Abstract
Extended-spectrum β-lactamases (ESBLs) are widespread in hospital settings worldwide. The present investigation was undertaken to assess the distribution and prevalence of ESBLs belonging to the TEM and SHV families in 448 ESBL-producing clinical isolates of Enterobacteriaceae collected from 10 different Italian hospitals. The natures of TEM and SHV determinants were identified by direct sequencing of PCR-amplified genes. TEM-52 and SHV-12 were the most common variants, and they were found in most hospitals and in several different species. Other less frequent variants included TEM-5, TEM-12, TEM-15, TEM-19, TEM-20, TEM-24, TEM-26, TEM-43, TEM-60, TEM-72, TEM-87, SHV-2a, SHV-5, and SHV-11. Proteus mirabilis was the most common producer of TEM-type ESBLs, while Klebsiella pneumoniae was the most common producer of SHV-type ESBLs. The distribution of TEM- and SHV-type ESBL variants in Enterobacteriaceae from Italian hospitals exhibited notable differences from those from other geographical settings.
The TEM-1/2 and SHV-1 broad-spectrum β-lactamases are the most prevalent secondary β-lactamases among clinical isolates of Enterobacteriaceae worldwide (1, 3, 9, 12, 16). Their evolutionary success is likely due to their efficient activities against penicillins and narrow- to intermediate-spectrum cephalosporins and to the fact that either the blaTEM or the blaSHV gene is often carried on self-transmissible or mobilizable plasmids capable of rapid horizontal spreading among different enterobacterial species (1, 2). The introduction in clinical practice of expanded-spectrum β-lactams resistant to the above enzymes, such as oxyimino cephalosporins and monobactams, was a major breakthrough in the antimicrobial chemotherapy of gram-negative infections. However, it was soon counteracted by the appearance of secondary β-lactamases with an extended spectrum of hydrolytic activity (expanded-spectrum β-lactamases [ESBLs]) that also includes many expanded-spectrum β-lactams. Among emerging ESBLs, those most commonly encountered in Enterobacteriaceae are derivatives of the TEM and SHV prototypes, in which one or more amino acid substitutions are responsible for an extension of the substrate specificity that may include oxyimino cephalosporins and monobactams (7, 8). To date, carbapenems and cephamycins have remained resistant to these TEM- and SHV-type ESBL variants, while mechanism-based β-lactamase inhibitors (such as clavulanate and tazobactam) have retained a very efficient inhibitory activity (2, 10).
Spreading of TEM- and SHV-type ESBLs among members of the family Enterobacteriaceae was originally reported in France (4, 8). More recently, the problem has emerged worldwide (19, 24). Several different enzyme variants have been identified (K. Bush and G. A. Jacoby, 1999, http://www.lahey.org/studies/webt.htm), and notable epidemiological differences have been reported in different geographic areas (11, 17, 18, 19, 20, 24, 25). In Italy, although ESBL-producing clinical isolates have been occasionally reported (6, 11, 16, 21) and resistance to expanded-spectrum β-lactams is increasing (16), the prevalence of ESBLs among nosocomial isolates of Enterobacteriaceae was not systematically studied until recently, when a large survey was conducted in representative Italian hospitals from different regions. In that survey, approximately 8,000 clinical isolates of Enterobacteriaceae were analyzed and ESBL producers were found in all of the participating institutions, with an overall prevalence of approximately 6% (23; T. Spanu, F. Luzzaro, M. Perilli, A. Toniolo, G. Amicosante, and G. Fadda, Clin. Microbiol. Infect. 6[Suppl. 1], p. 79, 2000).
In this study, the nature and diversity of TEM and SHV enzymes was investigated in a large number of ESBL-producing isolates collected during the above-mentioned survey.
(Part of this work was presented at the 40th Interscience Conference on Antimicrobial Agents and Chemotherapy, Toronto, Ontario, Canada, 17 to 20 September 2000, abstr. 1992 and 1993, p. 120.)
MATERIALS AND METHODS
Bacterial isolates.
