Abstract
Industrial biotechnology employs the controlled use of microorganisms for the production of synthetic chemicals or simple biomass that can further be used in a diverse array of applications that span the pharmaceutical, chemical and nutraceutical industries. Recent advances in metagenomics and in the incorporation of entire biosynthetic pathways into Saccharomyces cerevisiae have greatly expanded both the fitness and the repertoire of biochemicals that can be synthesized from this popular microorganism. Further, the availability of the S. cerevisiae entire genome sequence allows the application of systems biology approaches for improving its enormous biosynthetic potential. In this review, we will describe some of the efforts on using S. cerevisiae as a cell factory for the biosynthesis of high-value natural products that belong to the families of isoprenoids, flavonoids and long chain polyunsaturated fatty acids. As natural products are increasingly becoming the center of attention of the pharmaceutical and nutraceutical industries, the use of S. cerevisiae for their production is only expected to expand in the future, further allowing the biosynthesis of novel molecular structures with unique properties.
Introduction
Microbial biosynthesis of natural products is an emerging area of metabolic engineering and industrial biotechnology that offers significant advantages over conventional chemical methods or extraction from biomass. Lower energy requirements, lower CO2 emissions, less toxic waste in the form of solvents and metal catalysts, simpler purification schemes, renewable feed stocks such as corn or soybeans, and the general ability of enzymes to perform chiral synthesis are among the improvements to be gained by microbial synthesis.
Within this context, almost 10 years since the publication of its entire genome sequence [1], Saccharomyces cerevisiae, remains the key model eukaryotic organism, with several features that make it a useful tool for both industrial applications and research:
1. It grows easily and adapts well in controlled fermentation environments.
2. Powerful tools for genetic engineering have been extensively developed and are readily available [2-4].
3. It is nonpathogenic and has been used since ancient times in food technology.
4. Unlike bacteria that require extensive protein engineering [5,6], yeast can readily express functional type II P450 monooxygenases, required for the biosynthesis of higher eukaryotic secondary metabolites [7,8].
Moreover, the availability of the entire genome sequence has made possible the collection of comprehensive information that allows a systems biology approach towards strain development and optimization [9-11]. As such, S. cerevisiae truly appears to be an ideal production platform for important secondary metabolites, such as natural products.
Natural products tend to encompass a wide array of small molecules derived from both eukaryotic (plants, marine organisms) and prokaryotic species. Their chemical structure has been fine-tuned through billions of years of evolution to serve specific functions in living organisms. Many natural products are high-value chemicals, since only small quantities can be isolated from their native biosynthetic organisms with the majority of them utilized as therapeutic agents for (among others) infectious diseases and cancer. The advent of genomics and metagenomics has allowed not only the discovery of a huge number of new promising metabolites, but also the elucidation of their biosynthetic pathways. As a result, the use of well-characterized hosts for their efficient, large scale production is now possible.
Since presenting the entire range of natural product biosynthesis in S. cerevisiae would require an extensive amount of space, in the next few paragraphs we will only highlight some of the most characteristic work in this area.
Isoprenoid biosynthesis
Isoprenoids (also known as terpenoids) belong to a vast group of secondary metabolites [12] that include carotenoids, sterols, polyprenyl alcohols, ubiquinone (coenzyme Q), heme A and prenylated proteins. They are of valuable commercial interest as food colorants and antioxidants (carotenoids), aroma and flavor enhancers (terpenes), nutraceuticals (ubiquinone), and antiparasitic and anticarcinogenic compounds (taxol) [13-15].
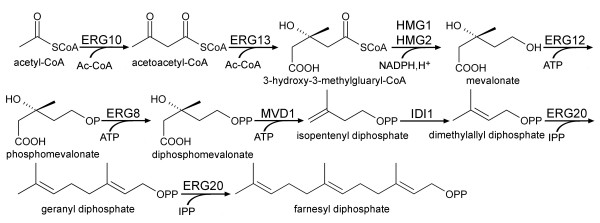
All isoprenoids are synthesized from a universal compound called isopentenyl diphosphate (IPP). The first pathway to IPP, the mevalonic acid (MVA) pathway, was first described by Bloch and Lynen [16,17]. In yeast, the mevalonate pathway (Figure 1) is chiefly employed to form ergosterol (provitamin D2) which is an essential part of the yeast membrane and provides membrane permeability and fluidity [18-21].
Figure 1.
Biosynthesis of isoprenoids by the mevalonate pathway. IPP: isopentenyl pyrophosphate; ERG10: acetoacetyl-CoA ligase; ERG13: 2-hydroxy-3-methylglutaryl-CoA synthase (HMG-CoA synthase); HMG1 and HMG2: 2-hydroxy-3-methylglutaryl-CoA reductases; ERG12: mevalonate kinase; ERG8: phosphomevalonate kinase; MVD1: diphosphomevalonate decarboxylase; IDI1: isopentenyl pyrophosphate:dimethylallyl diphosphate isomerase; ERG20: farnesyl diphosphate synthase.
Some of the most important isoprenoids and their engineered biosynthesis in S. cerevisiae will be further described.
Taxol
Taxol is a complex substituted terpenoid that was first isolated from the bark of Pacific yew (Taxus brevifolia). It is a potent anticancer agent that has been approved for the treatment of refractory ovarian and metastatic breast cancer. Currently, the demand for taxol greatly exceeds the supply that can be isolated from its natural sources [22]. Very elegant total chemical syntheses of taxol have been developed but, because of low yields and high costs, none of these approaches is suitable for its commercial production. Similarly, Taxus cell cultures have yet to reach commercialization due to low and unstable productivity [23].
