureus clones in Latin America from 1996 to 1998. J. Clin. Microbiol. 39:2197-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aires De Sousa, M., I. Santos-Sanches, M. L. Ferro, and H. de Lencastre. 2000. Epidemiological study of staphylococcal colonization and cross-infection in two West African Hospitals. Microb. Drug Resist. 6:133-141. [DOI] [PubMed] [Google Scholar]
- 3.Archer, G. L., D. M. Niemeyer, J. A. Thanassi, and M. J. Pucci. 1994. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob. Agents Chemother. 38:447-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, R. B., D. Cipriani, M. Schulte, A. Corl, and R. Pieczarka. 1994. Community-acquired bacteremias from tunneled central intravenous lines: results from studies of a single vendor. Am. J. Infect. Control 22:149-151. [DOI] [PubMed] [Google Scholar]
- 5.Burnie, J. P., M. Naderi-Nasab, K. W. Loudon, and R. C. Matthews. 1997. An epidemiological study of blood culture isolates of coagulase-negative staphylococci demonstrating hospital-acquired infection. J. Clin. Microbiol. 35:1746-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busch-Sørensen, C., N. Frimodt-Møller, G. H. Miller, and F. Espersen. 1996. Aminoglycoside resistance among Danish blood culture isolates of coagulase-negative staphylococci. APMIS 104:873-880. [DOI] [PubMed] [Google Scholar]
- 7.Cars, O., S. Molstad, and A. Melander. 2001. Variation in antibiotic use in the European Union. Lancet 357:1851-1853. [DOI] [PubMed] [Google Scholar]
- 8.Chung, M., H. de Lencastre, P. Matthews, A. Tomasz, I. Adamsson, M. Aires de Sousa, T. Camou, C. Cocuzza, A. Corso, I. Couto, A. Dominguez, M. Gniadkowski, R. Goering, A. Gomes, K. Kikuchi, A. Marchese, R. Mato, O. Melter, D. Oliveira, R. Palacio, R. Sá-Leão, I. Santos-Sanches, J. H. Song, P. T. Tassios, and P. Villari. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6:189-198. [DOI] [PubMed] [Google Scholar]
- 9.Couto, I., S. F. F. Pereira, M. Miragaia, I. Santos-Sanches, and H. de Lencastre. 2001. Identification of human clinical staphylococci by internal transcribed spacer PCR (ITS-PCR). J. Clin. Microbiol. 39:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crisóstomo, I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin susceptible and resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 98:9865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Lencastre, H., M. Chung, and H. Westh. 2000. Archaic strains of methicillin-resistant Staphylococcus aureus: molecular and microbiological properties of isolates from the 1960s in Denmark. Microb. Drug Resist. 6:1-10. [DOI] [PubMed] [Google Scholar]
- 12.Dominguez, M. A., J. Linares, A. Pulido, J. L. Perez, and H. de Lencastre. 1996. Molecular tracking of coagulase-negative staphylococcal isolates from catheter-related infections. Microb. Drug Resist. 2:423-429. [DOI] [PubMed] [Google Scholar]
- 13.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frimodt-Møller, N. F., F. Espersen, P. Skinhøj, and V. T. Rosdahl. 1997. Epidemiology of Staphylococcus aureus bacteremia in Denmark from 1957 to 1990. Clin. Microbiol. Infect. 3:297-305. [DOI] [PubMed] [Google Scholar]
- 15.Galdbart, J. O., A. Morvan, N. Desplaces, and N. el Solh. 1999. Phenotypic and genomic variation among Staphylococcus epidermidis strains infecting joint prostheses. J. Clin. Microbiol. 37:1306-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrett, D. O., E. Jochimsen, K. Murfitt, B. Hill, S. McAllister, P. Nelson, R. V. Spera, R. K. Sall, F. C. Tenover, J. Johnston, B. Zimmer, and W. R. Jarvis. 1999. The emergence of decreased susceptibility to vancomycin in Staphylococcus epidermidis. Infect. Control Hosp. Epidemiol. 20:167-170. [DOI] [PubMed] [Google Scholar]
- 17.George, C. G., and W. E. Kloos. 1994. Comparison of the SmaI-digested chromosomes of Staphylococcus epidermidis and the closely related species Staphylococcus capitis and Staphylococcus caprae. Int. J. Syst. Bacteriol. 44:404-409. [DOI] [PubMed] [Google Scholar]
- 18.Goldmann, D. A., and G. B. Pier. 1993. Pathogenesis of infections related to intravascular catheterization. Clin. Microbiol. Rev. 6:176-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hedin, G. 1993. Staphylococcus epidermidis—hospital epidemiology and the detection of methicillin resistance. Scand. J. Infect. Dis. Suppl. 90:1-59. [PubMed] [Google Scholar]
- 20.Heilmann, C., and G. Peters. 2000. Biology and pathogenesis of Staphylococcus epidermidis, p. 442-448. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens, 1st ed. American Society for Microbiology, Washington, D.C.
