Abstract
Neurofibromatosis type I (NF1; also known as von Recklinghausen’s disease) is a common autosomal-dominant condition primarily affecting neural crest–derived tissues. The disease gene, NF1, encodes neurofibromin, a protein of over 2,800 amino acids that contains a 216–amino acid domain with Ras–GTPase-activating protein (Ras-GAP) activity. Potential therapies for NF1 currently in development and being tested in clinical trials are designed to modify NF1 Ras-GAP activity or target downstream effectors of Ras signaling. Mice lacking the murine homolog (Nf1) have mid-gestation lethal cardiovascular defects due to a requirement for neurofibromin in embryonic endothelium. We sought to determine whether the GAP activity of neurofibromin is sufficient to rescue complete loss of function or whether other as yet unidentified functions of neurofibromin might also exist. Using cre-inducible ubiquitous and tissue-specific expression, we demonstrate that the isolated GAP-related domain (GRD) rescued cardiovascular development in Nf1–/– embryos, but overgrowth of neural crest–derived tissues persisted, leading to perinatal lethality. These results suggest that neurofibromin may possess activities outside of the GRD that modulate neural crest homeostasis and that therapeutic approaches solely aimed at targeting Ras activity may not be sufficient to treat tumors of neural crest origin in NF1.
Introduction
Neurofibromatosis type I (NF1) is one of the most common monogenetic disorders, affecting 1 in 3,500 individuals (1). The effects of NF1 are seen in nearly all parts of the body, such as learning disabilities (2, 3), skeletal dysplasias (4), vascular abnormalities such as moya-moya and renovascular hypertension (1, 5), and cardiac abnormalities such as valvar pulmonary stenosis (1, 6). However, the most prominent clinical symptoms affect tissues of neural crest origin (7), such as the peripheral nervous system and melanocytes, with clinical manifestations such as neurofibromas, axillary freckling, and café-au-lait spots. More ominously, NF1 patients are also at risk for the development of life-threatening malignancies such as malignant peripheral nerve sheath tumors (MPNSTs) (4, 8), central nervous system gliomas (9), and myeloid leukemia (10, 11).
Neurofibromin, the protein encoded by the disease gene NF1, is composed of 2,818 amino acids, most of which are of unknown function. A small, 216–amino acid domain of neurofibromin, the GTPase-activating protein–related (GAP-related) domain (GRD), bears strong sequence homology to mammalian and yeast Ras-GAPs (12) and is able to hydrolyze GTP-bound Ras to GDP-bound Ras, thus reducing Ras activity. This downregulation of active Ras is thought to account for the tumor suppressor function of neurofibromin (13). Mutations affecting the Ras-GAP activity of neurofibromin have been identified in NF1 patients and NF1-related malignancies (14–16), and downstream effectors of Ras signaling are activated in these cells. In particular, the demonstration of clinical NF1 in a family with a point mutation in the GRD leading to the specific loss of Ras-GAP activity without marked disruption of the protein structure suggests the necessity of this domain for tumor suppressor function (15). Further, reconstitution of Ras-GAP function through GRD expression rescues the growth abnormalities of Nf1–/– cultured hematopoietic stem cells, neural stem cells, and embryonic fibroblasts in vitro (17, 18).
On the other hand, large regions of the NF1 coding sequence have been conserved across millions of years of evolution from Drosophila to man, suggesting significant functions outside the GRD (19). In Drosophila, neurofibromin can function to modulate adenylate cyclase activity, and this function has been demonstrated in mammalian cell systems (19–22). Numerous mutations outside the GRD, which do not appear to affect protein stability or GAP function, have been correlated with human disease (23, 24). Neurofibromin has been found to associate with cytoskeletal proteins (25), and studies in cultured cells suggest that the GAP activity of neurofibromin does not account for NF1-dependent changes in cell adhesion and some aspects of MAPK signaling (22).
Despite these conflicting bodies of evidence concerning the relative importance of neurofibromin GAP activity, therapeutic approaches for the treatment of NF1-related tumors have focused on the Ras-GAP activity of neurofibromin and have sought to normalize elevated Ras activity levels. For example, clinical trials for the treatment of NF1-related malignancies, such as MPNSTs, with farnesyltransferase inhibitors are underway (26). However, initial results have not shown marked effects (27), and these approaches may not effectively target activated Ras (28–30).
Results
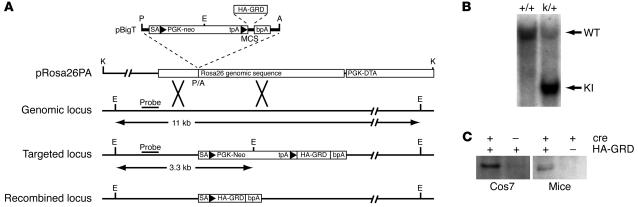
We have generated a tractable mouse model that allows for activation and functional expression of the isolated neurofibromin GRD in a tissue-specific fashion. Using homologous recombination in embryonic stem cells, we inserted a HA-tagged human NF1 GRD coding sequence (HA-GRD) into the ROSA26 locus (31, 32) downstream of a floxed PGK-neo cassette, designated HA-GRDk/+. The insert was not expressed unless cre-mediated recombination removed the neo cassette and allowed for HA-GRD expression (Figure 1A). We demonstrated appropriate targeting by Southern blotting (Figure 1B) and cre-dependent expression of HA-GRD in mice by Western blot analysis (Figure 1C).
Figure 1. Generation of HA-GRD ROSA26 knock-in (HA-GRDk/+) mice.