With the partial elucidation of the taxol biosynthetic pathway the possibility of taxadiene (a taxol biosynthetic intermediate) biosynthesis in Escherichia coli was successfully demonstrated [24]. One major limitation with that approach however is that the prokaryotic E. coli can not functionally express the P450 enzymes that widely participate in the taxol biosynthetic pathway. Hence, S. cerevisiae was chosen as an alternative, since it has been reported that yeast was successfully employed as host for the engineered, multi-step production of other (mevalonate-derived) terpenoids, including steroids and carotenoids [23].
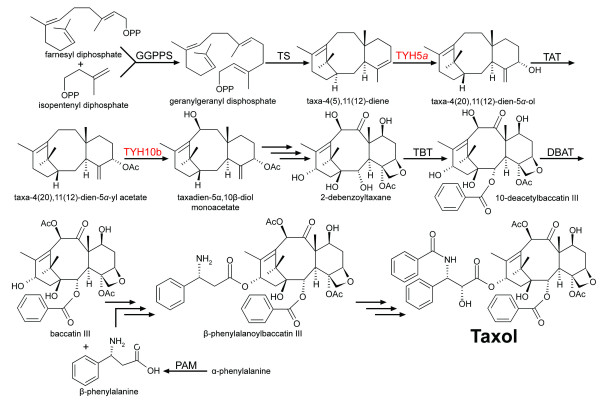
Recently, partial taxol biosynthetic pathway was constructed in S. cerevisiae by expressing five sequential pathway steps leading from primary isoprenoid metabolism to the intermediate taxadien-5α-acetoxy-10β-ol (Figure 2) [23]. The Taxus cuspidate genes the yeast host expressed included geranylgeranyl disphosphate synthase (GGPPS), taxadiene synthase (TS), cytochrome P450 taxadiene 5α-hydroxylase (TYH5a), taxadienol 5α-O-acetyl transferase (TAT) and taxoid 10β-hydroxylase (THY10b). The recombinant strain produced taxadiene at 1.0 mg/L while taxadien-5α-ol was produced in very small amounts (~25 μg/L). These results suggest that the first two enzymes (GGPPS and TS) cooperated well with each other and that the metabolic flux was reduced at the 5α-hydroxylation step, which is catalyzed by a cytochrome P450 hydroxylase [23]. It is anticipated that overexpressing Taxus P450 oxygenases with their corresponding P450 reductases in the yeast host would improve the overall production amounts [25].
Figure 2.
Taxol biosynthesis pathway. GGPPS: Genranylgenranyl diphosphate synthase; TS: taxadiene synthase; TYHa: taxadiene 5α-hydroxylase; TAT: taxa-4(20), 11(12)-dien-5a-ol-O-acetyltransferase; TYHb: taxane 10β-hydroxylase; TBT: taxane 2a-O-benzoyltransferase; DBAT: 10-deacetyl baccatin III-O-acetyltransferase. PAM: phenyalanine aminomutase. Multiple arrows indicate several as of yet undefined steps. Red text indicates a cytochrome P450 enzyme.
Although the metabolic engineering of taxol biosynthesis in yeast has only begun, the application of new protein engineering techniques to improve rate-controlling enzymes appears promising. As a result, S. cerevisiae recombinant strains not only provide a new approach for taxol production, but also create platforms that allow the synthesis of taxol analogues and other rare taxoids for clinical evaluation [15].
Carotenoids
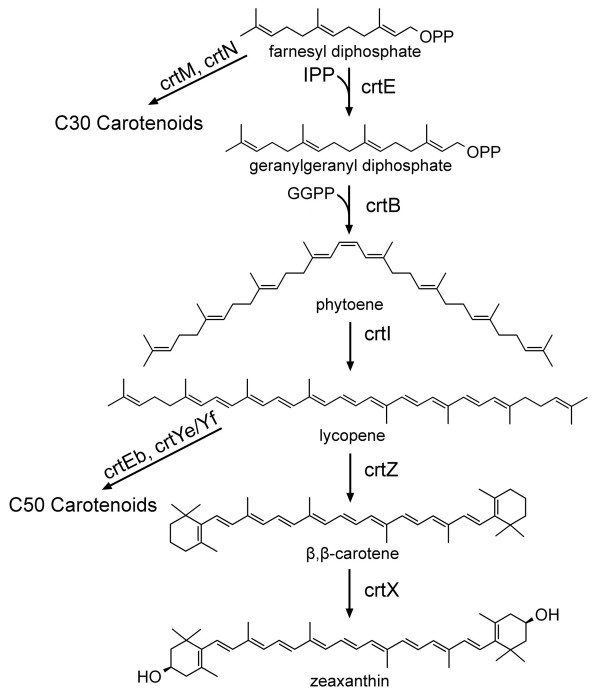
Carotenoids are a subfamily of isoprenoids that are the most widely distributed yellow, orange and red natural pigments synthesized in bacteria, algae, fungi [26]. Commercially carotenoids such as β-carotene and astaxanthin are used as food colorants, animal feed supplements and for nutritional and cosmetic purposes. More recently, carotenoids have received attention for their significant antioxidant activities and for playing an important role in inhibiting the onset of chronic diseases [27-30]. The majority of the 600 known structures of carotenoids are C40-carotenoids while a few bacterial carotenoids exist with 30, 45 or 50 carbon atoms (Figure 3).
Figure 3.
Carotenoid biosynthesis pathway. IPP: Isopentyl pyrophosphate; crtE: geranylgenranyl diphosphate synthase; crtB: phytoene synthase; crtI: phytoene desaturase; crtZ: β-carotene hydroxylase; crtX: zeaxanthin glucosylase; crtM: dehydrosqualene synthase; crtN: dehydrosqualene desaturase; crtEb: lycopene elongase; crtYe/Yf: heterodimeric decaprenoxanthin synthase.
