Abstract
The whole-genome response of Arabidopsis (Arabidopsis thaliana) exposed to different types and durations of abiotic stress has now been described by a wealth of publicly available microarray data. When combined with studies of how gene expression is affected in mutant and transgenic Arabidopsis with altered ability to transduce the low temperature signal, these data can be used to test the interactions between various low temperature-associated transcription factors and their regulons. We quantized a collection of Affymetrix microarray data so that each gene in a particular regulon could vote on whether a cis-element found in its promoter conferred induction (+1), repression (−1), or no transcriptional change (0) during cold stress. By statistically comparing these election results with the voting behavior of all genes on the same gene chip, we verified the bioactivity of novel cis-elements and defined whether they were inductive or repressive. Using in silico mutagenesis we identified functional binding consensus variants for the transcription factors studied. Our results suggest that the previously identified ICEr1 (induction of CBF expression region 1) consensus does not correlate with cold gene induction, while the ICEr3/ICEr4 consensuses identified using our algorithms are present in regulons of genes that were induced coordinate with observed ICE1 transcript accumulation and temporally preceding genes containing the dehydration response element. Statistical analysis of overlap and cis-element enrichment in the ICE1, CBF2, ZAT12, HOS9, and PHYA regulons enabled us to construct a regulatory network supported by multiple lines of evidence that can be used for future hypothesis testing.
The sequencing of the Arabidopsis (Arabidopsis thaliana) genome and the subsequent development of microarrays supporting near full-genome transcriptomic studies has resulted in the generation of large collections of experimental data describing the transcriptomes of wild-type, mutant, and transgenic plants under a variety of growth conditions, including exposure to biotic and abiotic stress. However, a clear challenge facing plant biologists is to devise methods to uncover connections between different stress regulons using the additional insights provided by the available Arabidopsis genomic map. Near full-genome array experiments lend themselves to promoter-content-based hypothesis testing in that they provide an open experimental architecture: No a priori knowledge of the plant's transcriptional response is needed to select the genomic population of genes sharing a common cis-element (cis-regulon) and almost all of the known genes containing a cis-element of interest will have corresponding expression data. The large number of data points generated by each new microarray experiment allows us to establish whether observed overlaps in regulon characteristics (e.g. gene identities or promoter elements) are occurring at rates significantly less or greater than random chance. Thus, we can statistically establish the links between transcription factors (TFs) identified by reverse genetics (and characterized in microarray studies) and the cis-elements shown to appear at frequencies greater than chance in the promoters of their regulon members. The wealth of data also facilitates the establishment of links between cis-elements identified based on binding assays or enrichment, and a statistically verified bioactivity based on patterns of the cis-regulon's expression in wild-type plants during time-course experiments, as well as between stress-associated, bioactive cis-elements and novel pathways/treatments by virtue of their statistically significant over- or underrepresentation in these differentially expressed gene groups of interest.
One very active area of current research is the transcriptional response of the model plant Arabidopsis to low temperatures. Recent reviews describing current models of the low temperature transcriptional responses show a linear cascade consisting of a nonphosphorylated form of the c-MYC-like basic helix-loop-helix Inducer of CBF Expression 1 (ICE1) protein that is phosphorylated following exposure to low temperatures and subsequently activates transcription of the CBF3 transcriptional activator (Chinnusamy et al., 2006; Nakashima and Yamaguchi-Shinozaki, 2006). CBF3, along with its low temperature-responsive paralogs CBF1 and CBF2, in turn activate the transcription of members of the CBF regulon, a group of genes containing the dehydration response element (DRE)/C-repeat element in their promoters. The CBF regulon(s) includes the transcriptional activators ZAT10, RAP2.1, and RAP2.6, along with many other well-known COR genes (Fowler and Thomashow, 2002). The ZAT12 transcriptional repressor, which has been placed outside the ICE1-directed cascade (Nakashima and Yamaguchi-Shinozaki, 2006), is responsible for repression of the CBFs and their downstream TFs, in addition to its own regulon (Vogel et al., 2005). HOS9, thought to be an ICE1-independent constitutively expressed transcriptional repressor, also modulates low temperature gene transcription and plant-freezing tolerance (Zhu et al., 2004). However, because the ICE1-directed cascade contains at least 52 cold-regulated TFs (Lee et al., 2005) and the ice1 mutation appears to affect plant hormonal homeostasis (Lee et al., 2005), there are still many more components to positionally map within the cascade and connect in terms of their interaction with other up- and downstream TFs.
In light of the combinatory nature of this task and the time-consuming lab bench procedures required to test each possible connection, it is unlikely that the entire ICE1-mediated TF cascade will be mapped using conventional methods. However, if we can bioinformatically reduce the number of bench-top verifications needed, this task becomes more feasible. Past studies have defined the gene regulons controlled by different ICE1 cascade TFs based on the subsets of genes that are both cold responsive in wild-type plants and differentially expressed in TF overexpressors/mutants (Chinnusamy et al., 2003; Lee et al., 2005; Vogel et al., 2005). An equally informative and complementary representation of a TF's regulon is the group of genes whose promoters contain the defined consensus binding sequence (cis-element) of that TF (its cis-regulon). Analysis of the behavior of the cis-regulon associated with a TF helps eliminate the biasing effect of its interconnected downstream regulons and is expected to be more robust across various experiments and between different experimenters. Because many of the TFs within the ICE1 cascade have been analyzed in Arabidopsis overexpressor and mutant plants using near full-genome microarrays (Boyce et al., 2003; Chinnusamy et al., 2003; Zhu et al., 2004; Lee et al., 2005; Vogel et al., 2005), many of the connections in the current model of the ICE1 cascade (Fig. 1) can now be tested using cis-regulon analysis.
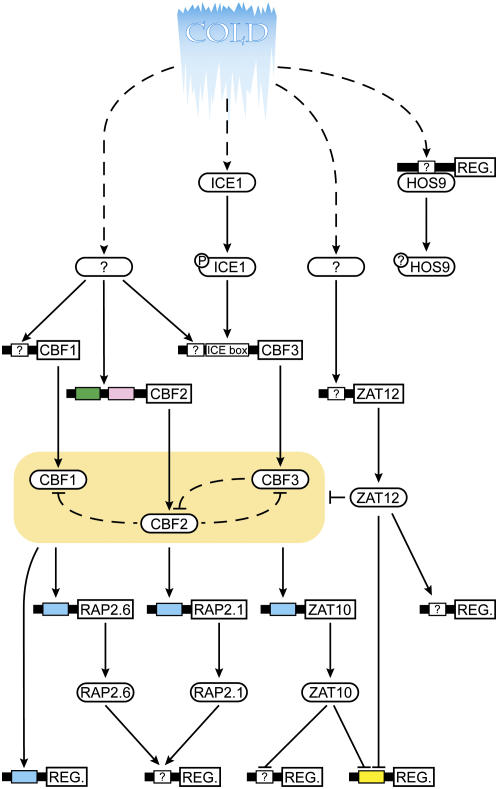
Figure 1.
Schematic representation of low temperature signal transduction assembled from reviews published by Chinnusamy et al. (2006) and Nakashima and Yamaguchi-Shinozaki (2006). Protein TFs are represented by ovals while TF regulons are represented by “REG” boxes. Where reported, the consensus sequences (cis-elements) bound by the upstream TF are shown as color-coded boxes within the promoter space (solid line) preceding the REG box or TF gene coding sequence (blue box, DRE; green box, ICEr1; pink box, ICEr2; and yellow box, EP2). Regulatory connections are mapped using triangular arrowheads to represent gene activation and flattened arrowheads to represent gene repression. Dashed arrows represent regulatory connections with experimental support but accomplished by unknown (and possibly posttranscriptional) mechanisms. Protein phosphorylation events, where known, are indicated by circles attached to the protein TFs. Genes modeled (and corresponding Arabidopsis Genome Initiative no.) are ICE1, AT3G26744; ZAT12, AT5G59820; HOS9, AT2G01500; CBF2, AT4G25470; CBF1, AT4G25490; CBF3, AT4G25480; ZAT10, AT1G27730; RAP2.1, AT1G46768; and RAP2.6, AT1G43160.
While the role of ICE1 in inducing CBF1 and CBF3 is well known, we were interested in examining the nature of the rest of the ICE1-controlled gene regulon. Specifically, we wanted to know how ICE1 interacts with the other reported low-temperature signaling transcriptional activators and repressors, and if the ICE1-mediated low temperature signaling cascade is a linear chain of transcriptional activations/repressions eventually leading to COR expression, or if the feedback mechanisms observed to occur among members of the CBF TFs also extend to ICE1 transcription. We present the ICE1-mediated transcriptional signaling cascade as a case study for our meta-analysis of microarray and genomic data. Our analysis shows that the previously identified ICEr1 consensus (Chinnusamy et al., 2003; Zarka et al., 2003) does not correlate with cold gene induction, while our bioinformatically identified ICE1-binding consensuses ICEr3 and ICEr4 are present in regulons of genes whose inductions are coordinate with observed ICE1 transcript accumulation and temporally precede that of genes containing the DRE. Additionally, the ICE1-binding consensus ICEr3 is similar to a cis-element previously reported to be phytochrome A (phyA) responsive (Hudson and Quail, 2003), potentially explaining the observed circadian gating and light responsiveness of the CBF-mediated/DRE regulon. We statistically link HOS9, CBF2, ZAT12, NAC072, and PHYA to the ICE1-mediated low temperature signaling pathway. Our set of correlation data was used to model a low temperature signaling framework that can be used for future hypothesis testing.