(A) Targeting strategy of HA-GRDk/+. NF1 HA-GRD was cloned into vector pBigT and subsequently into the targeting vector pRosa26PA. The expected sizes of the EcoRV (E) digests recognized by the indicated probe are shown. Arrowheads indicate loxP. A, AscI; P, PacI; K, KpnI. (B) Genomic Southern blot of wild-type (+/+) and HA-GRDk/+ (k/+) mice. Wild-type (WT; 11 kb) and knock-in (KI; 3.3 kb) bands are shown. (C) Anti-HA Western blot of transfected Cos7 cells and newborn mice. Cells were transfected with pCCALL–HA-GRD (HA-GRD) with or without pTurboCre (cre). The newborn mice were CMV-cre+ transgenic mice with or without HA-GRDk/+. In both instances HA-GRD was detected only with coexpression of cre recombinase.
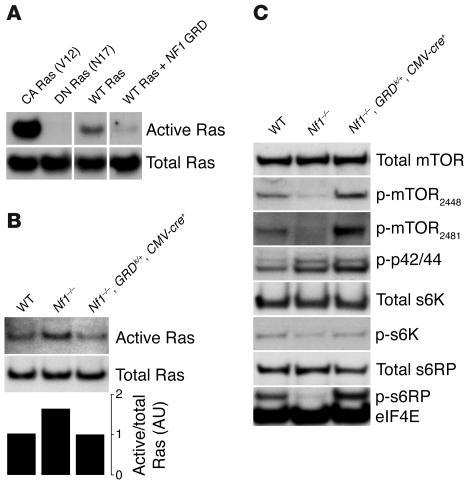
The isolated HA-GRD displays properties consistent with Ras-GAP activity both in vitro and in vivo. Using a related cre-activated plasmid to express HA-GRD in Cos cells (pCCALL2–HA-GRD), we showed that cre-mediated expression of GRD reduced Ras-GTP levels (Figure 2A). We crossed HA-GRDk/+ mice with CMV-cre+ mice, which express cre recombinase ubiquitously (33). Offspring were born in the expected Mendelian ratios and appeared normal, suggesting that overexpression of HA-GRD is well tolerated.
Figure 2. HA-GRD expression downregulates Ras activity and normalizes downstream effectors.
(A) HA-GRD expression downregulates activated Ras in cells. Cos7 cells were transfected with expression vectors for constitutively active (CA Ras, rasV12), dominant-negative (DN Ras, rasN17), and wild-type Ras (WT Ras). Cells with wild-type Ras were also transfected with an expression construct for HA-GRD (WT Ras + NF1 GRD). Protein samples were blotted with anti-Ras antibody before and after pulldown with the Ras-binding domain of raf-1. All bands in A were from a single scan of the same blot. (B) HA-GRD expression rescues abnormally activated Ras in Nf1–/– embryos. Ras pulldowns of protein lysates from E12.5 littermates: wild type; Nf1–/–; and Nf1–/–, HA-GRDk/+, CMV-cre+. The intensities of the bands were normalized to total Ras in the graph. (C) Embryonic HA-GRD expression rescues down-stream Ras effector abnormalities. Western blots of E12.5 embryos of wild-type; Nf1–/–; and Nf1–/–, HA-GRDk/+, CMV-cre+ genotypes. There was a reduction of phosphorylated mTOR (p-mTOR), phosphorylated S6K, and phosphorylated S6RP in Nf1–/– relative to wild type. CMV-cre–mediated expression of NF1 HA-GRD in Nf1–/– rescued this reduction in activated mTOR, s6K, and s6RP. eIF4E served as a loading control.
Next, we established appropriate matings to generate Nf1–/– mice that also expressed HA-GRD ubiquitously. We utilized CMV promoter–mediated cre expression by crossing Nf1+/–, HA-GRDk/+ with Nf1+/–, CMV-cre+ mice. Nf1–/– embryos die at mid-gestation with cardiovascular defects and are not identified beyond E13.5 (34, 35). Prior to this embryonic stage, we had not observed any alterations from expected Mendelian ratios. At E12.5, we harvested embryos and measured total and activated Ras. Active Ras was elevated in Nf1–/– embryos and returned to wild-type levels by expression of HA-GRD (Figure 2B).
Contrary to earlier reports using conditions vastly different from our own, including cultured cells and central nervous system tumors (36, 37), we found that Nf1 deficiency was associated with downregulation of phosphorylated (activated) mammalian target of rapamycin (mTOR) and its downstream signaling effectors s6 kinase (s6K) and s6 ribosomal protein (s6RP) in the developing embryo. In mice, these activities were returned to wild-type levels by expression of HA-GRD (Figure 2C). In cultured cells and some tissues, abnormal activation of mTOR and its downstream effectors has been reported upon loss of Nf1 (36, 37), while our results demonstrated abnormal downregulation of these same pathways. This discrepancy may reflect the differences between acute and chronic Ras dysregulation in some tissues. They may also be the result of differences in the nature of these pathways in oncologic and developmental contexts. Furthermore, our results could reflect secondary changes in tissues other than those primarily affected by loss of neurofibromin. Contextually divergent regulatory mechanisms and their resultant feedback controls may result in disparate findings in cell culture, in tumors, and in embryos. Outlining and understanding these differences will be important for future investigation.