Carotenoid pathways have been successfully introduced into noncarotenogenic microbes such as E. coli and S. cerevisiae [31-35]. Since S. cerevisiae produces ergosterol as its principal isoprenoid from farnesyl diphosphate (FPP), it would be possible to redirect the flux of FPP away from ergosterol into genranylgenranyl diphosphate (GGPP) and subsequent carotenoids. Indeed, the insertion of a plasmid containing Erwinia uredovora GGPP synthase (crtE), phytoene synthase (crtB) and phytoene desaturase (crtI) genes under the control of various S. cerevisiae promoters allowed S. cerevisiae to produce lycopene (113 μg/g dry weight) [35]. In addition, S. cerevisiae harboring a plasmid containing the additional E. uredovora lycopene cyclase (crtY) gene resulted in the production of β-carotene (103 μg/g dry weight) [35].
Sterols
Sterols are important for the physiology of eukaryotic organisms as they form part of the cellular membrane where they modulate their fluidity and function and participate as secondary messengers in developmental signaling [36]. In yeast, ergosterol is the main sterol and is analogous to cholesterol in mammalian cells [19]. Ergosterol is also an economically important metabolite, as it is the precursor for the production of vitamin D2. The complex production of sterols in yeast is tightly regulated at multiple levels. One of the major bottlenecks is the reaction catalyzed by HMG-CoA (Figure 1). Overexpression of a truncated Hmg1p led to a 40-fold increase of HMG-CoA activity and resulted in an increased accumulation of squalene, an intermediate towards ergosterol, of up to 5.5% of dry matter [37]. However, there was little effect on the overall ergosterol production, suggesting additional regulation further down the pathway to ergosterol. More recently, overexpression of the post-squalene genes led to the discovery of two additional bottlenecks in the pathway, namely squalene epoxidase (Erg1p) and sterol-14alpha-demethylase (Erg11p). Overexpression of Erg1p, Erg11p and a truncated Hmg1p led to a three-fold increase of sterol content over the wild-type [38]. However, 90% of the sterol content was esterfied sterol intermediates and not ergosterol. Obviously, further regulatory mechanisms are at play preventing the quantitative accumulation of ergosterol.
Besides ergosterol, several sterol intermediates are of biological importance including methylated sterol intermediates that activate meiosis [4,4-dimethyl-5alpha-cholesta-8,14,24-trienol (FF-MAS) and 4,4-dimethyl-5alpha-cholesta-8,24-dienol (T-MAS)] [39]. FF-MAS is the substrate of Erg24p and T-MAS is the substrate of Erg25p. While supplying exogenous ergosterol or cholesterol, engineered strains with abolished heme biosynthesis and deleted ERG25 gene accumulated T-MAS (1.3 mg/L) while deleting both ERG24 and ERG25 genes accumulated FF-MAS (5.4 mg/L) [40]. Zymosterol is another intermediate of interest as it is the precursor of cholesterol lowering substances. By deleting sterol transmethylase (ERG6) and 8-carbon sterol isomerase (ERG2), zymosterol accumulated as the major sterol [41].
Strains able to produce sterols not native to yeast have also been engineered. For example, a single mutation of Δ24(25)- to Δ24(28)-sterol methyl transferase (SMT) enzyme from S. cerevisiae resulted in a protein able to synthesize mono- and bis-C24-alkylated side chains of several Δ24(25)- and Δ24(28)-sterols [42]. Also, several 24-ethyl sterols and 24-ethylidene sterols were produced in engineered S. cerevisiae carrying a sterol 24-methyltransferase gene from the Arabidopsis thaliana [43].
Long chain Polyunsaturated Fatty Acids (PUFA) biosynthesis
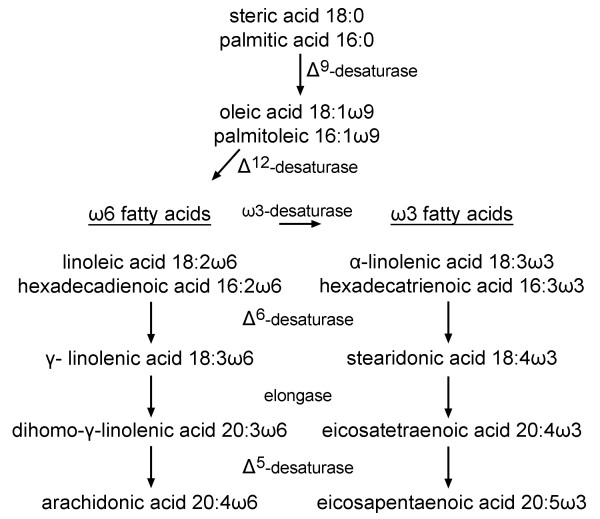
PUFAs are fatty acids of 16 or more carbon long chains with two or more double bonds (Figure 4). Normal cellular function is dependent upon PUFAs as they are signaling molecules and regulators of membrane fluidity [44]. In general, fatty acids can be synthesized endogenously; however, PUFAs are necessary part of a diet for normal growth and development because mammals are incapable of synthesizing the essential linoleic acid (18:2ω6) [45], and ω-3 and ω-6 fatty acids. Diets rich in PUFAs were shown in studies to protect infants from cardiovascular disease, and were beneficial for the development of retina and brain functions [44,46].
Figure 4.
Metabolic pathways of ω3 and ω6 fatty acids.
PUFAs are commercially obtained primarily from seeds, marine animals, microalgae and microheterotrophs. Over the past few years, progress has been made in engineering S. cerevisiae for fatty acid production. More specifically, in S. cerevisiae, only saturated (mainly 16:0 and 18:0) and mono-saturated (18:1ω9) fatty acids, are synthesized de novo [47,48]. Formation of PUFAs requires the expression of heterogeneous genes to introduce additional double bonds (eg. Δ6-, Δ12-desaturases) and produce enzymes for chain elongation (elongases) [18,49].