- 21.Jarløv, J. O. 1999. Phenotypic characteristics of coagulase-negative staphylococci: typing and antibiotic susceptibility. APMIS Suppl. 91:1-42. [PubMed] [Google Scholar]
- 22.Jessen, O., K. Rosendal, P. Bulow, V. Faber, and K. R. Eriksen. 1969. Changing staphylococci and staphylococcal infections; a ten-year study of bacteria and cases of bacteremia. N. Engl. J. Med. 281:627-635. [DOI] [PubMed] [Google Scholar]
- 23.Kloos, W. E., and T. L. Bannerman. 1994. Update on clinical significance of coagulase-negative staphylococci. Clin. Microbiol. Rev. 7:117-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kristinsson, K. G., M. A. Hjalmarsdottir, and O. Steingrimsson. 1992. Increasing penicillin resistance in pneumococci in Iceland. Lancet 339:1606-1607. [DOI] [PubMed] [Google Scholar]
- 25.Lina, B., F. Vandenesch, J. Etienne, B. Kreiswirth, and J. Fleurette. 1992. Comparison of coagulase-negative staphylococci by pulsed-field electrophoresis. FEMS Microbiol. Lett. 71:133-138. [DOI] [PubMed] [Google Scholar]
- 26.Melo-Cristino, J., A. F. Alves, J. S. Moreira, I. Calheiros, M. L. Felício, P. Lopes, L. Sobral, J. M. Diogo, M. Pinto, J. Marques, J. Piedade, L. Lito, M. J. Salgado, J. M. Amorim, E. Calado, M. H. Ramos, M. N. Costa, F. Martins, M. A. Pessanha, J. Correia da Fonseca, L. Ferro, J. Marques, D. Costa, G. Ribeiro, and C. Vieira. 1998. Antimicrobial resistance in staphylococci and enterococci in 10 Portuguese hospitals in 1996 and 1997. Microb. Drug Resist. 4:319-324. [DOI] [PubMed] [Google Scholar]
- 27.Melzer, M., H. Maiden, W. Gransden, J. Edgeworth, Y. Chan, and M. Kinirons. 1999. Community-acquired Staphylococcus epidermidis endocarditis complicated by splenic disease 9 years after aortic valve replacement. Scand. J. Infect. Dis. 31:595-596. [DOI] [PubMed] [Google Scholar]
- 28.National Committee for Clinical Laboratory Standards. 1995. Performance standards for antimicrobial disk susceptibility testing. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 29.Nouwen, J. L., A. van Belkum, S. de Marie, J. Sluijs, J. J. Wielenga, J. A. Kluytmans, and H. A. Verbrugh. 1998. Clonal expansion of Staphylococcus epidermidis strains causing Hickman catheter-related infections in a hemato-oncologic department. J. Clin. Microbiol. 36:2696-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paulsen, I. T., N. Firth, and R. A. Skurray. 1997. Resistance to antimicrobials other than β-lactams, p. 175-212. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease, 1st ed. Churchill Livingstone, New York, N.Y.