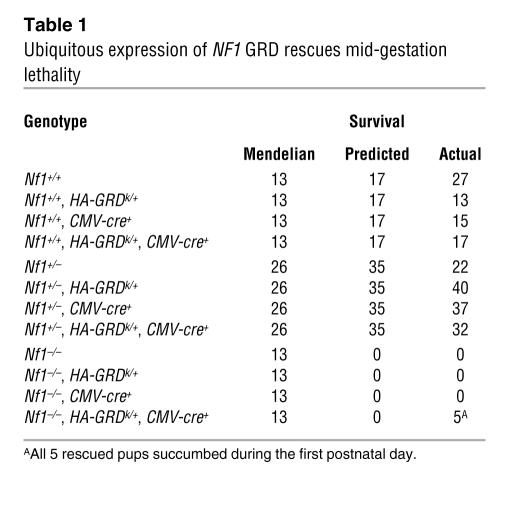
As expected, we did not identify any Nf1–/– pups at birth, nor did we identify any Nf1–/– pups that carried the CMV-cre transgene alone or 1 copy of the silent HA-GRDk/+ allele in the absence of cre (Table 1). However, out of 208 newborn pups examined, we identified 5 Nf1–/– pups that were CMV-cre+ and HA-GRDk/+ (Table 1). Thus, ubiquitous expression of HA-GRD is able to at least partially rescue mid-gestation lethality of Nf1–/– embryos.
Table 1 .
Ubiquitous expression of NF1 GRD rescues mid-gestation lethality
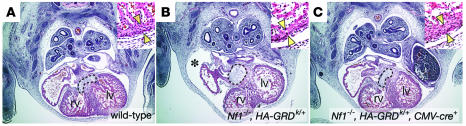
Rescued Nf1–/– pups, however, were not normal and succumbed in the perinatal period. Analysis of E12.5 embryos demonstrated normalization of cardiac development with HA-GRD expression in Nf1–/– (Figure 3). Histological examination of rescued newborns revealed normal cardiac anatomy (data not shown). All examined rescued pups, however, showed striking, tumor-like overgrowth of neural crest–derived structures. Compared with Nf1+/– littermates (Figure 4A), the neural crest–derived adrenal medulla in Nf1–/–, HA-GRDk/+, CMV-cre+ mice was significantly enlarged (Figure 4B). This enlargement was similar to what we have observed with mice with neural crest–specific Nf1 inactivation (Figure 4C) (38). Enormous paraspinal masses arising from the dorsal root ganglia were also evident in all rescued pups (compare Figure 4, E and G, with Figure 4D). These findings were almost identical to those that we have previously described in newborn pups with neural crest–specific inactivation of Nf1 (38). Like HA-GRD rescued Nf1–/– pups, neural crest–specific Nf1 knockout mice died at birth, with dramatic overgrowth of neural crest structures (Figure 4, C and F). Hence, ubiquitous expression of HA-GRD failed to rescue the phenotype of neurofibromin loss in neural crest derivatives during development.
Figure 3. HA-GRD expression rescues embryonic cardiovascular defects of Nf1–/–.
Representative cross-sections (magnification, ×4) of hearts through the developing endocardial cushions (dashed line) with high-power insets (magnification, ×20) of left ventricular wall from E12.5 wild-type (A); Nf1–/–, HA-GRDk/+ (B); and Nf1–/–, HA-GRDk/+, CMV-cre+ (C) littermates. Right ventricles (rv) and left ventricles (lv) are indicated. Arrowheads in the insets highlight thickness of the ventricular wall. The developing endocardial cushions of B were enlarged compared with those in A. In addition, there was a large pericardial effusion (asterisk in B), suggestive of cardiovascular failure, and a thinned ventricular myocardium (inset). Expression of HA-GRD mediated by a CMV-cre transgene (C) rescued these cardiovascular defects.
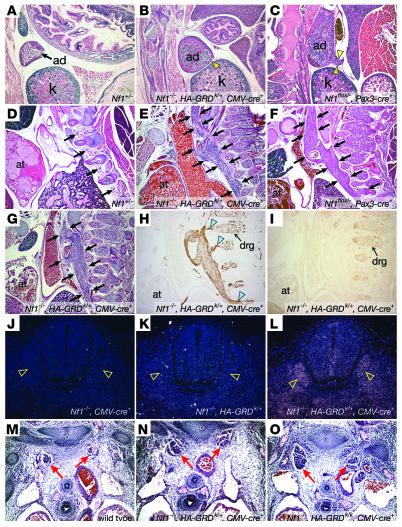
Figure 4. HA-GRD does not rescue abnormal neural crest–derived tissues lacking Nf1.
(A–C) Adrenal medullary enlargement in HA-GRD rescued newborns. Sagittal sections of adrenal glands (ad); Nf1–/–, HA-GRDk/+, CMV-cre+ (B) and the Nf1 neural crest–specific knockout, Nf1flox/–, Pax3-cre+ (C) had overgrown adrenal medullae compared with wild-type (A) with cortical effacement and tumor-like medullary protrusion (arrowheads). k, kidney. (D–I) Enlarged peripheral ganglia in HA-GRD rescued newborns. Sagittal sections of Nf1+/– (D); Nf1–/–, HA-GRDk/+, CMV-cre+ (E, G–I); and Nf1flox/–, Pax3-cre+ (F). at, cardiac atrium. Peripheral ganglia (arrows) were massively enlarged in Nf1–/–, HA-GRDk/+, CMV-cre+ newborns (E and G) compared with an Nf1+/– (D) littermate and similar the Nf1 neural crest–specific knockout (F). (H) Positive staining for neurofilament, with connections (arrowheads) to dorsal root ganglia (drg). (I) Negative staining for the glial marker S-100. (J–L) HA-GRD is expressed with CMV-cre. In situ hybridization of cross-sections of Nf1–/– E12.5 embryos with CMV-cre transgene (J), HA-GRD knock-in (K), or both (L) show expression of HA-GRD only with both. Dorsal root ganglia (arrowheads) had notably robust expression. (M–O) HA-GRD dose did not alter abnormalities of neural crest–derived tissue in Nf1–/– embryos. Cross-sections of wild-type (M); Nf1–/–, HA-GRDk/–, CMV-cre+ (N); and Nf1–/–, HA-GRDk/k, CMV-cre+ (O) E12.5 embryos highlighting sympathetic ganglia (red arrows), enlarged in both Nf1–/– genotypes and no smaller with 2 copies of HA-GRD (O) than with 1 copy (N). Magnification, ×4 (A–I); ×10 (J–O).