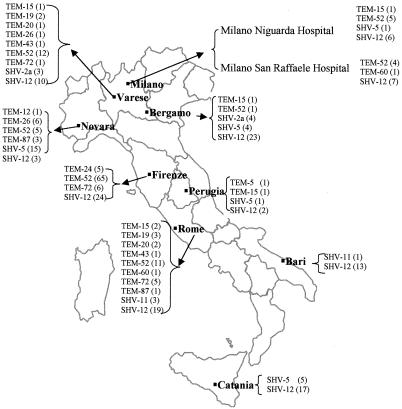
The isolates analyzed in this study were ESBL-producing Enterobacteriaceae collected during the first Italian nationwide survey of ESBLs (23; Spanu et al., Clin. Microbiol. Infect. 6[Suppl. 1], abstr. WEP67, 2000). Briefly, in that survey, 8,015 consecutive nonduplicate clinical isolates of Enterobacteriaceae were collected in 10 Italian hospitals from seven different regions (Fig. 1) from January to June 1999 and screened for ESBL production as previously described (23). Of the resulting 509 putative ESBL producers, 448 were found to carry either the blaTEM or the blaSHV gene (225 and 195 isolates, respectively) or both genes (28 isolates) by colony blot hybridization (23). Of those 448 isolates, 146 carrying the blaTEM gene, 157 carrying the blaSHV gene, and 26 carrying both genes, selected as representative of the various ESBL-producing species from the various hospitals, were included in the present study. Multiple isolates of the same species, showing the same resistance pattern and obtained from the same ward of the same hospital, were not taken into consideration. The sources of the isolates studied were as follows: 58% were from urine, 8% were from blood, 13% were from the respiratory tract, 13% were from skin and soft tissues, and 8% were from other specimens. Species identification was always double checked by the Vitek ID system (bioMérieux, Marcy l'Etoile, France).
FIG. 1.
Italian hospitals involved in the survey and distribution of TEM and SHV ESBL variants in various hospitals.
Molecular study.
The natures of the TEM- and SHV-type enzymes were determined by PCR amplification of the relevant determinants, followed by direct sequencing of the amplicons. The primers used for amplification of the blaTEM alleles were MAb/F (5"-GGGGAGCTCATAAAATTCTTGAAGAC) and MAb/R (5"-GGGGGA TCCTTACCAATGCTTAATCA) (modified from reference 13). Cycling conditions were as follows: 30 cycles of denaturation at 95°C for 30 s, annealing at 42°C for 1 min, and extension at 72°C for 1 min. The primers used for amplification of the blaSHV alleles were SHV/F (5"-GCCCGGGTTATTCTTATTTGTCGC) and SHV/R (5"-TCTTTCCGATGCCGCCGCCAGTCA). Cycling conditions were as follows: 30 cycles of denaturation at 95°C for 30 s, annealing at 58°C for 1 min, and extension at 72°C for 1 min. All reactions were performed in a 100-μl volume using 2.5 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer, Monza, Italy) in the reaction buffer provided by the manufacturer, containing 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphates, 80 pmol of each primer, and 2 μl of a crude bacterial lysate (obtained by boiling bacterial suspensions in distilled water for 10 min), as the template. The amplicons were directly sequenced on both strands by using a dRhodamina Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer), an ABI PRISM 377 DNA Sequencer (Perkin-Elmer), and custom sequencing primers. Alternatively, the Thermosequenase DNA sequencing kit (Amersham Pharmacia Biotech, Milan, Italy), a LICOR 4000 DNA sequencer (LI-COR Inc., Lincoln, Nebr.), and IRD 800-labeled custom sequencing primers (MWG-Biotech, Munich, Germany) were used. For each isolate, sequences were determined on two independent amplification products. The deduced amino acid sequences of the proteins were compared to those of known natural TEM and SHV variants (K. Bush and G. A. Jacoby, 1999, http://www.lahey.org/studies/webt.htm).
RESULTS
Diversity of TEM-type ESBLs in clinical isolates of Enterobacteriaceae from Italian hospitals.
The natures of the blaTEM variants carried by 146 ESBL-producing isolates of Enterobacteriaceae that were previously shown to carry blaTEM- but not blaSHV-related sequences were investigated by direct sequencing of the PCR-amplified blaTEM gene variants. The studied isolates included representatives of Proteus mirabilis (100 isolates), Klebsiella pneumoniae (11 isolates), Enterobacter aerogenes (8 isolates), Morganella morganii (8 isolates), Providencia stuartii (8 isolates), Escherichia coli (7 isolates), Klebsiella oxytoca (3 isolates), and Serratia marcescens (1 isolate).