RESULTS
The Current ICE1-Signaling Model Lacks Consensus
Our current understanding of the ICE1-mediated low temperature transcriptional cascade has been established through both forward and reverse genetic approaches coupled with microarray analyses. Based on recent reviews (Chinnusamy et al., 2006; Nakashima and Yamaguchi-Shinozaki, 2006) this cascade can be represented as a linear signaling path with several outliers that are known to be involved but with their linkages, if any, to other downstream targets of the cascade unclear (Fig. 1). The lack of well-defined consensus binding sequences (cis-elements) for many of these TFs makes identifying and mapping their target genes difficult. To date, fewer than a dozen cold-responsive cis-elements have been identified and reported to the PLACE cis-element database (http://www.dna.affrc.go.jp/PLACE). One way to fill in these gaps is to combine existing microarray results deposited into public databases with the genomic promoter data available online in a novel database that cross-references individual gene responsiveness to TF overexpression/mutation and low temperature with gene promoter content (i.e. the presence or absence of cis-elements of interest). However, before we can do this we need a collection of putative cis-element candidates to consider.
Identifying Election Candidates: Inclusive Promoter Sampling of TF Overexpressor/Mutant Regulons to Find Putative cis-Elements
Our first criteria for establishing a regulatory link between any single TF and its preferred cis-element is that Arabidopsis plants overexpressing the TF should induce or repress a regulon of genes containing that cis-element in their 500 bp promoter. Therefore, a motif search of the promoters of all genes differentially induced/repressed in the TF overexpressor should identify this cis-element as one of the nucleic acid consensuses overrepresented in the promoter population.
As the furthest upstream TF identified in the cold-signaling pathway to date, we began our analysis with the ICE1 microarray data reported by Chinnusamy et al. (2003) and Lee et al. (2005). Because constitutive ICE1 expression was insufficient to activate gene transcription in Arabidopsis (Chinnusamy et al., 2003), and because only a cold treatment induced CBF3-driven reporter and endogenous CBF expression in ICE1-overexpressing Arabidopsis, we submitted the promoters corresponding to the lists of 217 and 109 cold-responsive genes reported by Chinnusamy et al. (2003) and Lee et al. (2005) as less induced or nonresponsive in the ice1 mutants versus wild-type plants at 6 and 3 h cold treatment, respectively, to the Inclusive Motif Sampler (Coessens et al., 2003). These analyses identified CRCGT/CACGT as the potential ICE1-binding consensus core favored in vivo (Supplemental Fig. 1A), as judged by log likelihood score (Supplemental Table I). The ICEr1 element (CACATG) identified by Zarka et al. (2003) as responsible for CBF2 induction and shown to be bound by mutant and wild-type forms of the ICE1 protein in vitro (Chinnusamy et al., 2003) was not enriched in the ICE1-affected gene lists. To obtain sufficient statistical power for our bioinformatic analyses, we required consensus sequences containing a minimum of 6 bp, and therefore extended the novel element CACGT to include the preceding A/C base variation observed in our Inclusive Motif search (Supplemental Fig. 1A). Because the sequences ACACGT and CCACGT represented potentially novel cis-elements in the ICE1 regulon, we flagged them for further bioinformatic analyses. However, PatMatch queries for the list of all Arabidopsis genes containing ACACGT in the 500 bp promoter space returned a list that included genes containing the abscisic acid (ABA) response element (ABRE; GACACGT). Because ABRE was identified as enriched in the CBF3 regulon in its reverse complementary form (ACGTGTC; see Maruyama et al., 2004), we filtered the results of all further bioinformatic analyses of these novel ICE1 elements to exclude genes where A/CCACGT was preceded by G (the ABRE-containing subset). This was done by defining the new ICE1-related cis-elements as HACACGT and HCCACGT (where H is the bioinformatic representation of A or C or T), and we named these elements ICEr3 and ICEr4, respectively. Motif searching also uncovered an enrichment of the DRE cis-element in the 3 and 6 h ice1-affected gene promoter lists (Supplemental Table I), supporting the previous observation that the CBF TFs are ICE1 target genes and that ice1 mutation affects induction of the genes CBFs bind (i.e. DRE-containing genes).
To test the suggested exclusion of the HOS9 and ZAT12 TFs from the ICE1-directed cascade (Fig. 1), we needed to identify and bioinformatically confirm the bioactivity of their enriched cis-elements. A search of the 138 available promoter sequences for the genes reported to be differentially induced in hos9 mutants in the cold versus wild-type plants by Zhu et al. (2004) identified ACGCG(T), which we named HOS9r1, as the potential HOS9-binding consensus favored in vivo, as judged by log likelihood score (Supplemental Fig. 1B). Similarly, Vogel and colleagues (2005) identified the CATTGA core, which we refer to as ZAT12r1, as common to many genes induced by both cold and ZAT12 overexpression. Motif searches of the CBF2 (Vogel et al., 2005) and CBF3 (Maruyama et al., 2004) regulon promoters by other researchers had previously identified the DRE (RCCGAC) and ABRE (GACACGT) as enriched in their regulons.
Creating the Voter Registry: Assembly of MasterDB
We next needed to create a database (MasterDB) that would cross-reference all available transcriptional response data for each gene with the presence or absence of the identified candidate cis-elements in each gene's promoter. We first assembled the normalized Affymetrix microarray data for the cold-signaling TF overexpressors/mutants ice1 (Chinnusamy et al., 2003; Lee et al., 2005), 35S:CBF2 (Vogel et al., 2005), 35S:ZAT12 (Vogel et al., 2005), hos9 (Zhu et al., 2004), and 35S:NAC072 (Tran et al., 2004) from the supplementary material provided for the publications listed with the additional standardized stress treatment time-course data for wild-type plants reported by the AtGenExpress project authors (http://www.weigelworld.org/resources/microarray/AtGenExpress/), as well as selected mutant and ABA treatment experiments available at the Nottingham Arabidopsis Science Center (NASC) arrays (http://affymetrix.arabidopsis.info/narrays/experimentbrowse.pl) in MasterDB.
We intended to use MasterDB to determine whether our candidate cis-elements were bioactive by examining if their corresponding cis-regulons responded to cold in wild-type plants at a frequency greater than the chip-wide average. Our expectation was that the systematic selection of subpopulations of genes on the microarray using only genomic promoter data (i.e. cis-element content, not TF or cold responsiveness) would yield cis-regulons in which the distribution between the three bioinformatic states of induced, repressed, and nonresponsive was significantly different than the distribution of the whole array population. To perform this analysis, we needed to quantize all the transcriptomic fold-change comparison data in MasterDB into this set of discrete states. This allowed us to restrict our attention to the numerical relationships between the categories of genes with and without a particular cis-element in their 500 bp promoter and then mathematically examine their distribution using probability. Effectively, we wanted every gene on the genome chip to independently vote for its preferred transcriptional response to each particular treatment in question, be it induction, repression, or nonresponse. These responses were numerically represented as +1, −1, or 0, respectively, in MasterDB, and genes without corresponding expression data were designated numerically as 2 and omitted from further analyses.
Induction/repression calls in MasterDB were assigned using fold-change cutoffs that varied on a sliding scale to account for the fact that for highly expressed genes smaller fold changes represent large changes in absolute transcript numbers (and presumably transcriptional activity), and correspondingly, seemingly large fold changes in sparsely expressed genes represent relatively small changes in absolute transcript numbers and transcriptional activity. The four-tiered sliding scale had set cutoffs of 10-fold for genes whose normalized gene expression levels (NGELs) across all experimental controls were less than 10, 3-fold for genes with NGELs between 10 and 50, 2-fold for genes with NGELs between 50 to 500, and 1.5-fold for genes with NGELs >500 in the Affymetrix DNA-chip system. This meant that the majority of genes (51% or 12,144 of 23,714 genes in the MasterDB) in our sliding scale-rated system were assessed using a 2-fold cutoff (Supplemental Table II).