- 31.Pfaller, M. A., R. N. Jones, G. V. Doern, H. S. Sader, K. C. Kugler, M. L. Beach, and SENTRY. 1999. Survey of blood stream infections attributable to gram-positive cocci: frequency of occurrence and antimicrobial susceptibility of isolates collected in 1997 in the United States, Canada, and Latin America from the SENTRY Antimicrobial Surveillance Program. Diagn. Microbiol. Infect. Dis. 33:283-297. [DOI] [PubMed] [Google Scholar]
- 32.Rupp, M. E., and G. L. Archer. 1994. Coagulase-negative staphylococci: pathogens associated with medical progress. Clin. Infect. Dis. 19:231-243. [DOI] [PubMed] [Google Scholar]
- 33.Santos-Sanches, I., R. Mato, H. de Lencastre, A. Tomasz, CEM/NET Collaborators S. Nunes, C. R. Alves, M. Miragaia, J. Carriço, I. Couto, I. Bonfim, M. A. de Sousa, D. Oliveira, A. Gomes, M. Vaz, S. Fernandes, S. C. Verde, S. Ávila, F. Antunes, R. Sá-Leão, J. Almeida, O. Melter, and M. Chung, and International Collaborators M. C. Brandileone, E. Castañeda, I. Heitmann, M. Hortal, W. Hryniewicz, F. Jia, K. Kikuchi, K. G. Kristinsson, J. Liñares, A. Rossi, E. Z. Savov, J. Schindler, F. Solorzano-Santos, K. Totsuka, M. Venditti, P. Villari, H. Westh, J. S. Wu, and R. C. Zanella. 2000. Patterns of multidrug resistance among methicillin-resistant hospital isolates of coagulase-positive and coagulase-negative staphylococci collected in the international multicenter study RESIST in 1997 and 1998. Microb. Drug Resist. 6:199-211. [DOI] [PubMed] [Google Scholar]
- 34.Sieradzki, K., P. Villari, and A. Tomasz. 1998. Decreased susceptibilities to teicoplanin and vancomycin among coagulase-negative methicillin-resistant clinical isolates of staphylococci. Antimicrob. Agents Chemother. 42:100-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silva, F. R., E. M. Mattos, M. V. Coimbra, B. T. Ferreira-Carvalho, and A. M. Figueiredo. 2001. Isolation and molecular characterization of methicillin-resistant coagulase-negative staphylococci from nasal flora of healthy humans at three community institutions in Rio de Janeiro City. Epidemiol. Infect. 127:57-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy—the principles and practice of numerical classification. W. H. Freeman & Co., San Francisco, Calif.
- 37.Strausbaugh, L. J. 1999. Vancomycin-intermediate Staphylococcus epidermidis: curio or omen? Infect. Control Hosp. Epidemiol. 20:163-165. [DOI] [PubMed] [Google Scholar]
- 38.Struelens, M. J., and members of the European Study Group on Epidemiological Markers (ESGEM) of the European Society for Microbiology and Infectious Diseases (ESCMID). 1996. Consensus guidelines for appropriate use and evaluation of microbial typing systems. Clin. Microb. Infect. 2:2-11. [DOI] [PubMed] [Google Scholar]
- 39.Szewczyk, E. M., A. Piotrowski, and M. Rozalska. 2000. Predominant staphylococci in the intensive care unit of a paediatric hospital. J. Hosp. Infect. 45:145-154. [DOI] [PubMed] [Google Scholar]
- 40.Tammelin, A., P. Domicel, A. Hambraeus, and E. Stahle. 2000. Dispersal of methicillin-resistant Staphylococcus epidermidis by staff in an operating suite for thoracic and cardiovascular surgery: relation to skin carriage and clothing. J. Hosp. Infect. 44:119-126. [DOI] [PubMed] [Google Scholar]
- 41.Tan, T. Q., J. M. Musser, R. J. Shulman, E. O. Mason, Jr., D. H. Mahoney, Jr., and S. L. Kaplan. 1994. Molecular epidemiology of coagulase-negative Staphylococcus blood isolates from neonates with persistent bacteremia and children with central venous catheter infections. J. Infect. Dis. 169:1393-1397. [DOI] [PubMed] [Google Scholar]
- 42.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villari, P., C. Sarnataro, and L. Iacuzio. 2000. Molecular epidemiology of Staphylococcus epidermidis in a neonatal intensive care unit over a three-year period. J. Clin. Microbiol. 38:1740-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voss, A., D. Milatovic, C. Wallrauch-Schwarz, V. T. Rosdahl, and I. Braveny. 1994. Methicillin-resistant Staphylococcus aureus in Europe. Eur. J. Clin. Microbiol. Infect. Dis. 13:50-55. [DOI] [PubMed] [Google Scholar]
- 45.Wielders, C. L. C., M. R. Vriens, S. Brisse, L. A. M. de Graaf-Miltenburg, A. Troelstra, A. Fleer, F. J. Schmitz, J. Verhoof, and A. C. Fluit. 2001. Evidence for in vivo transfer of mecA DNA between strains of Staphylococcus aureus. Lancet 357:1674-1675. [DOI] [PubMed] [Google Scholar]
- 46.York, M. K., L. Gibbs, F. Chehab, and G. F. Brooks. 1996. Comparison of PCR detection of mecA with standard susceptibility testing methods to determine methicillin resistance in coagulase-negative staphylococci. J. Clin. Microbiol. 34:249-253. [DOI] [PMC free article] [PubMed] [Google Scholar]