Genes encoding TEM-type ESBLs were detected in 134 isolates and included the following variants: TEM-5, TEM-12, TEM-15, TEM-19, TEM-20, TEM-24, TEM-26, TEM-43, TEM-52, TEM-60, and two new variants identified for first time in this survey, namely, TEM-72 (20) and TEM-87 (21). In the remaining 12 isolates, either the blaTEM-1 or the blaTEM-2 gene was found, suggesting that the production of an additional non-TEM-type enzyme played a role in determining the ESBL phenotype in those cases. Table 1 shows the prevalence and distribution of the TEM-type ESBL determinants among different species. TEM-52 was by far the most common variant, being found in almost two-thirds of the isolates analyzed. Different determinants were variously distributed among enterobacterial species. The less common variants were apparently restricted to one or a few species, while TEM-52 was found in a broader species repertoire.
TABLE 1.
Distribution of TEM-type enzymes in different bacterial species
| Species (no. of isolates) | TEM-1 | TEM-2 | TEM-5 | TEM-12 | TEM-15 | TEM-19 | TEM-20 | TEM-24 | TEM-26 | TEM-43 | TEM-52 | TEM-60 | TEM-72 | TEM-87 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K. pneumoniae (23) | 6 | 1 | 3 (12)a | 1 | ||||||||||
| P. mirabilis (100) | 1 | 5 | 3 | 81 | 6 | 4 | ||||||||
| P. stuartii (8) | 3 | 1 | 4 | |||||||||||
| K. oxytoca (5) | 3 (2) | |||||||||||||
| E. coli (10) | 2 | 1 | 1 | 1 (3) | 1 | 1 | ||||||||
| E. aerogenes (8) | 5 | 1 | 2 | |||||||||||
| M. morganii (8) | 2 | 1 | 5 | |||||||||||
| S. marcescens (1) | 1 | |||||||||||||
| Total (163) | 11 | 1 | 1 | 1 | 6 | 3 (2) | 3 | 5 | 4 (3) | 2 | 91 (12) | 2 | 12 | 4 |
| % of total | 6.7 | 0.6 | 0.6 | 0.6 | 3.7 | 3.1 | 1.8 | 3.1 | 4.3 | 1.2 | 63 | 1.2 | 7.4 | 2.4 |
The number of isolates in which the ESBL determinant was associated with the presence of a blaSHV-1 determinant is in parentheses.
Although their prevalence differed among different hospitals, TEM-type ESBLs were detected in enterobacteria isolated from all of the hospitals participating to the survey (Fig. 1). TEM-52 was found in most of the hospitals, while the less prevalent variants had a more restricted distribution.
Diversity of SHV-type ESBLs in clinical isolates of Enterobacteriaceae from Italian hospitals.
The natures of the blaSHV variants carried by 157 ESBL-producing enterobacterial isolates that were previously shown to carry blaSHV- but not blaTEM-related sequences were investigated by direct sequencing of the PCR-amplified blaSHV gene variants. The isolates studied were representatives of K. pneumoniae (103 isolates), E. coli (22 isolates), K. oxytoca (8 isolates), Citrobacter freundii (6 isolates), E. aerogenes (7 isolates), Enterobacter cloacae (4 isolates), S. marcescens (3 isolates), Citrobacter koseri (3 isolates), and M. morganii (1 isolate).
Genes encoding SHV-type ESBL determinants were detected in 148 isolates and included the variants SHV-2a, SHV-5, and SHV-12. In the remaining nine isolates, a blaSHV-1 or a blaSHV-11 gene was found, suggesting that production of a different enzyme played a role in determining the ESBL phenotype in those cases. Figure 1 shows the prevalence and distribution of SHV-type ESBL determinants among different species. All three SHV variants were detected in K. pneumoniae and E. coli. Other species were shown to carry only the SHV-12 determinant.
Although their prevalence differed among different hospitals, SHV-type ESBLs were detected in isolates from all of the hospitals participating in the survey. SHV-12 was found in all of the hospitals, while the less prevalent variants had a more restricted distribution (Fig. 1).
TEM and SHV ESBLs in isolates carrying both types of genes.
The natures of the blaTEM and blaSHV variants carried by 26 ESBL-producing enterobacterial isolates that were previously shown to carry both blaTEM- and blaSHV-related sequences were investigated by direct sequencing of the amplified blaTEM and blaSHV genes. The isolates studied were representatives of K. oxytoca, E. coli, E. cloacae, and K. pneumoniae.
Genes encoding TEM-type ESBLs were detected in 17 isolates and included the variants TEM-19, TEM-26, and TEM-52; in these cases, the SHV gene simultaneously present was always blaSHV-1. Genes encoding SHV-type ESBLs were detected in nine isolates and included the variant SHV-12; in these cases, the TEM gene simultaneously present was blaTEM-1 or blaTEM-2. The prevalence and distribution of these TEM- and SHV-type ESBL determinants among different species are reported in Tables 1 and 2.