Tallying the Votes: Testing cis-Regulon Bioactivity and the Temporal Order of Regulon Induction/Repression in Wild-Type Plants
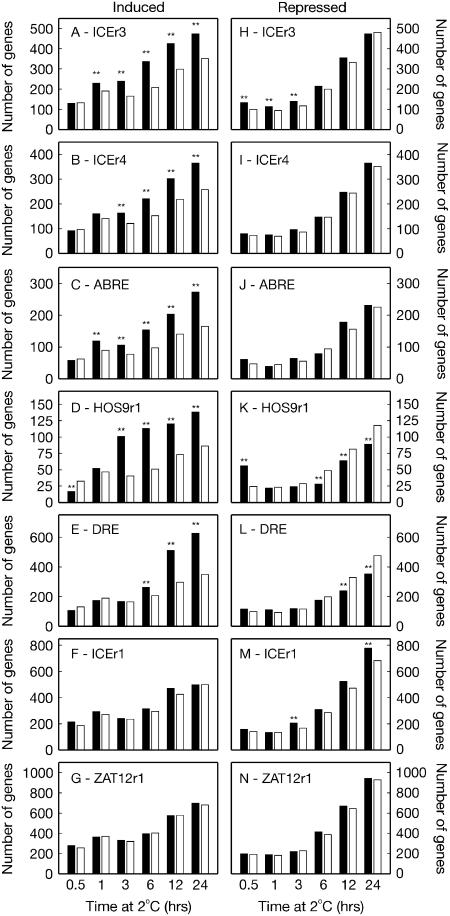
With the construction of the voter registry complete, it was time to tally the votes for the various candidate cis-elements. The voter participation rate for our elections was high, with approximately 70% of all genes encoded by the Arabidopsis genome containing the cis-elements of interest present on the array. When we compared the voting behavior of the ICEr3, DRE, ICEr1, ZAT12r1, HOS9r1, ABRE, and ICEr4 cis-regulons with that of all other genes in MasterDB, statistical analysis revealed that the ICEr1 and ZAT12r1 cis-regulons were no more likely to be induced than a regulon of randomly selected genes from the arrays examined at any point during the cold treatment time course (χ2, P < 0.01; Fig. 2, F and G). In contrast, the ICEr3 (Fig. 2A) and ABRE (Fig. 2C) cis-regulons were significantly more likely to be induced at 1 h (and subsequent time points of the cold treatment). This was followed by statistically significant ICEr4 and HOS9r1 cis-regulon induction at 3 h (Fig. 2, B and D), and DRE cis-regulon induction at 6 h (Fig. 2E). Examination of the cold repression of the various cis-regulons revealed a tendency for the ICEr3 (Fig. 2H) and HOS9r1 (Fig. 2K) cis-regulons to be repressed within the first 30 min of cold treatment. This preferential repression disappeared by 1 h for the HOS9r1 cis-regulon but continued for the ICEr3 cis-regulon until 3 h cold treatment. The 3 h time point was also the first interval at which significant repression of the ICEr1 cis-regulon was observed. The ICEr1 cis-regulon was also more likely to be repressed at 24 h of cold treatment.
Figure 2.
Observed transcriptional response of genes containing the ICEr3 (HACACGT), ICEr4 (HCCACGT), ABRE (GACACGT), HOS9r1 (ACGCGT), DRE (RCCGAC), ICEr1 (CACATG), and ZAT12r1 (CATTGA) elements in their 500 bp promoters to cold (2°C) treatment (using microarray data from WeigelWorld AtGenExpress Database). The number of genes induced (left column) and repressed (right column) by the treatment (black bars) was compared to the expected frequency (white bars), based on the percentage of all present cold-responding genes on the chip. Statistically significant difference was assessed by χ2 test: ** = P < 0.01. A, ICEr3-containing induced genes. B, ICEr4-containing induced genes. C, ABRE-containing induced genes. D, HOS9r1-containing induced genes. E, DRE-containing induced genes. F, ICEr1-containing induced genes. G, ZAT12r1-containing induced genes. H, ICEr3-containing repressed genes. I, ICEr4-containing repressed genes. J, ABRE-containing repressed genes. K, HOS9r1-containing repressed genes. L, DRE-containing repressed genes. M, ICEr1-containing repressed genes. N, ZAT12r1-containing repressed genes.
The temporal sequence of ICEr3, ICEr4, and DRE cis-regulon activity supported the conclusion that ICEr3 binds the ice1-affected transcriptional activator lying furthest upstream of the CBF TFs, while the lack of ICEr1 cis-regulon induction in wild-type plants argued against its role as a binding site for active ICE1. However, because of the sequence similarity between ICEr3 and the ABRE, and their similar cold responsiveness, we repeated the gene voting analysis used to produce Figure 2 over a wider variety of stresses and compared ABRE cis-regulon behavior with that of ICEr3 (as well as ICEr4 and ICEr1). ABRE-containing genes also responded to salt and mannitol in a manner similar to the ICEr3 cis-regulon (Table I). To determine whether the ABRE and ICEr3 elements acted independently (and therefore, if ICEr3 represented a unique cis-element, and not just an ABRE variant), we ran a principal component analysis (PCA)-partial least squares analysis of the observed minus expected gene counts used to create Table I and showed clear separation of the ICEr3 from the ABRE, ICEr4, and ICEr1 elements (Supplemental Fig. 2). Furthermore, while ICEr4 and ABRE did not form distinct clusters in our PCA-partial least squares analysis, the differential cold inducibility of the ICEr4 cis-regulon versus the ABRE cis-regulon in the ice1 mutant background (see below, and Fig. 3, B and F) after 3 and 6 h cold treatment led us to conclude that both the ICEr3 and ICEr4 sequences represented novel cis-elements.
Table I.
Comparison of the stress inducibility of ICEr3-, ICEr4-, ABRE-, and ICEr1-containing genes
χ2 P values for induced gene group enrichment versus predicted gene group induction shown; the P < 0.01 level of significance after Bonferroni correction for multiple comparisons is indicated in bold, P > 0.05 in italic. Induced gene group reduction (versus expected) is indicated with an asterisk.
| cis-Regulon | Time | UVB | Methyl Viologen | Drought | Salt | Cold | Mannitol | Wounding |
|---|---|---|---|---|---|---|---|---|
| h | ||||||||
| ICEr3-containing genes | 0.5 | 1.1E-06 | 9.0E-01 | 5.0E-11 | 6.8E-01 | 7.7E-01 | 1.4E-01 | 1.3E-11 |
| 1 | 9.6E-11 | 4.2E-01 | 1.1E-17 | 1.5E-01 | 4.3E-03 | 2.2E-09 | 6.9E-23 | |
| 3 | 5.6E-05 | 5.4E-01 | 1.1E-09 | 2.8E-21 | 3.3E-09 | 4.4E-40 | 1.7E-08 | |
| 6 | 5.2E-04 | 2.3E-01 | 4.2E-05 | 9.0E-22 | 3.1E-20 | 7.7E-52 | 9.4E-05 | |
| 12 | 1.1E-01 | 1.6E-08 | 1.5E-08 | 4.6E-30 | 1.1E-14 | 2.1E-47 | 9.4E-05 | |
| 24 | 7.9E-03 | 3.0E-07 | 3.3E-03 | 2.3E-26 | 4.8E-12 | 1.3E-37 | 8.7E-05 | |
| ICEr4-containing genes | 0.5 | 1.5E-01 | 4.7E-01 | 1.0E-04 | 8.5E-02 | 6.1E-01 | 6.7E-01 | 1.2E-02 |
| 1 | 1.9E-03 | 4.6E-01 | 1.9E-07 | 6.5E-01 | 7.9E-02 | 3.8E-02 | 5.3E-07 | |
| 3 | 4.2E-01 | 9.6E-01 | 3.9E-04 | 6.9E-11 | 8.6E-05 | 3.8E-16 | 3.0E-04 | |
| 6 | 3.0E-03 | 2.1E-01 | 1.1E-02 | 1.1E-11 | 8.0E-09 | 2.6E-17 | 2.2E-01 | |
| 12 | 6.5E-01 | 1.8E-02 | 3.5E-02 | 3.0E-11 | 2.9E-09 | 5.2E-17 | 2.2E-01 | |
| 24 | 1.5E-01 | 1.0E-03 | 2.0E-01 | 6.1E-11 | 1.5E-12 | 5.7E-18 | 3.4E-01 | |
| ABRE-containing genes | 0.5 | 2.0E-01 | 8.6E-01 | 3.3E-04 | 9.2E-01 | 5.9E-01 | 1.7E-01 | 1.4E-01 |
| 1 | 3.5E-03 | 3.7E-01 | 1.4E-09 | 3.2E-01 | 1.4E-03 | 2.9E-04 | 1.6E-05 | |
| 3 | 9.1E-02 | 1.9E-01 | 2.2E-02 | 1.1E-36 | 9.0E-04 | 8.8E-46 | 5.0E-01 | |
| 6 | 1.9E-03 | 6.8E-02 | 1.0E-01 | 1.7E-25 | 3.4E-09 | 2.1E-55 | 5.3E-01 | |
| 12 | 9.6E-01 | 2.3E-01 | 1.0E+00 | 3.0E-25 | 2.3E-08 | 2.3E-46 | 5.3E-01 | |
| 24 | 1.1E-01 | 1.8E-02 | 1.3E-01 | 9.6E-32 | 3.5E-19 | 7.8E-42 | 6.2E-01 | |
| ICEr1-containing genes | 0.5 | 9.7E-02 | 2.9E-01 | 1.7E-02 | 5.8E-01 | 5.3E-02 | 3.1E-01 | 4.4E-01 |
| 1 | 9.7E-02 | 8.8E-01 | 4.5E-02 | 4.9E-01 | 1.8E-01 | 2.8E-01 | 6.1E-03 | |
| 3 | 2.5E-01 | 9.0E-01 | 6.6E-02 | 7.1E-03 | 6.8E-01 | 2.2E-01 | 2.7E-02 | |
| 6 | 1.5E-02* | 3.2E-01 | 9.4E-01 | 1.6E-01 | 2.4E-01 | 3.5E-01 | 8.0E-01 | |
| 12 | 1.0E-01 | 7.0E-01 | 1.3E-01 | 2.5E-01 | 2.0E-02 | 1.1E-01 | 8.0E-01 | |
| 24 | 2.9E-01 | 1.7E-01 | 8.1E-02 | 2.1E-01 | 9.3E-01 | 2.0E-01 | 2.3E-01 |
Figure 3.