A number of desaturases from a variety of sources have been discovered and functionally expressed in S. cerevisiae to produce PUFAs. For example, expression of the Δ6-desaturase from Mucor rouxii in yeast resulted in an accumulation of γ-linolenic acid (18:3ω6) as 7.1% of the total fatty acids when fed with linoleic acid (18:2ω6) [50]. Introducing Δ12-desaturases from various sources allows for the accumulation of linoleic acid (18:2) and hexadecadienoic acid (16:2) fatty acids [51-55]. Co-expression of both a Δ6-desaturase and a Δ12-desaturase from M. alpine resulted in endogenous γ-linolenic acid (18:3ω6) as high as 8% of total fatty acid content [56].
The range and length of PUFAs synthesized in yeast have increased with the expression of various higher-eukaryote genes. For example in yeast, arachidonic acid and eicosapentaenoic acid can be produced by coexpression of Δ5- and Δ6-desaturases with a Δ6-elongase in the presence of exogenously supplied linoleic acid and linolenic acid, respectfully [57,58]. A desaturase from zebrafish (Danio rerio) displayed both Δ5 and Δ6-desaturase activity expressed in S. cerevisiae by converting linoleic acid (18:2ω6) and α-linolenic acid (18:3ω3) into their respective Δ6-desaturase products, γ-linolenic acid (18:3ω6) and stearidonic acid (18:4ω3) and by converting di-homo-γ-linoleic acid (20:3ω6) and eicosatetraenoic acid (20:4ω3) to their Δ5-desaturase products, arachionic acid (20:4ω6) and eicosapentaenoic acid (20:5ω3) [59]. The cDNAs encoding desaturase and elongase enzymes from various marine and fresh water fish, a primary source of fatty acids continue to be characterized [60,61].
Flavonoid biosynthesis
Flavonoids are a diverse group of plant secondary metabolites that contain a 15-carbon phenylpropanoid core, which is extensively modified by rearrangement, alkylation, oxidation and glycosylation [62]. These fascinating compounds possess extraordinary antioxidant activity and they also exhibit estrogenic, antiviral, antibacterial and anti-cancer activities [63].
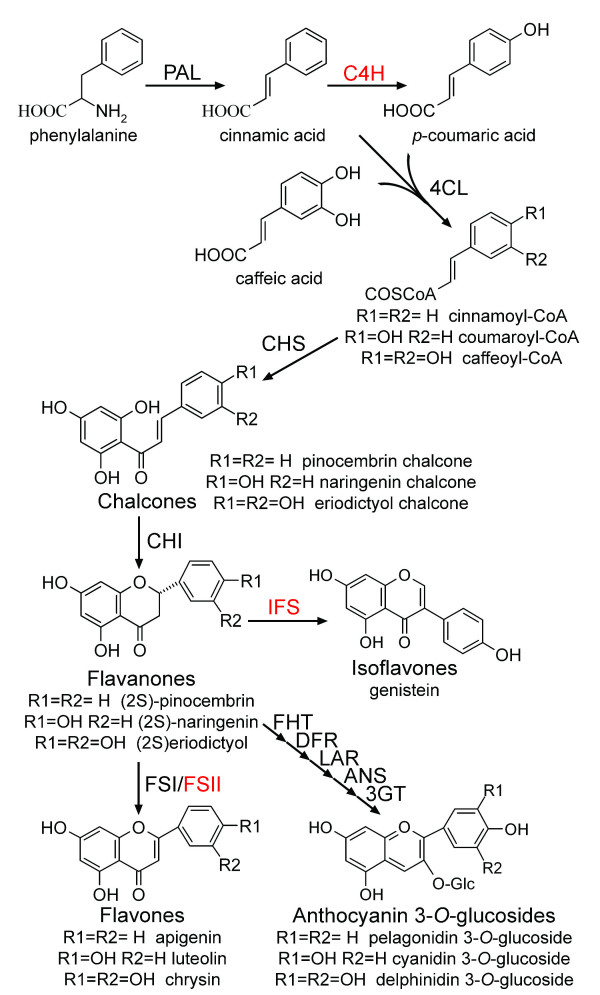
The health-protecting effects of flavonoids have stimulated significant research toward the elucidation of their biosynthetic networks (Figure 5), as well as the development of production platforms using well-characterized hosts, such as E. coli and S. cerevisiae [64]. The ability of yeast to express type II P450 hydroxylases, many of which are involved in flavonoid biosynthesis, makes it an attractive production platform, even though a recent report has demonstrated the functional expression of one such enzyme in E. coli [65]. Ro and Douglas [66] were the first to connect the two initial enzymes involved in phenylpropanoid pathway, namely phenylalanine ammonia lyase (PAL) and cinnamate 4-hydroxylase (C4H) in S. cerevisiae together with a cytochrome P450 reductase. The carbon flux through the multienzyme system from phenylalanine to p-coumaric acid in yeast was evaluated in their study. Later, two independent studies demonstrated the biosynthesis of flavanones, the common precursors of the vast majority of flavonoids, in S. cerevisiae. The first study by Jiang et al. [67] described the production of monohydroxylated naringenin and unhydroxylated pinocembrin at levels of 7 mg/L and 0.8 mg/L respectively. The second study by Yan et al. [68] reported the biosynthesis of flavanones in S. cerevisiae by constructing a gene cluster that included C4H from A. thaliana, 4cL-2 from parsley, and CHI-A (encoding for chalcone isomerase) and chs from petunia. The recombinant S. cerevisiae strain was fed with phenylpropanoid acids and produced naringenin (28.3 mg/L), pinocembrin (16.3 mg/L) and the trihydroxylated flavanone eriodictyol (6.5 mg/L). In a continuation of the previous work, Leonard et al demonstrated the biosynthesis of the flavone molecules apigenin, luteolin and chrysin by exploring the expression of a soluble flavone synthase I (FSI) and a membrane bound flavone synthase II (FSII) in yeast cells. The effect of the yeast P450 reductase overexpression on flavone biosynthesis was also investigated [8]. Recently, Ralston et al. [69] reported the partial reconstruction of isoflavonoid biosynthesis in S. cerevisiae by using different types of CHI and an isoflavone synthase (IFS) from soybean (Glycine max).