DISCUSSION
As in other countries (1, 4, 5, 9, 17, 18), rates of resistance to expanded-spectrum β-lactams among nosocomial isolates of Enterobacteriaceae are also increasing in Italy (reference 23 and Spanu et al., Clin. Microbiol. Infect. 6[Suppl. 1], abstr. WEP67, 2000). However, the nature and prevalence of the enzymes responsible for this phenomenon have not been investigated, except for a few cases in which the enzymes were characterized at the molecular level (11, 14, 16). In this report, we describe the first large-scale characterization of TEM- and SHV-type ESBL variants produced by Enterobacteriaceae circulating in Italian hospitals.
Results of this investigation revealed that both TEM- and SHV-type ESBLs are widespread in Italian hospitals, with TEM-52 and SHV-12 being the most prevalent ones.
TEM-52 was detected in seven hospitals, with a remarkable prevalence among P. mirabilis and occasional detection in P. stuartii, K. pneumoniae, E. aerogenes, and M. morganii. The first appearance of TEM-52 was reported in K. pneumoniae in France (22), where it was proposed, on the basis of its properties, to have been selected by the extensive use of cefotaxime and moxalactam. TEM-52 was also recently reported in Korea, where it appeared to represent the most prevalent TEM-derived ESBL (17). Also in this case, its high prevalence was associated with extensive clinical usage of moxalactam. Since this β-lactam has not been used in Italy, we suspect that in our area selection for this enzyme was preferentially driven by cefotaxime, a widely used drug. In fact, on the basis of kinetic evaluation of TEM-52, cefotaxime behaved as a good substrate, whereas moxalactam was not hydrolyzed (22). In addition to TEM-52, an abundant repertoire of other TEM-type ESBLs were detected in this survey, including TEM-5, TEM-12, TEM-15, TEM-19, TEM-20, TEM-24, TEM-26, TEM-43, TEM-52, and TEM-60, together with two new variants, TEM-72 and TEM-87, that were identified for the first time during this survey. TEM-72 is a natural TEM-2 derivative that was originally found in P. mirabilis and M. morganii in Florence (20) and was subsequently detected in isolates of the same species from other hospitals. In this study, TEM-72 represented the second most common TEM-type ESBL. TEM-87, found in P. mirabilis (21), resembles TEM-43 (25), except for the presence of a different Ω-loop substitution at position 164. The selection of TEM-72, which exhibits preferential activity for cefotaxime (20), and of TEM-87, which preferentially behaves as a ceftazidimase (21), is likely a consequence of the extensive clinical use of these drugs in our country. It is noteworthy that “old” TEM-derived cefotaximases, such as TEM-3, were not found in this survey.
SHV-12 was found in all of the participating hospitals and in a broad array of bacterial species, being most prevalent in K. pneumoniae and E. coli but also present in E. aerogenes, C. freundii, and K. oxytoca. SHV-12 was originally reported in K. pneumoniae and E. coli isolates from Switzerland (15) and subsequently found to be widespread among ESBL-producing K. pneumoniae isolates from Korea (9). The present data underscore the potential epidemiological relevance of this ESBL, which is able to efficiently hydrolyze cefotaxime, ceftazidime, and aztreonam (9). SHV-2a and SHV-5, on the other hand, were less prevalent and less widespread than SHV-12. Each of them was found only in some hospitals, mostly in Klebsiella spp.
The TEM- and SHV-type ESBL distribution found in Italian hospitals was different, overall, from that observed in other countries, confirming a notable variability in the epidemiology of these resistance determinants and underscoring the need to monitor them in different epidemiological settings. In particular, a notable feature was the widespread production of TEM-type ESBLs in P. mirabilis, a species that is not among those routinely screened for ESBLs. It would be interesting to investigate the genetic support and transferability of the various ESBL determinants and the clonal diversity of isolates to understand the role of vector versus clonal spread in the dissemination of these resistance genes.
It should be noted that the experimental approach adopted for this study, based on direct sequencing of PCR-amplified genes, could not reliably detect the presence of multiple TEM- or SHV-encoding genes in the same isolate, a possibility that has previously been reported (25) and that will be interesting to investigate further. It should also be noted that a number of isolates showing a phenotype suggestive of ESBL production contained blaTEM-1, blaTEM-2, or blaSHV-1 determinants, suggesting that ESBLs other than TEM or SHV types also occur among the clinical isolates of enterobacterial pathogens circulating in Italian hospitals. The nature of these enzymes is under investigation.