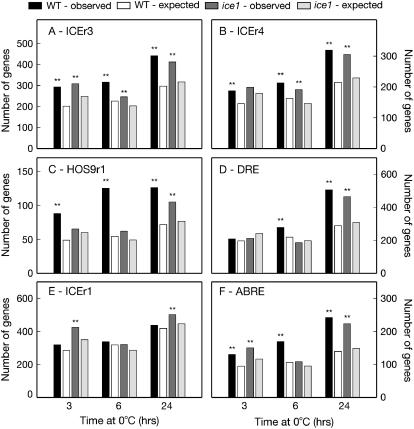
Observed transcriptional response of genes containing the ICEr3 (HACACGT), ICEr4 (HCCACGT), HOS9r1 (ACGCGT), DRE (RCCGAC), ICEr1 (CACATG), and ABRE (GACACGT) elements in their 500 bp promoters to cold (0°C) treatment in wild-type and ice1 Arabidopsis backgrounds (using microarray data published in Lee et al., 2005). The number of genes induced by the treatment was compared to the expected frequency, based on the percentage of all present cold-responding genes on the chip. Statistically significant difference was assessed by χ2 test: ** = P < 0.01. A, ICEr3-containing induced genes. B, ICEr4-containing induced genes. C, HOS9r1-containing induced genes. D, DRE-containing induced genes. E, ICEr1-containing induced genes. F, ABRE-containing induced genes.
Assessing Regional Voting Trends: cis-Regulon Responsiveness in the ice1 Mutant Background
After statistically associating single TFs with their corresponding cis-element(s), the next step in constructing a regulatory map is to determine which TFs are predicted to belong to the same transcriptional cascade, and how they are connected within that cascade. To determine which cis-regulons were affected by the ice1 mutation during cold stress (and therefore which regulons were ICE1 dependent), we repeated our analysis of cis-regulon cold inducibility in the ice1 mutant background (Fig. 3). The ice1 mutation had obvious effects on the cold inducibility of the DRE cis-regulon: While DRE cis-regulon inducibility was evident in wild-type plants at 6 and 24 h, regulon inducibility was negligible at 6 h in the ice1 plants (Fig. 3D). Similarly, the significant HOS9r1 cis-regulon induction at 3 and 6 h in wild-type plants was not mirrored in ice1 mutants at these time points (Fig. 3B). The ICEr4 cis-regulon also demonstrated delayed induction in ice1 mutants (at 6 h) versus wild-type plants (Fig. 3B). In contrast, the ICEr1 cis-regulon became significantly cold inducible in ice1 plants (as opposed to noninducible in wild-type plants) at both 3 and 24 h (Fig. 3C). The ICEr3 cis-regulon was cold responsive at 3, 6, and 24 h in both wild-type and ice1 plants, but the strength of the enrichment of ICEr3-containing genes in the total induced gene list was stronger in the wild-type than in ice1 mutant plants (Fig. 3A). For the ABRE cis-regulon, which was cold responsive at all three time points in wild-type plants, the ice1 mutation only affected regulon inducibility at 6 h. These results suggested that the TFs binding to the DRE, HOS9r1, ICEr4, ICEr1, ICEr3, and ABRE cis-elements were part of the ICE1-mediated TF cascade, and demonstrated the functional independence of the ICEr3, ICEr4, and ABRE elements.
TF Regulon Overlap
When examined in the proper temporal window, downstream TFs belonging to the same transcriptional cascade as upstream TFs should demonstrate statistically significant TF regulon overlap, because upstream TFs should induce a regulon of genes that includes both the downstream TFs and their respective regulons. χ2 comparisons between the 1,475 genes repressed at 3 h cold in the ice1 mutant (versus wild type) and the regulons induced or repressed by the TF or phytochrome mutants/overexpressors hos9_1, phyA, phyB, sfr6, CBF2-OX, and ZAT12-OX in MasterDB were performed to determine which TF regulons demonstrated statistically significant overlap (Table II
Table II.
ICE1 TF regulon overlap with selected low temperature signaling TFs
χ2 P values for induced/repressed gene group enrichment/reduction versus expected are shown; the P < 0.05 level of significance after Bonferroni correction is indicated in bold. For full comparisons, see Supplemental Table III.
| First Regulon | Second Regulon | Observed No. Genes | Expected No. Genes | χ2P Values |
|---|---|---|---|---|
| ice1 repressed (1475) at 3 h cold (versus wild type) | hos9_1 induced | 5 | 9 | 1.94E-01 |
| hos9_1 repressed | 9 | 2 | 4.96E-06 | |
| CBF2 induced | 85 | 78 | 4.07E-01 | |
| CBF2 repressed | 142 | 59 | 2.78E-28 | |
| phyB induced | 107 | 235 | 6.89E-20 | |
| phyB repressed | 86 | 145 | 2.48E-07 | |
| phyA induced | 54 | 65 | 1.66E-01 | |
| phyA repressed | 105 | 53 | 1.63E-13 | |
| phyA + FR induced | 325 | 136 | 9.92E-65 | |
| phyA + FR repressed | 86 | 73 | 1.33E-01 | |
| NAC072 induced | 13 | 31 | 1.03 E-03 | |
| NAC072 repressed | 79 | 65 | 8.03E-02 | |
| ice1 induced (2012) at 3 h cold (versus wild type) | hos9_1 induced | 41 | 12 | 6.77E-17 |
| hos9_1 repressed | 15 | 3 | 5.12E-12 | |
| CBF2 induced | 307 | 106 | 4.14E-89 | |
| CBF2 repressed | 139 | 80 | 2.78E-11 | |
| ZAT12 induced | 200 | 55 | 9.36E-89 | |
| ZAT12 repressed | 414 | 659 | 2.73E-31 | |
| phyB induced | 156 | 321 | 9.08E-24 | |
| phyB repressed | 82 | 198 | 4.33E-18 | |
| phyA induced | 249 | 89 | 3.87E-68 | |
| phyA repressed | 132 | 72 | 3.76E-13 | |
| phyA + FR induced | 249 | 186 | 1.09E-06 | |
| phyA + FR repressed | 256 | 100 | 2.15E-57 | |
| NAC072 induced | 27 | 42 | 1.65E-02 | |
| NAC072 repressed | 7 | 89 | 6.30E-19 |
TF Regulon cis-Element Enrichment
While inclusive motif sampling of TF regulons uncovered the most highly overrepresented cis-elements in the individual TF regulons, we wanted to expand our TF regulon analysis to include all of our candidate cis-elements (Table III; note that regulon sizes differ from those reported by the original authors because of the sliding-scale cutoff used in MasterDB). The NAC072-OX induced and repressed TF regulons were included as negative controls for this overlap analysis, since NAC072 is strongly down-regulated during the first 5 h of cold treatment (Tran et al., 2004). The ice1 cold-repressed (normally ICE1 cold induced) genes were found to be more likely to be repressed in hos9_1 mutants (i.e. normally HOS9 induced/derepressed), and to be repressed in 35S∷CBF2-OX plants (Table II), suggesting regulatory connection between the ICE1, HOS9, and CBF2 TFs. Furthermore, a statistically significant fraction of the genes belonging to the ice1 cold-repressed regulon were also misregulated in phyA mutants (versus wild type) preconditioned with far-red (FR) light. In contrast, the genes misregulated in phyB mutants (versus wild type) were underrepresented in the ice1 cold repressed regulon. These data indicated cross talk and/or a regulatory connection between the PHYA and ICE1 signaling cascades that were lacking for PHYB. As expected, the NAC072 induced regulon was underrepresented in the ice1 cold-repressed regulon. Analysis of the ice1 cold-induced (normally ICE1 cold repressed) regulon expanded the list of statistically significant ICE1 coregulatory relationships to include the HOS9 induced/repressed regulons, the CBF2 induced/repressed regulons, the ZAT12 induced regulon, the PHYA induced/repressed regulons, and the PHYA + FR induced/repressed regulons. Similar analysis of the regulon overlaps for TFs reported to be downstream of ICE1 (see Supplemental Table III) also showed a strong enrichment for PHYA induced/repressed genes. As reported previously by Vogel et al. (2005), using a uniform 2.5-fold-change cutoff, the CBF2 induced and ZAT12 induced regulons also showed statistically significant overlap (Supplemental Table III). Contrary to previous reports (Knight et al., 1999; Boyce et al., 2003), the sfr6 induced and repressed regulons lacked significant overlap with both the CBF2 regulon and the other investigated TF regulons (Supplemental Table III). Considering the substantial experimental evidence for sfr6 effects on DRE inducibility (Knight et al., 1999; Boyce et al., 2003), we interpreted this as an indication that the SFR6 TF regulons examined were either temporally too far removed to demonstrate overlap, or that the sfr6 mutational effect is low temperature dependent.) to support or refute regulatory connections already indicated by earlier bioinformatic analyses. Consistent with the proposed binding of HOS9 to HOS9r1, the HOS9-repressed (hos9 induced) TF regulon was enriched for the HOS9r1 cis-element versus the genomic average but no other cold-responsive element was significantly enriched in this regulon. Similarly, the CBF2 induced TF regulon was enriched for the DRE cis-element as well as ICEr3 and ABRE. The ZAT12 repressed TF regulon was also enriched for the ABRE but not ZAT12r1. ZAT12r1 was also not enriched in the ZAT12 induced TF regulon, once again indicating that ZAT12r1 is unlikely to be the cis-element that ZAT12 binds. While the 3 h ICE1 induced (ice1 repressed) regulon was enriched for ICEr1, ICEr4, ABRE, and PhyAr1 versus their respective genomic average frequencies, the 3 h ICE1 repressed (ice1 induced) TF regulon was enriched for ICEr3 and ABRE. We found the lack of DRE enrichment in the 3 h ICE1-regulated regulons surprising in light of the fact that the ice1 mutation affects the 3 h cold induction of CBF1 and CBF3 (Chinnusamy et al., 2003) and the fact that the DRE was identified by the original inclusive search of the ice1 3 h repressed regulon (Supplemental Table I). The expected enrichment of DRE and ICEr3 was observed only when the ICE1 induced and repressed TF regulons were filtered to leave only genes induced by cold in wild type at 3 h (versus wild-type control). Nevertheless, the presence of the ICEr4 consensus variant in the 3 h ice1 repressed (ICE1 induced) list before (and after) filtering suggested that it is the ICEr variant most strongly affected by the ice1 mutation. The NAC072 repressed TF regulon narrowly missed the P < 0.05 significance level for ICEr1 enrichment but this was likely due to the small number of genes in this regulon. Investigation of the SFR6 induced and repressed TF regulons in both the light and dark showed no significant enrichment of any of the cis-elements except the PhyAr1 in the SFR6 repressed (in light) TF regulons.