Figure 5.
Flavonoid biosynthesis pathway. PAL: Phenylalanine ammonia-lyase; C4H: cinnamate-4-hydroxylase; 4CL: 4-coumaroyl:CoA-ligase; CHS: chalcone synthase; CHI: chalcone isomerase; FSI: flavone synthase; FSII: cytochrome P450 flavone synthase; IFS: cytochrome P450 isoflavone synthase; FHT: flavanone 3β-hydroxylase; DFR: dihydroflavonol 4-reductase; LAR: leucoanthocyanidin synthase; ANS: anthocyanidin synthase; 3GT: UDPG-flavonoid 3-O-glucosyl transferase. Red text indicates cytochrome P450 enzymes.
With the rapid unraveling of the flavonoid biosynthetic pathways a wide array of flavonoid compounds, natural and unnatural, is expected to be produced from S. cerevisiae in the near future. In addition, the natural coloration of some of the flavonoid molecules (anthocyanins) opens up the possibility of using directed evolution for protein activity fine tuning and thus further improvement of the overall productivity [64].
Conclusion
Ever since the era of recombinant DNA technology for natural product biosynthesis emerged [70], S. cerevisiae has remained the production platform of choice for many fine chemicals, including natural products. The rapid elucidation of biosynthetic pathways made possible through advanced genomic tools, has made natural product again the molecules of choice for drug development. Indeed, half of the drugs currently in clinical use are natural products and it is expected that the market size of biotech-derived small molecules will exceed 100 billion US$ in 2010 and 400 billion US$ in 2030 [71,72]. As such, we believe that S. cerevisiae will not only remain the cell factory of choice but that the application of powerful systems biology approaches will facilitate its expanded role in industrial applications [73-75].
Authors' contributions
MAGK: Contributing author.
YY: Contributing author.
JAC: Contributing author and designer of all figures.
Contributor Information
Joseph A Chemler, Email: jchemler@buffalo.edu.
Yajun Yan, Email: yajunyan@buffalo.edu.
Mattheos AG Koffas, Email: mkoffas@buffalo.edu.
References
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. Life with 6000 genes. Science. 1996;274:546–&. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng WT, Zhang W, Yang X, Pootoolal J, Chua G, Lopez A, Trochesset M, Morse D, Krogan NJ, Hiley SL, Li Z, Morris Q, Grigull J, Mitsakakis N, Roberts CJ, Greenblatt JF, Boone C, Kaiser CA, Andrews BJ, Hughes TR. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Hotze M, Schroder G, Schroder J. Cinnamate 4-hydroxylase from Catharanthus roseus, and a strategy for the functional expression of plant cytochrome P-450 proteins as translational fusions with P-450 reductase in Escherichia coli. Febs Letters. 1995;374:345–350. doi: 10.1016/0014-5793(95)01141-Z. [DOI] [PubMed] [Google Scholar]
- Sieber V, Martinez CA, Arnold FH. Libraries of hybrid proteins from distantly related sequences. Nature Biotechnology. 2001;19:456–460. doi: 10.1038/88129. [DOI] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P. Yeast expression of animal and plant P450s in optimized redox environments. Cytochrome P450, Pt B. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- Leonard E, Yan YJ, Lim KH, Koffas MAG. Investigation of two distinct flavone synthases for plant-specific flavone biosynthesis in Saccharomyces cerevisiae. Applied and Environmental Microbiology. 2005;71:8241–8248. doi: 10.1128/AEM.71.12.8241-8248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster J, Famili I, Fu P, Palsson BO, Nielsen J. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Research. 2003;13:244–253. doi: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil KR, Akesson M, Nielsen J. Use of genome-scale microbial models for metabolic engineering. Current Opinion in Biotechnology. 2004;15:64–69. doi: 10.1016/j.copbio.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Daran-Lapujade P, Jansen MLA, Daran JM, van Gulik W, de Winde JH, Pronk JT. Role of transcriptional regulation in controlling fluxes in central carbon metabolism of Saccharomyces cerevisiae - A chemostat culture study. J Biol Chem J Biol Chem. 2004;279:9125–9138. doi: 10.1074/jbc.M309578200. [DOI] [PubMed] [Google Scholar]
- Sacchettini JC, Poulter CD. Creating isoprenoid diversity. Science. 1997;277:1788–1789. doi: 10.1126/science.277.5333.1788. [DOI] [PubMed] [Google Scholar]
- Watts KT, Mijts BN, Schmidt-Dannert C. Current and emerging approaches for natural product biosynthesis in microbial cells. Advanced Synthesis & Catalysis. 2005;347:927–940. doi: 10.1002/adsc.200505062. [DOI] [Google Scholar]
- Haynes RK, Krishna S. Artemisinins: activities and actions. Microbes and Infection. 2004;6:1339–1346. doi: 10.1016/j.micinf.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Jennewein S, Croteau R. Taxol: biosynthesis, molecular genetics, and biotechnological applications. Applied Microbiology and Biotechnology. 2001;57:13–19. doi: 10.1007/s002530100757. [DOI] [PubMed] [Google Scholar]