TABLE 2.
Distribution of SHV-type enzymes in different bacterial species
| Species (no. of isolates) | SHV-1 | SHV-2a | SHV-5 | SHV-11 | SHV-12 |
|---|---|---|---|---|---|
| C. freundii (6) | 6 | ||||
| C. koserii (3) | 1 | 1 | 1 | ||
| E. aerogenes (7) | 7 | ||||
| E. cloacae (5) | 4 (1)a | ||||
| E. coli (24) | 1 | 1 | 3 | 17 (2) | |
| K. oxytoca (9) | 1 | 7 (1) | |||
| K. pneumoniae (108) | 2 | 5 | 23 | 3 | 70 (5) |
| S. marcescens (3) | 3 | ||||
| M. morganii (1) | 1 | ||||
| Total (166) | 5 | 7 | 26 | 4 | 115 (9) |
| % of total | 3.0 | 4.2 | 15.7 | 2.4 | 74.7 |
The number of isolates in which the ESBL determinant was associated with the presence of a blaTEM-1 or blaTEM-2 determinant is in parentheses.
Acknowledgments
This work was supported in part by a grant from MURST-PRIN 99 to G.A. and partially by a grant from Wyeth-Lederle (Italy).
We acknowledge the help from the Italian ESBL Study Group with this study.
REFERENCES
- 1.Bradford, P. A., C. E. Cherubin, V. Idemyor, B. A. Rasmussen, and K. Bush. 1994. Multiply resistant Klebsiella pneumoniae strains from two Chicago hospitals: identification of the extended-spectrum TEM-12 and TEM-10 ceftazidime-hydrolyzing β-lactamases in a single isolate. Antimicrob. Agents Chemother. 38:761-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cha|$$|Ad|fibi, E. B., D. Sirot, G. Paul, and R. Labia. 1999. Inhibitor-resistant TEM β-lactamases: phenotypic, genetic and biochemical characteristics. J. Antimicrob. Chemother. 43:447-458. [DOI] [PubMed] [Google Scholar]
- 3.Chanal, C., D. Sirot, J. P. Romaszko, L. Bret, and J. Sirot. 1996. Survey of prevalence of extended spectrum β-lactamases among Enterobacteriaceae. J. Antimicrob. Chemother. 38:127-132. [DOI] [PubMed] [Google Scholar]
- 4.De Champs, C., D. Sirot, C. Chanal, M. C. Puopart, M. P. Dumas, and J. Sirot. 1991. Concomitant dissemination of three extended-spectrum β-lactamases among different Enterobacteriaceae isolated in a French hospital. J. Antimicrob. Chemother. 27:441-457. [DOI] [PubMed] [Google Scholar]
- 5.De Champs, C., D. Sirot, C. Chanal, R. Bonnet, J. Sirot, and The French Study Group. 2000. A 1998 survey of extended-spectrum β-lactamases in Enterobacteriaceae in France. Antimicrob. Agents Chemother. 44:3177-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franceschini, N., M. Perilli, B. Segatore, D. Setacci, G. Amicosante, A. Mazzariol, and G. Cornaglia. 1998. Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived extended-spectrum β-lactamase, TEM-60. Antimicrob. Agents Chemother. 42:1459-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacoby, G. A. 1997. Extended-spectrum β-lactamases and other enzymes providing resistance to oxyimino-β-lactams. Infect. Dis. Clin. N. Am. 11:875-887. [DOI] [PubMed] [Google Scholar]
- 8.Jarlier, V., M. H. Nicholas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 9.Kim, J., J. Kwon, H. Pai, J.-W. Kim, and D.-T. Cho. 1998. Survey of Klebsiella pneumoniae strains producing extended-spectrum β-lactamases: prevalence of SHV-12 and SHV-2a in Korea. J. Clin. Microbiol. 36:1446-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knox, J. R. 1995. Extended-spectrum and inhibitor-resistant TEM-type β-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob. Agents Chemother. 39:2593-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laksai, Y., M. Severino, M. Perilli, G. Amicosante, G. Bonfiglio, and S. Stefani. 2000. First identification of an SHV-12 extended-spectrum β-lactamase in Klebsiella pneumoniae isolated in Italy. J. Antimicrob. Chemother. 45:349-351. [DOI] [PubMed] [Google Scholar]
- 12.Livermore, D. M. 1995. β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mabilat, C., S. Goussard, W. Sougakoff, R. C. Spencer, and P. Courvalin. 1990. Direct sequencing of the amplified structural gene and promoter for the extended-broad-spectrum β-lactamase TEM-9 (RHH-1) of Klebsiella pneumoniae. Plasmid 23:27-34. [DOI] [PubMed] [Google Scholar]