Table III.
Cis-element frequency in cold-signaling induced and repressed TF regulons
Genomic frequency in 500 bp promoters is shown in parentheses. Bold text represents a statistically significant (χ2 P < 0.05) difference between the genomic and regulon cis-element frequencies after Bonferroni correction for multiple comparisons.
| Regulon | No. Genes | HOS9r1 (6.5) | ICEr3 (14.2) | ICEr1 (21.3) | DRE (13.9) | ABRE (6.1) | ZAT12 (29.6) | PhyAr1 (8.5) | ICEr4 (10.0) |
|---|---|---|---|---|---|---|---|---|---|
| % | % | % | % | % | % | % | % | ||
| Wild type induced 3 h | 1,429 | 6.2 | 20.5 | 22.3 | 14.6 | 9.1 | 27.8 | 10.4 | 13.1 |
| Wild type repressed 3 h | 157 | 19.1 | 19.1 | 21.7 | 8.3 | 5.1 | 31.2 | 7.0 | 10.8 |
| hos9 induced | 138 | 19.6 | 17.4 | 15.9 | 15.2 | 6.5 | 29.7 | 7.2 | 14.5 |
| ice1 induced 3 h versus wild type | 2,012 | 3.9 | 16.0 | 22.0 | 11.9 | 7.4 | 28.9 | 7.3 | 7.3 |
| ice1 repressed 3 h versus wild type | 1,475 | 3.3 | 15.5 | 25.6 | 13.9 | 9.6 | 27.1 | 10.2 | 12.1 |
| ice1 induced + wild-type cold repressed | 72 | 1.4 | 27.8 | 20.8 | 6.9 | 4.2 | 29.2 | 12.5 | 15.3 |
| ice1 repressed + wild-type cold induced | 190 | 6.3 | 27.4 | 25.3 | 21.1 | 14.2 | 22.1 | 15.8 | 17.4 |
| CBF2 induced | 1,098 | 2.8 | 17.5 | 21.6 | 23.2 | 7.9 | 27.4 | 9.4 | 12.2 |
| CBF2 repressed | 836 | 3.7 | 15.0 | 25.1 | 10.2 | 5.9 | 26.0 | 5.4 | 11.6 |
| ZAT12 induced | 577 | 3.8 | 13.9 | 19.6 | 13.2 | 6.9 | 28.2 | 6.8 | 10.2 |
| ZAT12 repressed | 6,962 | 3.8 | 13.4 | 17.7 | 14.4 | 7.0 | 27.3 | 8.3 | 10.1 |
| sfr6 induced (light) | 1,323 | 2.6 | 14.2 | 19.8 | 13.5 | 7.5 | 27.1 | 8.8 | 10.7 |
| sfr6 repressed (light) | 892 | 3.9 | 15.8 | 20.4 | 13.2 | 7.5 | 26.1 | 7.3 | 9.3 |
| sfr6 induced (dark) | 1,349 | 2.5 | 14.3 | 20.6 | 13.7 | 7.1 | 28.5 | 8.1 | 9.4 |
| sfr6 repressed (dark) | 838 | 2.6 | 14.8 | 20.5 | 15.5 | 5.0 | 25.1 | 6.9 | 9.8 |
| NAC72 induced | 101 | 2.0 | 23.8 | 17.8 | 14.9 | 7.9 | 21.8 | 11.9 | 17.8 |
| NAC72 repressed | 212 | 3.8 | 19.8 | 24.5 | 14.6 | 9.0 | 22.6 | 11.3 | 13.7 |
| phyA induced | 943 | 2.8 | 15.0 | 20.5 | 12.7 | 5.5 | 29.4 | 5.1 | 11.2 |
| phyA repressed | 763 | 3.8 | 16.8 | 22.8 | 15.7 | 7.6 | 26.3 | 8.7 | 12.5 |
| phyB induced | 1,161 | 2.2 | 13.5 | 22.4 | 13.9 | 5.1 | 26.6 | 5.9 | 9.0 |
| phyB repressed | 715 | 3.5 | 19.9 | 18.3 | 13.6 | 11.2 | 26.0 | 12.3 | 11.0 |
| phyA + FR induced | 1,976 | 2.2 | 14.9 | 25.5 | 12.9 | 6.9 | 27.4 | 7.1 | 10.1 |
| phyA + FR repressed | 1,066 | 4.1 | 17.8 | 17.8 | 16.0 | 7.6 | 25.6 | 9.0 | 13.5 |
| Dusk induced | 117 | 1.7 | 3.4 | 19.7 | 4.3 | 1.7 | 26.5 | 0.9 | 6.8 |
| Dusk repressed | 198 | 3.0 | 24.7 | 27.3 | 13.1 | 13.6 | 22.7 | 17.7 | 15.2 |
Because reports have identified variable inducibility of DRE-containing genes depending on time of day (Kim et al., 2002; Fowler et al., 2005), we examined the cis-element enrichment in genes responsive to phytochrome A and B mutation as well as the PHYA-dependent FR-conditioned gene expression and the groups of genes differentially induced and repressed at dusk (in a circadian time course) versus expression throughout the rest of the day. We observed a statistically significant reduction in the frequency of the HOS9r1, ICEr3, DRE, ABRE, and PhyAr1 cis-elements in the group of genes induced at dusk. In contrast, ICEr3, ICEr1, ABRE, ICEr4, and PhyAr1 cis-elements were enriched in the promoters of genes repressed at dusk. These data support our hypothesis that time of day affects DRE-cis-regulon inducibility through phytochrome A repression of the ICE1-mediated transcriptional cascade (which includes the DRE-binding CBF TFs).
In Silico Mutagenesis
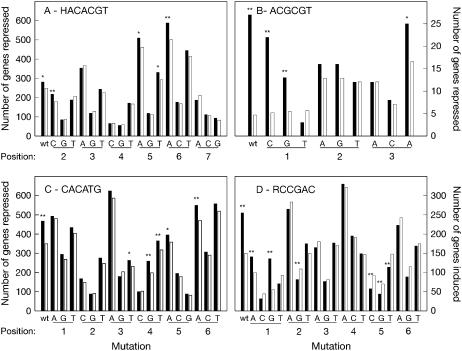
The most conclusive piece of evidence establishing signaling connectivity is the presence of an upstream TF's preferred cis-element in the 500 bp promoter of a candidate downstream TF. However, TF binding to cis-elements is often promiscuous, with several permitted consensus variants capable of binding the same TF. We therefore used in silico mutagenesis (Geisler et al., 2006) to compare the behavior of the ICEr3 cis-regulon with the behavior of the populations of genes naturally containing different single-mismatch versions of the ICEr3 consensus in their 500 bp promoter (Fig. 4A). Single-base mutations of ICEr3 at positions 2 (A to C, yielding the ICEr4 element), 5 (C to A or T), and 6 (G to A) did not result in a loss of significant cis-regulon cold repression in the ice1 mutant. All 12 possible combinations of these permitted positional variations were then examined to determine whether their cis-regulons were preferentially induced during cold, ABA, salt, and dehydration treatments in wild-type plants (Table IV, only bioactive variants shown). This analysis revealed that the reported ICE1 induction detected by northern blotting at 3 h cold, 3 h ABA (100 μm), and 5 h salt (Chinnusamy et al., 2003) corresponded to the preferential induction of genes containing the consensus HACACGT (ICEr3) and HCCACGT (ICEr4) by 3 h cold, 3 h ABA (100 μm), and 6 h salt. Genes containing these two consensus variants in their 500 bp promoters were all also preferentially induced on Affymetrix microarrays by a 30 min dehydration treatment. Surprisingly, the permitted mutation of the ICEr3 element from G to A at position 6 in ice1 mutant plants (Fig. 4A) did not result in any ICE element variant that was functional during these treatments in wild-type plants, even when combined with all other permitted mutations at positions 2 and 5. This observation was interesting in light of the fact that such a mutation creates an ICEr1-like element [i.e. HACACAT(G)] known to be bound by both mutant and wild-type forms of the ICE1 protein (Chinnusamy et al., 2003). Similar in silico mutagenesis of the palindromic HOS9r1 consensus, using cis-regulon induction in the hos9 genetic background as the baseline, revealed that HOS9r1 could vary at positions 1 (A, C, or G) or 3 (G or T) without loss of cis-regulon induction (Fig. 4B). The cold, ABA, and salt responsiveness of these consensus sequences were also examined (Table IV) and unlike ICEr3, neither HOS9r1 nor its variant CCGCGT (that we named HOS9r2) was salt responsive. In silico mutagenesis of ICEr1 using cold repression in ice1 mutants as the reference showed that the bioactivity of ICEr1 (Fig. 4C) was unaffected by mutations at positions 4 (A, G, or T), 5 (T or A), or 6 (G or A). DRE mutagenesis reported previously in cold-stressed wild-type plants (Geisler et al., 2006) was repeated in the CBF2 overexpressor background, confirming previous observations of the functionality of the A/GCCGAC variants (Fig. 4D).