- Katsuki HBK. Studies on the biosynthesis of ergosterol in yeast. Formation of methylated internediates. Journal of Biological Chemistry. 1967;242:222–227. [PubMed] [Google Scholar]
- Lynen F. Biosynthetic pathways from acetate to natural products. Pure and Applied Chemistry. 1967;14:137–167. doi: 10.1351/pac196714010137. [DOI] [PubMed] [Google Scholar]
- Veen M, Lang C. Production of lipid compounds in the yeast Saccharomyces cerevisiae. Applied Microbiology and Biotechnology. 2004;63:635–646. doi: 10.1007/s00253-003-1456-7. [DOI] [PubMed] [Google Scholar]
- Bloch KE. Sterol structure and membrane function. CRC Critical Reviews in Biochemistry. 1983;14:47–92. doi: 10.3109/10409238309102790. [DOI] [PubMed] [Google Scholar]
- Parks LW, Casey WM. Physiological implications of sterol biosynthesis in yeast. Annual Review of Microbiology. 1995;49:95–116. doi: 10.1146/annurev.mi.49.100195.000523. [DOI] [PubMed] [Google Scholar]
- Parks LW, Smith SJ, Crowley JH. Biochemical and physiological effects of sterol alterations in yeast - A review. Lipids. 1995;30:227–230. doi: 10.1007/BF02537825. [DOI] [PubMed] [Google Scholar]
- Hezari M, Lewis NG, Croteau R. Purification and characterization of taxa-4(5),11(12)-diene synthase from Pacific Yew (Taxus brevifolia) that catalyzes the first committed step of taxol biosynthesis. Archives of Biochemistry and Biophysics. 1995;322:437–444. doi: 10.1006/abbi.1995.1486. [DOI] [PubMed] [Google Scholar]
- Dejong JM, Liu Y, Bollon AP, Long RM, Jennewein S, Williams D, Croteau RB. Genetic engineering of taxol biosynthetic genes in Saccharomyces cerevisiae. Biotechnol Bioeng. 2006;93:212–224. doi: 10.1002/bit.20694. [DOI] [PubMed] [Google Scholar]
- Yang CS, Landau JM, Huang MT, Newmark HL. Inhibition of carcinogenesis by dietary polyphenolic compounds. Annual Review of Nutrition. 2001;21:381–406. doi: 10.1146/annurev.nutr.21.1.381. [DOI] [PubMed] [Google Scholar]
- Jennewein S, Park H, DeJong JM, Long RM, Bollon AP, Croteau RB. Coexpression in yeast of Taxus cytochrome P450 reductase with cytochrome P450 oxygenases involved in Taxol biosynthesis. Biotechnol Bioeng. 2005;89:588–598. doi: 10.1002/bit.20390. [DOI] [PubMed] [Google Scholar]
- Goodwin TW, Britton G. Distribution and analysis of carotenoids. In: Goodwin TW, editor. Plant Pigments. London , Academic Press; 1988. pp. 61–132. [Google Scholar]
- Bertram JS. Carotenoids and gene regulation. Nutrition Reviews. 1999;57:182–191. doi: 10.1111/j.1753-4887.1999.tb06941.x. [DOI] [PubMed] [Google Scholar]
- Edge R, McGarvey DJ, Truscott TG. The carotenoids as anti-oxidants - a review. J Photoch Photobio B J Photoch Photobio B. 1997;41:189–200. doi: 10.1016/S1011-1344(97)00092-4. [DOI] [PubMed] [Google Scholar]
- Giuliano A. Cancer chemoprevention - Part 1: Retinoids and carotenoids and other classic antioxidants - The Singh/Lippman article reviewed. Oncology-New York. 1998;12:1659–1660. [PubMed] [Google Scholar]
- Smith TAD. Carotenoids and cancer: prevention and potential therapy. Brit J Biomed Sci Brit J Biomed Sci. 1998;55:268–275. [PubMed] [Google Scholar]
- Misawa N, Nakagawa M, Kobayashi K, Yamano S, Izawa Y, Nakamura K, Harashima K. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. Journal of Bacteriology. 1990;172:6704–6712. doi: 10.1128/jb.172.12.6704-6712.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa N, Yamano S, Ikenaga H. Production of beta-carotene in Zymomonas mobilis and Agrobacterium tumefaciens by introduction of the biosynthesis genes from Erwinia uredovora. Applied and Environmental Microbiology. 1991;57:1847–1849. doi: 10.1128/aem.57.6.1847-1849.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa N, Yamano S, Linden H, de Felipe MR, Lucas M, Ikenaga H, Sandmann G. Functional expression of the Erwinia uredovora carotenoid biosynthesis gene Crtl in transgenic plants showing an increase of beta-carotene biosynthesis activity and resistance to the bleaching herbicide Norflurazon. Plant Journal. 1993;4:833–840. doi: 10.1046/j.1365-313X.1993.04050833.x. [DOI] [PubMed] [Google Scholar]
- Ausich RL. Production of carotenoids by recombinant-DNA technology. Pure Appl Chem. 1994;66:1057–1062. [Google Scholar]
- Yamano S, Ishii T, Nakagawa M, Ikenaga H, Misawa N. Metabolic engineering for production of beta-carotene and lycopene in Saccharomyces cerevisiae. Biosci Biotechnol Biochem. 1994;58:1112–1114. doi: 10.1271/bbb.58.1112. [DOI] [PubMed] [Google Scholar]
- Mansour MP, Volkman JK, Blackburn SI. The effect of growth phase on the lipid class, fatty acid and sterol composition in the marine dinoflagellate, Gymnodinium sp in batch culture. Phytochemistry. 2003;63:145–153. doi: 10.1016/S0031-9422(03)00052-9. [DOI] [PubMed] [Google Scholar]
- Polakowski T, Stahl U, Lang C. Overexpression of a cytosolic hydroxymethylglutaryl-CoA reductase leads to squalene accumulation in yeast. Appl Microbiol Biotechnol. 1998;49:66–71. doi: 10.1007/s002530051138. [DOI] [PubMed] [Google Scholar]