- 14.Marchese, A., G. Arlet, G. C. Schito, P. H. Lagrange, and A. Philippon. 1996. Detection of SHV-5 extended spectrum β-lactamase in Klebsiella pneumoniae strains isolated in Italy. Eur. J. Clin. Microbiol. Infect. Dis. 15:245-248. [DOI] [PubMed] [Google Scholar]
- 15.Nüesch-Inderbinen, M. T., F. H. Kayser, and H. Hächler. 1997. Survey and molecular genetics of SHV β-lactamases in Enterobacteriaceae in Switzerland: two novel enzymes, SHV-11 and SHV-12. Antimicrob. Agents Chemother. 41:943-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pagani, L., M. Perilli, R. Migliavacca, F. Luzzaro, and G. Amicosante. 2000. Extended-spectrum TEM- and SHV-type β-lactamase-producing Klebsiella pneumoniae strains causing outbreaks in intensive care units in Italy. Eur. J. Clin. Microbiol. Infect. Dis. 19:765-772. [DOI] [PubMed] [Google Scholar]
- 17.Pai, H., S. Lyu, J. H. Lee, J. Kim, Y. Kwon, J.-W. Kim, and K. W. Choe. 1999. Survey of extended-spectrum β-lactamases in clinical isolates of Escherichia coli and Klebsiella pneumoniae: prevalence of TEM-52 in Korea. J. Clin. Microbiol. 37:1758-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palucha, A., B. Mikiewicz, W. Hryniewiewicz, and M. Gniadkowski. 1999. Concurrent outbreaks of extended-spectrum β-lactamase-producing organisms of the family Enterobacteriaceae in a Warsaw hospital. J. Antimicrob. Chemother. 44:489-499. [DOI] [PubMed] [Google Scholar]
- 19.Perilli, M., A. Felici, N. Franceschini, A. De Santis, L. Pagani, F. Luzzaro, A. Oratore, G. M. Rossolini, J. R. Knox, and G. Amicosante. 1997. Characterization of a new TEM-derived β-lactamase produced in a Serratia marcescens strain. Antimicrob. Agents Chemother. 41:2374-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perilli, M., B. Segatore, M. R. De Massis, M. L. Riccio, C. Bianchi, A. Zollo, G. M. Rossolini, and G. Amicosante. 2000. TEM-72, a new extended-spectrum β-lactamase detected in Proteus mirabilis and Morganella morganii in Italy. Antimicrob. Agents Chemother. 44:2537-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perilli, M., B. Segatore, M. R. De Massis, N. Franceschini, C. Bianchi, G. M. Rossolini, and G. Amicosante. Characterization of a new extended-spectrum β-lactamase (TEM-87) isolated in Proteus mirabilis during an Italian survey. Antimicrob. Agents Chemother., in press. [DOI] [PMC free article] [PubMed]
- 22.Poyart, C., P. Mugnier, G. Quesne, P. Berche, and P. Trieu-Cuot. 1998. A novel extended-spectrum TEM-type β-lactamase (TEM-52) associated with decreased susceptibility to moxalactam in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 42:108-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spanu, T., F. Luzzaro, M. Perilli, G. Amicosante, A. Toniolo, G. Fadda, and The Italian ESBL Study Group. 2002. Occurrence of extended-spectrum β-lactamases in members of the family Enterobacteriaceae in Italy: implications for resistance to β-lactams and other antimicrobial drugs. Antimicrob. Agents Chemother. 46:196-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tenover, F. C., M. J. Mohammed, T. S. Gorton, and Z. F. Dembek. 1999. Detection and reporting of organisms producing extended-spectrum β-lactamases: survey of laboratories in Connecticut. J. Clin. Microbiol. 37:4065-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang, Y., N. Bhachech, P. A. Bradford, B. D. Jett, D. F. Saham, and K. Bush. 1998. Ceftazidime-resistant Klebsiella pneumoniae and Escherichia coli isolates producing TEM-10 and TEM-43 β-lactamases from St. Louis, Missouri. Antimicrob. Agents Chemother. 42:1671-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]