Figure 4.
In silico mutagenesis of the ICEr3 (HACACGT)-, HOS9r1 (ACGCGT)-, ICEr1 (CACATG)-, and DRE (RCCGAC)-binding consensus sequences. Identity and position of each nucleic acid mutation in the corresponding consensus sequence are indicated. The number of genes induced or repressed in the genetic background/treatment indicated (black bars) was compared to the expected frequency (white bars), based on the percentage of all present and responding genes on the chip. Statistically significant difference was assessed by χ2 test: * = P < 0.05, ** = P < 0.01. A, ICEr3-containing genes repressed in 6 h cold-treated ice1 plants (versus wild type). B, HOS9r1-containing genes repressed in 24 h cold-treated hos9-1 plants (versus wild type). C, ICEr1-containing genes repressed in 6 h cold-treated ice1 plants (versus wild type). D, DRE-containing genes induced in 35S:CBF2 overexpressors at growth temperatures (versus wild type).
Table IV.
Frequency and enrichment (versus frequency predicted by chance) of stress-responsive ICE1 and HOS9 consensus variants in the 500 bp promoters of all genes in the Arabidopsis genome
χ2 P values for induced gene group enrichment versus predicted gene group frequency for stress and hormonal treatments shown. Bold indicates P < 0.05 after Bonferroni correction for multiple comparisons.
| TF | Element | No. Genes | Element Probability | Expected No. Genes | Genomic Enrichment | Cold 3 h | ABA 3 h | Salt 6 h | Dehydration 0.5 h |
|---|---|---|---|---|---|---|---|---|---|
| ICE1 | HACACGT | 4,014 | 1.4E-04 | 3,921 | 1.02 | 4.3E-09 | 6.0E-42 | 4.1E-22 | 1.9E-10 |
| HCCACGT | 2,805 | 6.4E-05 | 1,910 | 1.47 | 9.9E-05 | 3.6E-15 | 3.6E-12 | 1.3E-04 | |
| HACAAGT | 8,861 | 2.9E-04 | 7,750 | 1.14 | 7.3E-03 | 6.3E-02 | 7.3E-01 | 8.4E-02 | |
| HOS9 | ACGCGT | 2,038 | 7.6E-05 | 2,261 | 0.90 | 1.3E-22 | 2.4E-04 | 8.8E-01 | 6.8E-06 |
| CCGCGT | 1,139 | 3.6E-05 | 1,085 | 1.05 | 6.9E-21 | 3.0E-05 | 8.1E-01 | 1.1E-05 | |
| ACGCGC | 1,273 | 3.6E-05 | 1,085 | 1.17 | 2.3E-07 | 4.8E-01 | 2.5E-01 | 9.8E-02 |
Using TF Promoter Data to Create a Map of Cold-Stress Regulation
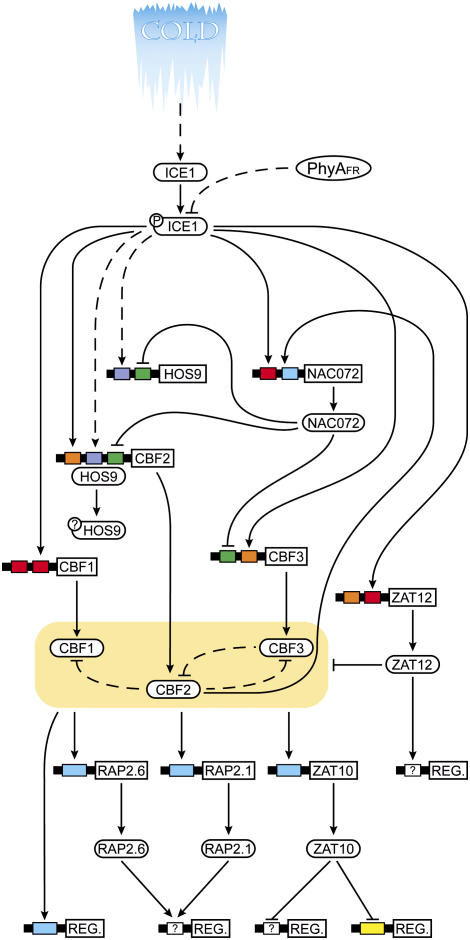
With these bioinformatic analyses complete, it was possible to revise the current model of the ICE1 transcriptional cascade (Fig. 1), this time incorporating both the results of our bioinformatic analyses and TF promoter element searches that considered cis-element variants identified by in silico mutagenesis (Fig. 5). From this analysis, HOS9 was placed within the ICE1 signaling cascade, downstream of ICE1, based on the following evidence: (1) the induction of the HOS9r1 cis-regulon temporally follows the induction of the ICEr3 cis-regulon (Fig. 2); (2) the induction of the HOS9r1 cis-regulon is reduced and delayed in ice1 mutant plants responding to cold (Fig. 3C); (3) the HOS9 TF regulon shares significant overlap with the ICE1 TF regulon (Table II); and (4) the HOS9 promoter contains elements shown to be affected by ICE1 mutation, namely ICEr1 and HOS9r1 (Fig. 3, C and E). Similarly, though previously thought to lie outside the ICE1 signaling cascade, we include ZAT12 in the ICE1-mediated cascade based on the presence of ICEr3 and ICEr4 sites in its 500 bp promoter (that are significantly affected in their cold induction by the ice1 mutation; Fig. 3), and the significant overlap of both the ice1 induced and CBF2 induced TF regulons with the ZAT12 induced TF regulon (Table II; Supplemental Table V). Our bioinformatic analyses provide no support for the suggestion that ZAT12 acts through the ZAT12r1 element (CATTGA; see Vogel et al., 2005). The ZAT12r1 element is not enriched in any examined cold-responsive regulons (Table III) nor is the ZAT12r1 cis-regulon cold responsive (Fig. 2, G and N). Because further independent inclusive motif searches failed to uncover any bioactive enriched elements within the ZAT12 induced or repressed TF regulons, we conclude that the ZAT12-binding consensus is either larger than the 8 bp windows examined, or it is not sufficient to confer cold regulation. Although the EP2 element [ACTX(3–4)AGT or AGTX(3–4)ACT; Sakamoto et al., 2004] represents a larger potential binding consensus sequence, our analyses determined that it also lacked both bioactivity and ZAT12 TF regulon enrichment (data not shown). The experimentally established connections between ICE1 and the CBF TFs were supported by our bioinformatics analyses. Elements affected by the ice1 mutation and potentially bound by ICE1 were present in all three CBF promoters. However, the number and identity of the ICEr variants present differed between the CBF paralogs (Fig. 5), possibly explaining why the ice1 mutation differentially affects the cold induction of the different CBF paralogs. The A/C variation at position 2 of our proposed ICE1-binding consensus, yielding ICEr3 and ICEr4, respectively, had important consequences for bioactivity with respect to ICE1 (Fig. 4A). Of the two ICEr variants, ICEr4 was more strongly affected by the ice1 mutation after 3 h cold (Fig. 3, A versus B; Table III) but the presence of additional neighboring elements (Fig. 5) also contributed to paralog inducibility. The cold inducibility of cis-element bound by CBF1, 2, and 3, DRE (RCCGAC) was suppressed by ice1 mutation (Fig. 3D) and was underrepresented in the ice1 induced regulon (Table III). NAC072 was placed within the ICE1 cascade based on the fact that its promoter contains ICEr3 and DRE sites. The NAC072 repressed TF regulon is enriched (χ2, P < 0.06) for ICEr1 (Table III), and the statistically significant tendency for ICEr1 cis-regulon members to be repressed late in the cold treatment time course (Fig. 2M) agrees with the experimental observation that NAC072 transcripts are induced after 10 h of cold treatment (Tran et al., 2004).
Figure 5.
Schematic representation of low temperature signal transduction. Solid arrows represent connections with at least two pieces of bioinformatic support (triangular arrowhead represents gene activation, flattened arrowhead represents gene repression). Dashed arrows represent regulatory connections with experimental/bioinformatic support but accomplished by unknown (and possibly posttranscriptional) mechanisms. Boxes represent cis-elements within the 500 bp promoters of the genes listed (ICE1, AT3G26744; ZAT12, AT5G59820; HOS9, AT2G01500; CBF2, AT4G25470; NAC072, AT4G27410; CBF1, AT4G25490; and CBF3, AT4G25480) and are color coded according to cis-element identity (red box, ICEr3; orange box, ICEr4; blue box, DRE; green box, ICEr1; purple box, I HOS9r1; and yellow box, EP2). Protein phosphorylation events, where known, are indicated by circles containing “P” attached to the protein TFs. Unknown types of posttranslational modification are indicated by circles containing “?” attached to the protein TFs. The ICEr4 element present in the CBF3 promoter contains the CCACGT core, but lacks an “H” in the first position.