- Veen M, Stahl U, Lang C. Combined overexpression of genes of the ergosterol biosynthetic pathway leads to accumulation of sterols in Saccharomyces cerevisiae. Fems Yeast Research. 2003;4:87–95. doi: 10.1016/S1567-1356(03)00126-0. [DOI] [PubMed] [Google Scholar]
- Byskov AG, Andersen CY, Leonardsen L, Baltsen M. Meiosis activating sterols (MAS) and fertility in mammals and man. J Exp Zool. 1999;285:237–242. doi: 10.1002/(SICI)1097-010X(19991015)285:3<237::AID-JEZ6>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Xu R, Wilson WK, Matsuda SPT. Production of melosis-activating sterols from metabolically engineered yeast. J Am Chem Soc J Am Chem Soc. 2002;124:918–919. doi: 10.1021/ja0175542. [DOI] [PubMed] [Google Scholar]
- Heidepriem RW, Livant PD, Parish EJ, Barbuch RJ, Broaddus MG, Bard M. A simple method for the isolation of zymosterol from a sterol mutant of Saccharomyces cerevisiae. J Steroid Biochem J Steroid Biochem. 1992;43:741–743. doi: 10.1016/0960-0760(92)90300-8. [DOI] [PubMed] [Google Scholar]
- Nes WD, McCourt BS, Marshall JA, Ma JZ, Dennis AL, Lopez M, Li HX, He L. Site-directed mutagenesis of the sterol methyl transferase active site from Saccharomyces cerevisiae results in formation of novel 24-ethyl sterols. J Org Chem J Org Chem. 1999;64:1535–1542. doi: 10.1021/jo9819943. [DOI] [PubMed] [Google Scholar]
- Husselstein T, Gachotte D, Desprez T, Bard M, Benveniste P. Transformation of Saccharomyces cerevisiae with a cDNA encoding a sterol C-methyltransferase from Arabidopsis thaliana results in the synthesis of 24-ethyl sterols. FEBS Lett. 1996;381:87–92. doi: 10.1016/0014-5793(96)00089-0. [DOI] [PubMed] [Google Scholar]
- von Schacky C, Dyerberg J. Omega 3 fatty acids. From eskimos to clinical cardiology - What took us so long? World Rev Nutr. 2001;88:90–99. doi: 10.1159/000059772. [DOI] [PubMed] [Google Scholar]
- Broun P, Gettner S, Somerville C. Genetic engineering of plant lipids. Annual Review of Nutrition. 1999;19:197–216. doi: 10.1146/annurev.nutr.19.1.197. [DOI] [PubMed] [Google Scholar]
- Damsgaard CT, Schack-Nielsen L, Michaelsen KF, Fruekilde MB, Hels O, Lauritzen L. Fish oil affects blood pressure and the plasma lipid profile in healthy Danish infants. J Nutr. 2006;136:94–99. doi: 10.1093/jn/136.1.94. [DOI] [PubMed] [Google Scholar]
- Daum G, Vance JE. Import of lipids into mitochondria. Progress in Lipid Research. 1997;36:103–130. doi: 10.1016/S0163-7827(97)00006-4. [DOI] [PubMed] [Google Scholar]
- Martin CE, Oh CS, Kandasamy P, Chellapa R, Vemula M. Yeast desaturases. Biochem Soc T Biochem Soc T. 2002;30:1080–1082. doi: 10.1042/BST0301080. [DOI] [PubMed] [Google Scholar]
- Dyal SD, Narine SS. Implications for the use of Mortierella fungi in the industrial production of essential fatty acids. Food Res Int Food Res Int. 2005;38:445–467. [Google Scholar]
- Laoteng K, Mannontarat R, Tanticharoen M, Cheevadhanarak S. Δ6-Desaturase of Mucor rouxii with high similarity to plant Δ6-desaturase and its heterologous expression in Saccharomyces cerevisiae. Biochem Bioph Res Co Biochem Bioph Res Co. 2000;279:17–22. doi: 10.1006/bbrc.2000.3856. [DOI] [PubMed] [Google Scholar]
- Covello PS, Reed DW. Functional expression of the extraplastidial Arabidopsis thaliana oleate desaturase gene (FAD2) in Saccharomyces cerevisiae. Plant Physiology. 1996;111:223–226. doi: 10.1104/pp.111.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuradani E, Kobayashi M, Ashikari T, Shimizu S. Identification of Δ12-fatty acid desaturase from arachidonic acid-producing Mortierella fungus by heterologous expression in the yeast Saccharomyces cerevisiae and the fungus Aspergillus oryzae. Eur J Biochem Eur J Biochem. 1999;261:812–820. doi: 10.1046/j.1432-1327.1999.00333.x. [DOI] [PubMed] [Google Scholar]
- Kajiwara S, Shirai A, Fujii T, Toguri T, Nakamura K, Ohtaguchi K. Polyunsaturated fatty acid biosynthesis in Saccharomyces cerevisiae: Expression of ethanol tolerance and the FAD2 gene from Arabidopsis thaliana. Applied and Environmental Microbiology. 1996;62:4309–4313. doi: 10.1128/aem.62.12.4309-4313.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyou-Ndi MM, Watts JL, Browse J. Identification and characterization of an animal Δ12 fatty acid desaturase gene by heterologous expression in Saccharomyces cerevisiae. Arch Biochem Biophys. 2000;376:399–408. doi: 10.1006/abbi.2000.1733. [DOI] [PubMed] [Google Scholar]
- Suga K, Honjoh K, Furuya N, Shimizu H, Nishi K, Shinohara F, Hirabaru Y, Maruyama I, Miyamoto T, Hatano S, Iio M. Two low-temperature-inducible Chlorella genes for Δ12 and ω-3 fatty acid desaturase (FAD): Isolation of Δ12 and ω-3 FAD cDNA clones, expression of Δ12 fad in Saccharomyces cerevisiae, and expression of ω-3 fad in Nicotiana tabacum. Bioscience Biotechnology and Biochemistry. 2002;66:1314–1327. doi: 10.1271/bbb.66.1314. [DOI] [PubMed] [Google Scholar]
- Huang YS, Chaudhary S, Thurmond JM, Bobik EG, Yuan L, Chan GM, Kirchner SJ, Mukerji P, Knutzon DS. Cloning of Δ12-and Δ6-desaturases from Mortierella alpina and recombinant production of γ-linolenic acid in Saccharomyces cerevisiae. Lipids. 1999;34:649–659. doi: 10.1007/s11745-999-0410-8. [DOI] [PubMed] [Google Scholar]