This abnormal tissue is neural in origin, as demonstrated by expression of neurofilament (Figure 4H). Furthermore, it appears to be contiguous with the developing dorsal root ganglia, whose overgrowth is a particular pathological finding in Nf1–/– embryos. Interestingly, it did not stain for the glial marker S-100 (Figure 4I), which is usually positive in neurofibromas, but often negative in cells isolated from MPNSTs (39, 40). Hence, the tissue overgrowth seen in this model of Nf1 deficiency and GAP rescue had a phenotype that was at least in some respects similar to the malignant tumors found in NF1.
While it remains possible that failure to rescue this neural crest aspect of Nf1 deficiency was due to insufficient GAP activity resulting from HA-GRD expression, this is unlikely. First, our examinations have shown that HA-GRD was expressed in developing neural crest–derived tissues, such as the dorsal root ganglia (Figure 4, J–L). We also found expression of HA-GRD via RT-PCR of tissue isolated from microscopic slides of Nf1–/–, HA-GRDk/+, CMV-cre+ newborns (data not shown). In addition, overall levels of active Ras were returned to normal (Figure 2B). Finally, we examined E12.5 Nf1–/– embryos with 2 copies of the HA-GRD knock-in allele activated by CMV-cre (Nf1–/–, HA-GRDk/k, CMV-cre+), comparing them with wild-type littermates and Nf1–/– embryos with a single copy of HA-GRD activated by CMV-cre (Nf1–/–, HA-GRDk/+, CMV-cre+; Figure 4, M–O). There was enlargement of the neural crest–derived sympathetic ganglia in both Nf1–/– embryos, with no difference in the degree of overgrowth in the presence of 1 or 2 copies of HA-GRD knock-in.
Our previous studies have described the importance of neurofibromin modulation of Ras signaling during the formation of cardiac valves in early and mid-gestation (34, 37). Epithelial-mesenchymal transformation during endocardial cushion formation is abnormal in Nf1–/– embryos in a fashion that can be mimicked by Ras activation and blocked by dominant-negative Ras (41). Endothelial-specific inactivation of Nf1 recapitulates the embryonic cardiac defects seen in Nf1–/– embryos (38). The endocardial overgrowth seen in Nf1-deficient embryos resembles that seen in mouse models of several related disease syndromes, such as Noonan syndrome (42), Costello syndrome (43), and cardio-facial-cutaneous syndrome (CFC) (44). The cardiac defects of all of these disorders involve derivatives of the endocardial cushions, such as the outflow tract valves. NF1, Noonan syndrome, CFC, and Costello syndrome have diverse sets of abnormalities, but share a marked incidence of valvar pulmonic stenosis in humans.
The former 2 disorders also share some interesting aspects in their clinical presentation. First, there is a clearly recognized condition with an apparent overlap of NF1 and Noonan syndrome, the neurofibromatosis–Noonan syndrome (45). This disease is characterized as neurofibromatosis combined with clinical features of Noonan syndrome such as short stature, midface hypoplasia, and webbed neck. Some cases of this syndrome have been noted to be due to unique mutations in the GRD of NF1 (46). Second, NF1 and Noonan syndrome also share a propensity for juvenile myelomonocytic leukemia (JMML), and we have suggested that the cardiovascular and hematologic abnormalities in these syndromes may represent a defect of the hemangioblast, a common precursor of blood and endothelium (47).
The role of Ras activation in the process of epithelial to mesenchymal transformation, and in particular the development of the endocardial cushions has been recently demonstrated in both animal models (41) and humans. For example, Costello syndrome has been found to be the result of activation of H-Ras (43). In addition, some cases of CFC and Noonan syndrome have activating mutations in K-Ras (44, 48). Further, some models of NF1 pathogenesis suggest activation of K-Ras (49). Taken together, these data suggest that abnormal Ras activation may account for cardiovascular and blood disorders in these related human syndromes. Hence, we sought to determine whether HA-GRD expression in endothelial cells rescues cardiovascular development in Nf1-deficient mice.
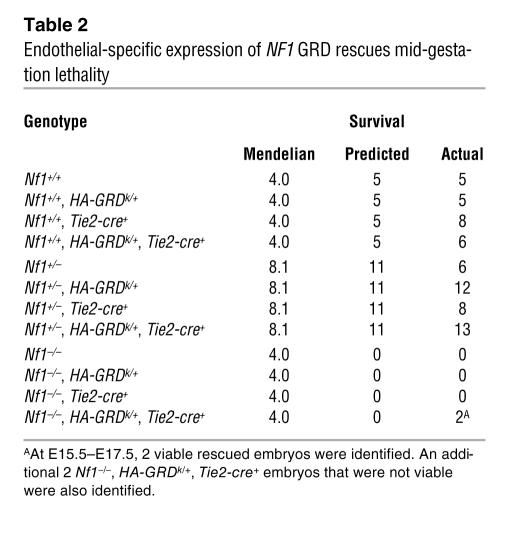
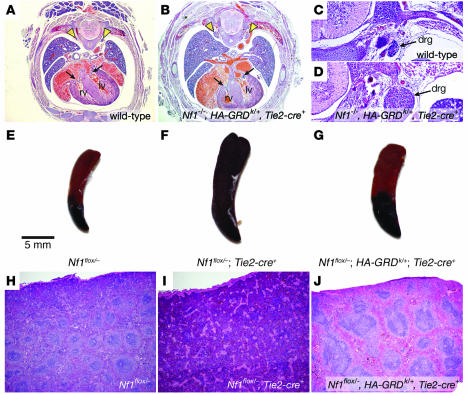
Using Tie2-cre (50) to drive HA-GRD expression, we collected late-gestation embryos several stages past expected lethality from cardiovascular failure in Nf1–/–. We found 2 embryos at late stages after crossing Nf1+/–, HA-GRDk/+ with Nf1+/–, Tie2-cre+, in which endothelial expression of HA-GRD was able to rescue the loss of Nf1 (Table 2). Analysis of the late-gestation embryos showed normal cardiac anatomy, indistinguishable from wild-type littermates (Figure 5, A and B). However, as in the HA-GRD rescued newborns, there was a marked enlargement of neural crest–derived tissues, such as dorsal root ganglia, compared with wild type (Figure 5, C and D). Hence, endothelial expression of HA-GRD partially rescued mid-gestation cardiovascular lethality, but failed to rescue the neonatal lethality associated with overgrowth of neural crest–derived structures.