DISCUSSION
Near full-genome transcriptomic studies of response of Arabidopsis to low temperature have generated complex data sets that are difficult to analyze because of their scale and variability. This necessitates the use of mathematical and statistical approaches to resolve questions about the interconnectivity of gene regulatory networks, just as statistical approaches were necessary in Mendel's discovery of the particulate model of genetic inheritance (see Janick, 1989). The advantage of modeling the low temperature transcriptional signaling cascade from a genomic perspective (documenting regulon connectivity) instead of a gene perspective (documenting connectivity gene by gene) is that observing the behavior of an entire cis-regulon of hundreds to thousands of genes in vivo during a particular stress or in a mutant background lends the statistical power to resolve our experimental observations and minimizes the risk that the chosen experimental subject/system represents a behavioral outlier. The large number of data points, much like voting in a democracy, smoothes away the effect of individual nonrepresentative genes.
Meta-analyses of publicly available microarray data allowed us to construct an ICE1 signaling model (Fig. 5) that predicted regulatory connections already established by conventional bench-top methods (e.g. ICE1 transcriptional activation of the DRE-binding CBF TFs) and identified previously unknown regulatory connections between ICE1 and HOS9, ZAT12, NAC072, and PHYA that can be tested in future experiments. Evidence generated from our meta-analyses of TF regulon cis-element enrichment (Supplemental Fig. 1; Table III), cis-regulon induction order (Figs. 2 and 3), TF regulon overlap (Table II), and in silico mutagensis (Fig. 4) provided the anchors we used to create our bioinformatically generated regulatory map. Our observation that the ICEr3 cis-element was both enriched in the promoters of genes affected by the ice1 mutation (Supplemental Fig. 1; Table III) and associated with a cis-regulon whose induction preceded the induction of the DRE cis-regulon in wild-type plants (Fig. 2) supports our conclusion that ICE1 binds ICEr3 (and/or ICEr4) to induce expression of the DRE-binding transcriptional activators CBF1, CBF2, and CBF3 (along with the ZAT12 and NAC072 transcriptional repressors). Conversely, the nonfunctionality of the ICEr1 cis-element in wild-type plants (Fig. 2F) argues against it playing a role in cold gene activation in vivo, though its acquired cis-regulon inducibility in ice1 plants (Fig. 3E) may explain why ICEr1 is enriched in the ice1 3 h cold-repressed TF regulon (Table III).
In agreement with previous experiments implicating circadian-gating/phytochrome regulation of CBF inducibility (Kim et al., 2002; Fowler et al., 2005), we were able to detect statistically significant depletion of ICEr3-containing genes (and the downstream signaling elements DRE, HOS9r1, ABRE, and ZAT12r1) from the list of all genes induced at dusk (relative to the rest of the circadian day; Table III). Although DRE-reporter constructs have been shown to be cold inducible in a phyA mutant background (Kim et al., 2002) and to be unresponsive in phyB mutants (suggesting PHYB mediation of the light signal; Kim et al., 2002), we were unable to establish the link between PHYB- and the ICE1-induced regulon through regulon overlap (Table II). In fact, there was a statistically significant exclusion of the ICE1- and CBF2-controlled regulon members from the phyB regulon. In contrast, the regulon of genes differentially regulated in FR light preconditioned phyA mutant plants (versus wild type) shows significant overlap with the ICE1-repressed regulon and is enriched for the ICEr3, DRE, and ICEr4 elements (Table III). While our results cannot exclude the influence of PHYB on the ICE1-mediated low temperature signaling pathway, they clearly implicate PHYA as an ICE1 transduction pathway mediator. This observation is consistent with reports that decreased temperature can modulate the functional relationships between phytochromes, and that PHYA, PHYD, and PHYE play greater roles with respect to PHYB in controlling flowering at lower temperatures (Halliday et al., 2003; Halliday and Whitelam, 2003). The exact nature of the interaction between ICE1 and PHYA needs to be determined, but it is noteworthy that the ICE1 gene promoter does not contain the previously reported PHYA regulatory site PhyAr1 (Hudson and Quail, 2003) and transcript levels of ICE1 are not affected in phyA mutants (as measured by microarray). The PHYA-ICE1 coupling may therefore be caused either via direct interaction with ICE1 (plausible, as PHYA is known to interact with basic helix-loop-helix TFs) via PHYA-mediated phosphorylation of ICE1, or through modulation of ICE1 proteolysis via a PHYA-mediated COP1-like ubiquitin-ligase degradation pathway such as HOS1 (Lee et al., 2001).
Because the individual microarray experiments used to construct our model represent snapshots of gene expression at single points in time, when interpreting the results it is necessary to be cognizant of the hidden temporal influences on the data and resulting model. For example, based on current evidence it appears that a phosphorylation event triggered after a drop in temperature converts the constitutively expressed ICE1 protein into a transcriptional activator (Gilmour et al., 1988; Chinnusamy et al., 2003). Looking at the behavior of the ICEr3 cis-regulon at a time point before this phosphorylation event occurred during an experimental time course would introduce the possibility of spurious conclusions being drawn about ICE1 bioactivity and connectivity with downstream TFs. A similar example drawn from our analysis is the bioinformatic and experimental data collected to position NAC072/RD26 in our model (Fig. 5). NAC072 is induced only after long period of low temperature treatment (10 h according to northern blotting; Tran et al., 2004), its promoter contains the ICEr3 consensus, and NAC072 is known to bind an element containing the ICEr1 consensus core (in its reverse complementary form CATGTG; Tran et al., 2004). Our meta-analysis of microarray data demonstrated that ICEr1-containing genes are repressed at 24 h in the cold (Fig. 2), suggesting that ICE1 may induce NAC072, leading to repression of ICEr1-containing genes. This implicated NAC072 in the longer-term repression of the CBF TFs and their downstream regulons via the ICEr1 sites present in the CBF2 and CBF3 promoters. We observed no overlap between the NAC072 induced and 3 h ice1 repressed TF regulons (Table II) despite enrichment of the NAC072 TF regulon promoters for the ICEr3 sites (Table III) due to the fact that the ICE1 regulon used for the TF regulon overlap analysis was composed of genes differentially regulated at a time point when NAC072 is not detectable by northern blotting (Tran et al., 2004). When we checked for ICE1 TF regulon overlap with the NAC072 repressed regulon after 24 h of cold treatment, significant enrichment was found (χ2, P < 0.05; Supplemental Table III).
The complexity of our proposed low temperature signaling model (Fig. 5) will likely increase as more low temperature signaling TFs are isolated and more time points are added to the microarray databases. Nevertheless, our model highlights several important considerations. First, the ICE-/CBF-mediated low temperature signaling pathway may be ABA independent (Gilmour and Thomashow, 1991), but ABA can act to amplify the ICE-/CBF-mediated signal via ICE1 transcript and ICEr3 cis-regulon induction (Chinnusamy et al., 2003; Knight et al., 2004; Table I). Second, the ICE-/CBF-mediated low temperature signaling pathway contains both positive and negative feedback loops. Expression of the NAC072/RD26 gene, which could positively feed back on ICE1-mediated signaling by increasing ABA sensitivity (Fujita et al., 2004), is activated by CBF TFs via the DRE element in its promoter, and we propose that it feeds back negatively on CBF expression via the ICEr1 site. This proposal is supported by gel-shift studies (Tran et al., 2004) and the agreement between the accumulation of NAC072 transcripts (at 10 h cold; Tran et al., 2004) and time of response for the ICEr1 regulon (24 h cold). Third, as suggested by our analysis and the experimental results of other authors (Gong et al., 2002; Kim et al., 2002), light affects the transcriptional activity of ICE1 and CBFs, with the ICEr3 and DRE element being less inducible than expected at dusk (versus the whole day) and strong interactions between phytochrome- and ICE1-mediated signaling pathways. Finally, while our analysis has focused on the ICE-meditated cold-signaling pathway, the gene-voting principle can be applied to a wide range of biological questions and should facilitate the mapping of bioactive linkages within complex signaling cascades, developing models that can be tested and verified by fewer bench-top experiments.
MATERIALS AND METHODS
Searching for Arabidopsis cis-Regulon Members
The annotated Arabidopsis (Arabidopsis thaliana) genome database (The Arabidopsis Information Resource; www.arabidopsis.org) was searched to determine the number of genes with at least one copy of each candidate element within their 500 bp promoter (5′ upstream region) using the online PATMATCH string-search tool. This tool returns a list that includes gene identity, number of elements, position(s), and element sequence (where degenerate base codes are used). The probability of the cis-element occurring by random chance was calculated based on average promoter nucleotide frequencies (68% AT, 32% GC) in Arabidopsis, assuming that nucleotides are arranged at random in the promoter, and that no evolutionary selection pressure has operated on that sequence. The expected number of genes containing the element was calculated by multiplying the expected frequency by the total number of genes searched. Cis-element enrichment was calculated as the ratio of observed to expected number of times that cis-element occurred in the annotated Arabidopsis genome.
cis-Regulon Correlation Analysis
We took the list of all Arabidopsis genes belonging to each cis-regulon and identified the behavior of those regulon members in different microarray experiments. We compared the number of induced/suppressed genes in each cis-regulon to the number of genes expected to be induced/suppressed based on the fraction of all genes that responded to that treatment (see “Creating the Voter Registry” section of the “Results” for full cutoff description). Our null hypothesis predicts that a nonfunctional cis-element produces a cis-regulon whose number of induced/suppressed genes is not significantly different than a similarly sized regulon chosen randomly from the genome. When the cis-regulon is significantly more induced or suppressed using a χ2 test (P < 0.01), the corresponding cis-element is deemed to have functionally correlated with regulation.