- Beaudoin F, Michaelson LV, Hey SJ, Lewis MJ, Shewry PR, Sayanova O, Napier JA. Heterologous reconstitution in yeast of the polyunsaturated fatty acid biosynthetic pathway. Proc Natl Acad Sci U S A. 2000;97:6421–6426. doi: 10.1073/pnas.110140197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domergue F, Abbadi A, Ott C, Zank TK, Zahringer U, Heinz E. Acyl carriers used as substrates by the desaturases and elongases involved in very long-chain polyunsaturated fatty acids biosynthesis reconstituted in yeast. J Biol Chem. 2003;278:35115–35126. doi: 10.1074/jbc.M305990200. [DOI] [PubMed] [Google Scholar]
- Hastings N, Agaba M, Tocher DR, Leaver MJ, Dick JR, Sargent JR, Teale AJ. A vertebrate fatty acid desaturase with Δ5 and Δ6 activities. P Natl Acad Sci USA P Natl Acad Sci USA. 2001;98:14304–14309. doi: 10.1073/pnas.251516598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agaba M, Tocher DR, Dickson CA, Dick JR, Teale AJ. Zebrafish cDNA encoding multifunctional fatty acid elongase involved in production of eicosapentaenoic (20 : 5n-3) and docosahexaenoic (22 : 6n-3) acids. Marine Biotechnology. 2004;6:251–261. doi: 10.1007/s10126-003-0029-1. [DOI] [PubMed] [Google Scholar]
- Agaba MK, Tocher DR, Zheng XZ, Dickson CA, Dick JR, Teale AJ. Cloning and functional characterisation of polyunsaturated fatty acid elongases of marine and freshwater teleost fish. Comp Biochem Phys B Comp Biochem Phys B. 2005;142:342–352. doi: 10.1016/j.cbpb.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Turnbull JJ, Nakajima J, Welford RWD, Yamazaki M, Saito K, Schofield CJ. Mechanistic studies on three 2-oxoglutarate-dependent oxygenases of flavonoid biosynthesis - Anthocyanidin synthase, flavonol synthase, and flavanone 3 β-hydroxylase. J of Biol Chem. 2004;279:1206–1216. doi: 10.1074/jbc.M309228200. [DOI] [PubMed] [Google Scholar]
- Forkmann G, Martens S. Metabolic engineering and applications of flavonoids. Current Opinion in Biotechnology. 2001;12:155–160. doi: 10.1016/S0958-1669(00)00192-0. [DOI] [PubMed] [Google Scholar]
- Yan Y, Chemler J, Huang L, Martens S, Koffas MA. Metabolic engineering of anthocyanin biosynthesis in Escherichia coli. Appl Environ Microbiol. 2005;71:3617–3623. doi: 10.1128/AEM.71.7.3617-3623.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard E, Yan Y, Koffas MA. Functional expression of a P450 flavonoid hydroxylase for the biosynthesis of plant-specific hydroxylated flavonols in Escherichia coli. Metab Eng. 2006;8:172–181. doi: 10.1016/j.ymben.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Ro DK, Douglas CJ. Reconstitution of the entry point of plant phenylpropanoid metabolism in yeast (Saccharomyces cerevisiae) - Implications for control of metabolic flux into the phenylpropanoid pathway. J of Biol Chem. 2004;279:2600–2607. doi: 10.1074/jbc.M309951200. [DOI] [PubMed] [Google Scholar]
- Jiang HX, Wood KV, Morgan JA. Metabolic engineering of the phenylpropanoid pathway in Saccharomyces cerevisiae. Applied and Environmental Microbiology. 2005;71:2962–2969. doi: 10.1128/AEM.71.6.2962-2969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Kohli A, Koffas MA. Biosynthesis of natural flavanones in Saccharomyces cerevisiae. Applied and Environmental Microbiology. 2005;71:5610–5613. doi: 10.1128/AEM.71.9.5610-5613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston L, Subramanian S, Matsuno M, Yu O. Partial reconstruction of flavonoid and isoflavonoid biosynthesis in yeast using soybean type I and type II chalcone isomerases. Plant Physiology. 2005;137:1375–1388. doi: 10.1104/pp.104.054502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood DA, Malpartida F, Kieser HM, Ikeda H, Duncan J, Fujii I, Rudd BA, Floss HG, Omura S. Production of 'hybrid' antibiotics by genetic engineering. Nature. 1985;314:642–644. doi: 10.1038/314642a0. [DOI] [PubMed] [Google Scholar]
- Maury J, Asadollahi MA, Moller K, Clark A, Nielsen J. Microbial isoprenoid production: An example of green chemistry through metabolic engineering. Biotechnology for the Future. 2005;100:19–51. doi: 10.1007/b136410. [DOI] [PubMed] [Google Scholar]
- Paterson I, Anderson EA. Chemistry. The renaissance of natural products as drug candidates. Science. 2005;310:451–453. doi: 10.1126/science.1116364. [DOI] [PubMed] [Google Scholar]
- Koffas M, Stephanopoulos G. Strain improvement by metabolic engineering: lysine production as a case study for systems biology. Current Opinion in Biotechnology. 2005;16:361–366. doi: 10.1016/j.copbio.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Raab RM, Tyo K, Stephanopoulos G. Metabolic engineering. Adv Biochem Eng Biotechnol. 2005;100:1–17. doi: 10.1007/b136411. [DOI] [PubMed] [Google Scholar]
- Bro C, Regenberg B, Forster J, Nielsen J. In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab Eng Metab Eng. 2006;8:102–111. doi: 10.1016/j.ymben.2005.09.007. [DOI] [PubMed] [Google Scholar]