Table 2.
Endothelial-specific expression of NF1 GRD rescues mid-gestation lethality
Figure 5. Tie2-cre –mediated expression of HA-GRD rescues cardiac development and hematologic abnormalities.
(A and B) Cardiac rescue with endothelial expression of HA-GRD. Representative low-power photomicrographs (magnification, ×4) of E17.5 wild-type (A) and Nf1–/–, HA-GRDk/+, Tie2-cre+ (B) littermates. There was normal cardiac anatomy, and the inflow valves (arrows) were comparable. However, there was marked enlargement of the dorsal root ganglia (arrowheads) in B compared with wild type. Higher-power images (magnification, ×10) of the dorsal root ganglia for wild-type (C) and Nf1–/–, HA-GRDk/+, Tie2-cre+ (D) littermates. (E–J) HA-GRD expression rescues hematologic abnormalities of the Tie2-cre–mediated loss of Nf1. Gross (E–G) and low-power hematoxylin and eosin photomicrographs (H–J; magnification, ×4) of spleens from affected mice and controls. A few Nf1flox/–, Tie2-cre+ mice survived embryonic development, but went on to develop a myeloproliferative disorder ultimately resembling juvenile myelomonocytic leukemia. This disease was manifest by gross splenic enlargement (F) and loss of normal architecture (I) compared with Nf1flox/– (E and H). In a Nf1flox/–, HA-GRDk/+, Tie2-cre+ littermate (G and J), in which HA-GRD was expressed under the same cre, there was less severe splenic enlargement (G) and a partial rescue of splenic tissue architecture (J).
Mice lacking Nf1 in the endothelial compartment have the same sorts of developmental cardiovascular abnormalities seen in the complete Nf1-null embryo (38). However, in rare instances, these mice survive through embryonic development, only to later develop myeloproliferative disease similar to the juvenile myelomonocytic leukemia seen in NF1 (47). Pathologically, they develop an enlarged spleen with loss of normal tissue architecture due to abnormal overgrowth of granulocytic precursors. In littermates expressing HA-GRD, we found that they had less splenic enlargement, and partial rescue of splenic architecture, with return of germinal centers and loss of fibrosis (Figure 5, E–J). These data provide additional evidence for the importance of the Ras-GAP activity of neurofibromin in endothelial and hematopoietic precursors.
Discussion
Our data suggest that the GAP activity of neurofibromin is sufficient for embryonic endothelial function, a result consistent with prior reports (41). Restoration of the Ras-GAP activity through expression of the human NF1 GRD was able to normalize cardiovascular development. We have previously described how loss of Nf1 from the endothelial compartment can recapitulate the cardiovascular defects of the complete null genotype (38). The present work demonstrates that the mechanism for this abnormal cardiovascular development stems from hyperactivation of Ras in those tissues. Furthermore, our work also suggests that mTOR may be an important downstream mediator of this effect, although the detailed analysis of this potential downstream effector pathway requires clarification (36, 37).
However, not all of the pathologies associated with loss of Nf1 could be attributed solely to dysregulation of Ras and downstream targets. We found that the neural crest pathologies in Nf1–/– embryos and mice with ubiquitous expression of HA-GRD and reconstitution of NF1 Ras-GAP activity were identical to those in tissue-specific neural crest knockout mice (38). Further, restoration of ubiquitous NF1 Ras-GAP activity resulted in a neonatal lethal phenotype that was no less severe in terms of neural crest overgrowth than that seen in neonates with only endothelial reconstitution of Ras-GAP activity or with neural crest–specific deletion of Nf1. These results strongly argue that a function or functions outside the GAP domain of Nf1 are critical for proper regulation of neural crest growth and homeostasis. In other words, reconstitution of NF1 Ras-GAP activity is not sufficient to rescue the loss of neurofibromin in neural crest derivatives. While modulation of Ras activity may contribute to NF1 tumor formation or progression, restoration of normal Ras activity does not appear to be sufficient to restore normal behavior in this tissue. This finding has important implications for NF1-related cancer therapy, as current trials have targeted activated Ras for the prevention and treatment of NF1-related malignancies (26). It should be noted that the experimental approach we have chosen has several limitations, and that a negative result (lack of rescue) is inherently difficult to interpret. Subcellular localization, posttranslational processing, and tissue-specific expression levels of the isolated GRD may be different from the full-length protein. While we have attempted to control for as many of these variables as possible, conclusive demonstration of critical non-GRD functions of neurofibromin awaits the identification and characterization of these domains. However, our results should encourage this further investigation.