Searching for New Arabidopsis cis-Elements
The 500 bp promoters of all genes induced (or separately suppressed) by the specific cold-signaling TF mutation or overexpression in question were assembled and searched using a Gibbs sampling program called MotifSampler2.0, online at http://www.esat.kuleuven.ac.be/∼thijs/Work/MotifSampler.html (Thijs et al., 2002). We used the Arabidopsis background model (in Motifsampler) and looked for motifs eight nucleotides in length on either DNA strand. Prior probability was set to 0.5 and a maximum overlap of two nucleotides was allowed.
Microarray Data Sources
Normalized microarrays were taken from the NASC (http://affymetrix.arabidopsis.info/narrays/experimentbrowse.pl), The Arabidopsis Information Resource public databases, or WeigelWorld (http://www.weigelworld.org/resources/microarray/AtGenExpress/). Experimental statistics and normalization were carried out by the original authors or the database provider.
Meta-analysis of cis-element responsiveness in wild-type Arabidopsis to cold (4°C) and other abiotic stresses (UVB, methyl viologen, drought, salt, mannitol, and wounding) was carried out on the WeigelWorld abiotic stress treatment set. These experiments are fully described at the project Web site (URL above). NASC experiments were carried out on the Affymetrix ATH1 Arabidopsis genome array. Ecotype Columbia-0 (Col-0) cell culture was transferred to a high-light growth cabinet (240 μmol photons m−2 s−1; 16-h light/8-h dark) for 24 h prior to start of treatments. After this time, these wild-type/sfr6 plants either remained in this high-light treatment or were subjected to dark treatment by being wrapped in aluminum foil for 3 h before being harvested (sfr6; H. Knight, M. Knight, NASC experiment identification no. 194, 2005). Ler-0 ecotype plants were grown at 20°C (16 h d) and rosette and cauline leaf RNA extracted (phyB, phyAphyBcry1, and cry2; J. Casal, NASC experiment identification number 21, 2002). Col-0 ecotype seedlings on Murashige and Skoog media (no Suc) were preconditioned in the dark or FR light before transfer to continuous white light. Whole-seedling RNA was extracted (phyA and phyA + FR; A. McCormac, M. Terry, NASC experiment identification number 89, 2003). Ecotype Wassilewskija of Arabidopsis-2 was grown in long days and treated with ABA solution for 3 h, whole-plant RNA extracted (ABA; H. Okamoto, M. Knight, NASC experiment identification number 57, 2002). Col-0 seeds were sown on Murashige and Skoog agar plates (3% Suc), imbibed at 4°C for 96 h. Seeds were then entrained for 7 d at 22°C, in cycles of 12 h white light, 12 h darkness. After 7 d they were transferred to constant white light at 22°C (this is time 0 h). Tissue was harvested at 4 h intervals after time 0 (Dusk; K. Edwards, A. Millar, NASC experiment identification number 108, 2004).
These experiments were entered into MasterDB so that the expression of each gene (in rows) could quickly be read across to find induction/suppression calls for all treatments (in columns). In MasterDB, gene induction was annotated as 1, suppression as −1, no expression change as 0, and no data available as 2. We used fold-change cutoffs for induction/suppression calls that decreased with increasing gene expression level as described in the “Results” section of this article (Supplemental Table II). Our normalized microarray dataset and filtering-statistics formula spreadsheets (in Microsoft Excel) are available upon request (currently a 70 Mb file).
PCA
PCA is a multivariate projection method designed to extract and display the systematic variation in a data matrix X (Jackson, 1991). PCA is often used to get an overview of a data table X, detect clusters, and identify anomalies and outliers. The first two principal components define a plane that approximates all the variation in X. The coordinates of the points projected down onto this hyperplane are called scores (tan, a = 1,2,3,…A). The direction of each dimension in the hyperplane is its loading (pak, a = 1,2,3,…A). The part of X that is not explained by the model forms the residuals (Enk). The scores, loadings, and residuals together describe all of the variation in X.
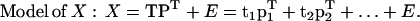 |
The PCA model calculations and visualization were performed using the SIMCA-P+ 11.0 software from Umetrics AB, Umeå, Sweden (www.umetrics.com).
Acknowledgments
We thank Professor Detlef Weigel, Max Planck Institute for Developmental Biology, Tübingen, and the AtGenExpress expression atlas project for the use of their microarray data sets, as well as the NASC database contributors listed above. We also thank Dr. Åsa Strand and Jan Eklöf, Umeå Plant Science Centre, for help revising the manuscript.
This work was supported by grants from the Swedish Forestry and Agricultural Research Council (to V.H.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Catherine Benedict (catherine.benedict@plantphys.umu.se).
The online version of this article contains Web-only data.
References
- Boyce JM, Knight H, Deyholos M, Openshaw MR, Galbraith DW, Warren G, Knight MR (2003) The sfr6 mutant of Arabidopsis is defective in transcriptional activation via CBF/DREB1 and DREB2 and shows sensitivity to osmotic stress. Plant J 34: 395–406 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong XH, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu J-K (2006) Gene regulation during cold acclimation in plants. Physiol Plant 126: 52–61 [Google Scholar]
- Coessens B, Thijs G, Aerts S, Marchal K, De Smet F, Engelen K, Glenisson P, Moreau Y, Mathys J, De Moor B (2003) INCLUSive: a web portal and service registry for microarray and regulatory sequence analysis. Nucleic Acids Res 31: 3468–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler SG, Cook D, Thomashow ME (2005) Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol 137: 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LSP, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39: 863–876 [DOI] [PubMed] [Google Scholar]
- Geisler M, Kleczkowski LA, Karpinski S (2006) A universal algorithm for genome-wide in silico identification of biologically significant gene promoter putative cis-regulatory-elements; identification of new elements for reactive oxygen species and sucrose signaling in Arabidopsis. Plant J 45: 384–398 [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Hajela RK, Thomashow MF (1988) Cold acclimation in Arabidopsis thaliana. Plant Physiol 87: 745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Thomashow MF (1991) Cold acclimation and cold regulated gene expression in ABA mutants of Arabidopsis thaliana. Plant Mol Biol 17: 1233–1240 [DOI] [PubMed] [Google Scholar]
- Gong ZZ, Lee H, Xiong LM, Jagendorf A, Stevenson B, Zhu JK (2002) RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc Natl Acad Sci USA 99: 11507–11512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday KJ, Salter MG, Thingnaes E, Whitelam GC (2003) Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J 33: 875–885 [DOI] [PubMed] [Google Scholar]
- Halliday KJ, Whitelam GC (2003) Changes in photoperiod or temperature alter the functional relationships between phytochromes and reveal roles for phyD and phyE. Plant Physiol 131: 1913–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson ME, Quail PH (2003) Identification of promoter motifs involved in the network of phytochrome A-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol 133: 1605–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JE (1991) A Users Guide to Principle Components. Wiley, New York
- Janick J (1989) Gregor Mendel. In J Janick, ed, Classic Papers in Horticultural Science. Prentice Hall, Englewood Cliffs, NJ, pp 406–412
- Kim HJ, Kim YK, Park JY, Kim J (2002) Light signalling mediated by phytochrome plays an important role in cold-induced gene expression through the C-repeat/dehydration responsive element (C/DRE) in Arabidopsis thaliana. Plant J 29: 693–704 [DOI] [PubMed] [Google Scholar]
- Knight H, Veale EL, Warren GJ, Knight MR (1999) The sfr6 mutation in Arabidopsis suppresses low-temperature induction of genes dependent on the CRT DRE sequence motif. Plant Cell 11: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Zarka DG, Okamoto H, Thomashow ME, Knight MR (2004) Abscisic acid induces CBF gene transcription and subsequent induction of cold-regulated genes via the CRT promoter element. Plant Physiol 135: 1710–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Henderson DA, Zhu JK (2005) The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17: 3155–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Xiong LM, Gong ZZ, Ishitani M, Stevenson B, Zhu JK (2001) The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev 15: 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K (2004) Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J 38: 982–993 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K (2006) Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiol Plant 126: 62–71 [Google Scholar]
- Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol 136: 2734–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thijs G, Moreau Y, De Smet F, Mathys J, Lescot M, Rombauts S, Rouze P, De Moor B, Marchal K (2002) INCLUSive: INtegrated clustering, upstream of sequence retrieval and motif sampling. Bioinformatics 18: 331–332 [DOI] [PubMed] [Google Scholar]
- Tran LSP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41: 195–211 [DOI] [PubMed] [Google Scholar]
- Zarka DG, Vogel JT, Cook D, Thomashow MF (2003) Cold induction of Arabidopsis CBF genes involves multiple ICE (Inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol 133: 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JH, Shi HZ, Lee BH, Damsz B, Cheng S, Stirm V, Zhu JK, Hasegawa PM, Bressan RA (2004) An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc Natl Acad Sci USA 101: 9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]