Malignant pathologies in NF1 exist in many tissues; however, some of the most prominent clinical symptoms are seen in neural crest–derived tissues, such as neurofibromas and MPNSTs (8). Our findings suggest that therapeutic modalities aimed solely at restoration of normal Ras activity or its downstream effectors may not be sufficient in the clinical treatment of these pathologies associated with NF1. Further understanding of the biological functions of neurofibromin in mammalian development will be needed to develop better treatments for NF1.
Methods
Mouse breeding and genotyping.
Rosa–HA-GRD mice were generated following a protocol similar to that described previously (31). Mice were bred on a mixed C57BL/6 and 129/Sv background. Genotyping of Nf1, Nf1flox, and Tie2-cre mice was performed as previously described (34, 38, 50, 51). Genotyping of HA-GRDk/+ mice was slightly modified from that for Gt(ROSA)26Sortm1Sor (32). The primers used were as follows: R26F2, 5′-AAAGTCGCTCTGAGTTGTTAT-3′; R1295, 5′-GCGAAGAGTTTGTCCTCAACC-3′; and R3307, 5′-ACGGGGAGAAATGGATATGAA-3′. Primers for CMV-cre (33) were as follows: forward, 5′-ATTCTCCCACCGTCAGTACG-3′; reverse, 5′-CGTTTTCTGAGCATACCTGGA-3′. All protocols conformed to the guidelines established by the Association for the Assessment and Accreditation of Laboratory Animal Care and were approved by the University of Pennsylvania Animal Care and Use Committees.
Histology.
Tissue collection, fixation, and staining protocols were as previously described (38). Briefly, embryos were collected in ice-cold PBS, fixed overnight in 4% paraformaldehyde at 4°C, washed in PBS, and dehydrated through an ethanol series prior to paraffin embedding. Late-stage embryos and newborns were decalcified in 0.15 M EDTA, pH 6.6, for 48–72 hours prior to dehydration.
Immunohistochemistry protocols were as previously described (38). A 2H3-neurofilament monoclonal mouse antibody (Developmental Studies Hybridoma Bank) was used at 1:100 concentration, and S-100 polyclonal rabbit antibody (Vector Laboratories) was used at 1:50, with secondary detection by HRP activity.
In situ hybridization was performed as previously described (52). Briefly, the 35S-labeled antisense riboprobe to HA-GRD was synthesized with T7 RNA polymerase and 35S-UTP. Hybridization was performed at 55°C overnight. Slides were dipped in NTB-2 emulsion (Kodak), exposed for 7 days at 4°C, and developed and fixed in Dektol Developer and Fixer (Kodak). Cell nuclei were counterstained with Hoechst 33258 (Sigma-Aldrich) and mounted in Canada balsam/methyl salicylate.
Western blots.
Tissue lysates were prepared in magnesium lysis buffer (MLB), consisting of 2.5 mM HEPES, pH 7.5, 150 mM NaCl, 1% Igepal CA-630, 10% glycerol, 10 mM MgCl2, 1 mM EDTA, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mM sodium orthovanidate. Samples were separated by SDS-PAGE and transferred to PVDF membranes. Antibodies for eIF4E, mTOR, phosphorylated mTOR (serine 2448), phosphorylated mTOR (serine 2481), phosphorylated p42/44 MAPK, s6K, phosphorylated s6K, s6RP, and phosphorylated s6RP (Cell Signaling Technology) were used at concentrations as suggested by the manufacturer.
Ras activation assays.
Tissue homogenates of cells and embryos were prepared in MLB, precleared with glutathione agarose, incubated with raf-1 RBD (Ras binding domain) agarose beads (Upstate USA Inc.), and blotted with anti-Ras monoclonal antibody (Upstate USA Inc.) following SDS-PAGE and transfer to PVDF membranes. These blots of activated Ras (Ras-GTP) were compared with blots of the same samples for total Ras prior to raf-1 RBD pulldown to quantify Ras activation. Comparison of band intensity was performed with NIH ImageJ software version 1.33u (http://rsb.info.nih.gov/ij/).
Cell culture expression.
Cos7 cells were transfected using Lipofectin (Invitrogen) with a cre-dependent mammalian expression plasmid, pCCALL2–HA-GRD, based upon the pCCALL2 vector (53). Briefly, the HA-tagged NF1 GRD used in our knock-in mice was cloned into pCCALL2, allowing for expression following cre-mediated recombination. For expression of HA-GRD in cells, they were cotransfected with a mammalian expression vector for cre recombinase, pTurboCre (GenBank accession no. AF334827).
Acknowledgments
The NF1 HA-GRD construct was kindly provided by N. Ratner. The vectors pBigT and pRosa26PA were provided by F. Constantini. This work was supported by grants RO1 HL062974 (to J.A. Epstein) and K08 HL075179 (to F.A. Ismat) from the NIH. F.A. Ismat was additionally supported by a Young Investigator Award from the Children’s Tumor Foundation.
Footnotes
Nonstandard abbreviations used: GAP, GTPase-activating protein; GRD, GAP-related domain; MPNST, malignant peripheral nerve sheath tumor; mTOR, mammalian target of rapamycin; NF1, neurofibromatosis type I; s6K, s6 kinase; s6RP, s6 ribosomal protein.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 116:2378–2384 (2006). doi:10.1172/JCI28341.
References
- 1.Friedman J.M. Epidemiology of neurofibromatosis type 1. Am. J. Med. Genet. 1999;89:1–6. [PubMed] [Google Scholar]
- 2.Ozonoff S. Cognitive impairment in neurofibromatosis type 1. Am. J. Med. Genet. 1999;89:45–52. [PubMed] [Google Scholar]
- 3.North K. Neurofibromatosis type 1. Am. J. Med. Genet. 2000;97:119–127. doi: 10.1002/1096-8628(200022)97:2<119::aid-ajmg3>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 4.Korf B.R. Clinical features and pathobiology of neurofibromatosis 1. J. Child Neurol. 2002;17:573–577; discussion 602–604, 646–651. doi: 10.1177/088307380201700806. [DOI] [PubMed] [Google Scholar]
- 5.Fossali E., et al. Renovascular disease and hypertension in children with neurofibromatosis. Pediatr. Nephrol. 2000;14:806–810. doi: 10.1007/s004679900260. [DOI] [PubMed] [Google Scholar]
- 6.Lin A.E., et al. Cardiovascular malformations and other cardiovascular abnormalities in neurofibromatosis 1. Am. J. Med. Genet. 2000;95:108–117. doi: 10.1002/1096-8628(20001113)95:2<108::aid-ajmg4>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 7.Riccardi V.M. Neurofibromatosis update. Neurofibromatosis. 1989;2:284–291. [PubMed] [Google Scholar]
- 8.Gutmann D.H., et al. The diagnostic evaluation and multidisciplinary management of neurofibromatosis 1 and neurofibromatosis 2. Jama. 1997;278:51–57. [PubMed] [Google Scholar]
- 9.North K.N. Clinical aspects of neurofibromatosis 1. Eur. J. Paediatr. Neurol. 1998;2:223–231. doi: 10.1016/s1090-3798(98)80035-4. [DOI] [PubMed] [Google Scholar]
- 10.Birnbaum R.A., et al. Nf1 and Gmcsf interact in myeloid leukemogenesis. Mol. Cell. 2000;5:189–195. doi: 10.1016/s1097-2765(00)80415-3. [DOI] [PubMed] [Google Scholar]
- 11.Side L., et al. Homozygous inactivation of the NF1 gene in bone marrow cells from children with neurofibromatosis type 1 and malignant myeloid disorders. N. Engl. J. Med. 1997;336:1713–1720. doi: 10.1056/NEJM199706123362404. [DOI] [PubMed] [Google Scholar]
- 12.Cichowski K., Jacks T. NF1 tumor suppressor gene function: narrowing the GAP. Cell. 2001;104:593–604. doi: 10.1016/s0092-8674(01)00245-8. [DOI] [PubMed] [Google Scholar]
- 13.DeClue J.E., et al. Abnormal regulation of mammalian p21ras contributes to malignant tumor growth in von Recklinghausen (type 1) neurofibromatosis. Cell. 1992;69:265–273. doi: 10.1016/0092-8674(92)90407-4. [DOI] [PubMed] [Google Scholar]
- 14.Garicochea B., Giorgi R., Odone V.F., Dorlhiac-Llacer P.E., Bendit I. Mutational analysis of N-RAS and GAP-related domain of the neurofibromatosis type 1 gene in chronic myelogenous leukemia. Leuk. Res. 1998;22:1003–1007. doi: 10.1016/s0145-2126(98)00076-9. [DOI] [PubMed] [Google Scholar]
- 15.Klose A., et al. Selective disactivation of neurofibromin GAP activity in neurofibromatosis type 1. . Hum. Mol. Genet. 1998;7:1261–1268. doi: 10.1093/hmg/7.8.1261. [DOI] [PubMed] [Google Scholar]
- 16.Upadhyaya M., et al. Mutational and functional analysis of the neurofibromatosis type 1 (NF1) gene. Hum. Genet. 1997;99:88–92. doi: 10.1007/s004390050317. [DOI] [PubMed] [Google Scholar]
- 17.Hiatt K.K., Ingram D.A., Zhang Y., Bollag G., Clapp D.W. Neurofibromin GTPase-activating protein-related domains restore normal growth in Nf1–/– cells. J. Biol. Chem. 2001;276:7240–7245. doi: 10.1074/jbc.M009202200. [DOI] [PubMed] [Google Scholar]
- 18.Dasgupta B., Gutmann D.H. Neurofibromin regulates neural stem cell proliferation, survival, and astroglial differentiation in vitro and in vivo. J. Neurosci. 2005;25:5584–5594. doi: 10.1523/JNEUROSCI.4693-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The I., et al. Rescue of a Drosophila NF1 mutant phenotype by protein kinase A. Science. 1997;276:791–794. doi: 10.1126/science.276.5313.791. [DOI] [PubMed] [Google Scholar]
- 20.Guo H.F., The I., Hannan F., Bernards A., Zhong Y. Requirement of Drosophila NF1 for activation of adenylyl cyclase by PACAP38-like neuropeptides. Science. 1997;276:795–798. doi: 10.1126/science.276.5313.795. [DOI] [PubMed] [Google Scholar]
- 21.Guo H.F., Tong J., Hannan F., Luo L., Zhong Y. A neurofibromatosis-1-regulated pathway is required for learning in Drosophila. Nature. 2000;403:895–898. doi: 10.1038/35002593. [DOI] [PubMed] [Google Scholar]
- 22.Corral T., Jimenez M., Hernandez-Munoz I., Perez de Castro I., Pellicer A. NF1 modulates the effects of Ras oncogenes: evidence of other NF1 function besides its GAP activity. J. Cell. Physiol. 2003;197:214–224. doi: 10.1002/jcp.10349. [DOI] [PubMed] [Google Scholar]
- 23.Abernathy C.R., et al. NF1 mutation analysis using a combined heteroduplex/SSCP approach. Hum. Mutat. 1997;9:548–554. doi: 10.1002/(SICI)1098-1004(1997)9:6<548::AID-HUMU8>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 24.Fahsold R., et al. Minor lesion mutational spectrum of the entire NF1 gene does not explain its high mutability but points to a functional domain upstream of the GAP-related domain. Am. J. Hum. Genet. 2000;66:790–818. doi: 10.1086/302809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu H., Gutmann D.H. Mutations in the GAP-related domain impair the ability of neurofibromin to associate with microtubules. Brain Res. 1997;759:149–152. doi: 10.1016/s0006-8993(97)00328-4. [DOI] [PubMed] [Google Scholar]
- 26.Widemann B.C., et al. Phase I trial and pharmacokinetic study of the farnesyltransferase inhibitor tipifarnib in children with refractory solid tumors or neurofibromatosis type I and plexiform neurofibromas. J. Clin. Oncol. 2006;24:507–516. doi: 10.1200/JCO.2005.03.8638. [DOI] [PubMed] [Google Scholar]
- 27.Packer R.J., et al. Plexiform neurofibromas in NF1: toward biologic-based therapy. Neurology. 2002;58:1461–1470. doi: 10.1212/wnl.58.10.1461. [DOI] [PubMed] [Google Scholar]
- 28.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 29.Efuet E.T., Keyomarsi K. Farnesyl and geranylgeranyl transferase inhibitors induce G1 arrest by targeting the proteasome. Cancer Res. 2006;66:1040–1051. doi: 10.1158/0008-5472.CAN-05-3416. [DOI] [PubMed] [Google Scholar]
- 30.Pan J., Yeung S.C. Recent advances in understanding the antineoplastic mechanisms of farnesyltransferase inhibitors. Cancer Res. 2005;65:9109–9112. doi: 10.1158/0008-5472.CAN-05-2635. [DOI] [PubMed] [Google Scholar]
- 31.Srinivas S., et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 33.Su H., Mills A.A., Wang X., Bradley A. A targeted X-linked CMV-Cre line. Genesis. 2002;32:187–188. doi: 10.1002/gene.10043. [DOI] [PubMed] [Google Scholar]
- 34.Brannan C.I., et al. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- 35.Jacks T., et al. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat. Genet. 1994;7:353–361. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- 36.Johannessen C.M., et al. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8573–8578. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dasgupta B., Yi Y., Chen D.Y., Weber J.D., Gutmann D.H. Proteomic analysis reveals hyperactivation of the mammalian target of rapamycin pathway in neurofibromatosis 1-associated human and mouse brain tumors. Cancer Res. 2005;65:2755–2760. doi: 10.1158/0008-5472.CAN-04-4058. [DOI] [PubMed] [Google Scholar]
- 38.Gitler A.D., et al. Nf1 has an essential role in endothelial cells. Nat. Genet. 2003;33:75–79. doi: 10.1038/ng1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiss S.W., Langloss J.M., Enzinger F.M. Value of S-100 protein in the diagnosis of soft tissue tumors with particular reference to benign and malignant Schwann cell tumors. Lab. Invest. 1983;49:299–308. [PubMed] [Google Scholar]
- 40.Frahm S., et al. Genetic and phenotypic characterization of tumor cells derived from malignant peripheral nerve sheath tumors of neurofibromatosis type 1 patients. Neurobiol. Dis. 2004;16:85–91. doi: 10.1016/j.nbd.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 41.Lakkis M.M., Epstein J.A. Neurofibromin modulation of ras activity is required for normal endocardial-mesenchymal transformation in the developing heart. Development. 1998;125:4359–4367. doi: 10.1242/dev.125.22.4359. [DOI] [PubMed] [Google Scholar]
- 42.Araki T., et al. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat. Med. 2004;10:849–857. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- 43.Aoki Y., et al. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat. Genet. 2005;37:1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- 44.Niihori T., et al. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat. Genet. 2006;38:294–296. doi: 10.1038/ng1749. [DOI] [PubMed] [Google Scholar]
- 45.Allanson J.E., Hall J.G., Van Allen M.I. Noonan phenotype associated with neurofibromatosis. Am. J. Med. Genet. 1985;21:457–462. doi: 10.1002/ajmg.1320210307. [DOI] [PubMed] [Google Scholar]
- 46.Baralle D., et al. Different mutations in the NF1 gene are associated with Neurofibromatosis-Noonan syndrome (NFNS). Am. J. Med. Genet. A. 2003;119:1–8. doi: 10.1002/ajmg.a.20023. [DOI] [PubMed] [Google Scholar]
- 47.Gitler A.D., et al. Tie2-Cre-induced inactivation of a conditional mutant Nf1 allele in mouse results in a myeloproliferative disorder that models juvenile myelomonocytic leukemia. Pediatr. Res. 2004;55:581–584. doi: 10.1203/01.PDR.0000113462.98851.2E. [DOI] [PubMed] [Google Scholar]
- 48.Schubbert S., et al. Germline KRAS mutations cause Noonan syndrome. Nat. Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- 49.Dasgupta B., Li W., Perry A., Gutmann D.H. Glioma formation in neurofibromatosis 1 reflects preferential activation of K-RAS in astrocytes. . Cancer Res. 2005;65:236–245. [PubMed] [Google Scholar]
- 50.Kisanuki Y.Y., et al. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. . Dev. Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 51.Zhu Y., et al. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15:859–876. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gitler A.D., Lu M.M., Jiang Y.Q., Epstein J.A., Gruber P.J. Molecular markers of cardiac endocardial cushion development. Dev. Dyn. 2003;228:643–650. doi: 10.1002/dvdy.10418. [DOI] [PubMed] [Google Scholar]
- 53.Novak A., Guo C., Yang W., Nagy A., Lobe C.G. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]