Abstract
The wheat (Triticum aestivum) and rye (Secale cereale) β-d-glucosidases hydrolyze hydroxamic acid-glucose conjugates, exist as different types of isozyme, and function as oligomers. In this study, three cDNAs encoding β-d-glucosidases (TaGlu1a, TaGlu1b, and TaGlu1c) were isolated from young wheat shoots. Although the TaGlu1s share very high sequence homology, the mRNA level of Taglu1c was much lower than the other two genes in 48- and 96-h-old wheat shoots. The expression ratio of each gene was different between two wheat cultivars. Recombinant TaGlu1b expressed in Escherichia coli was electrophoretically distinct fromTaGlu1a and TaGlu1c. Furthermore, coexpression of TaGlu1a and TaGlu1b gave seven bands on a native-PAGE gel, indicating the formation of both homo- and heterohexamers. One distinctive property of the wheat and rye glucosidases is that they function as hexamers but lose activity when dissociated into smaller oligomers or monomers. The crystal structure of hexameric TaGlu1b was determined at a resolution of 1.8 Å. The N-terminal region was located at the dimer-dimer interface and plays a crucial role in hexamer formation. Mutational analyses revealed that the aromatic side chain at position 378, which is located at the entrance to the catalytic center, plays an important role in substrate binding. Additionally, serine-464 and leucine-465 of TaGlu1a were shown to be critical in the relative specificity for DIMBOA-glucose (2-O-β-d-glucopyranosyl-4-hydroxy-7-methoxy-1,4-benzoxazin-3-one) over DIBOA-glucose (7-demethoxy-DIMBOA-glucose).
β-d-Glucosidase (EC. 3.2.1.21) is a major member of the family GH1 and GH3 glycoside hydrolases and is responsible for hydrolysis of terminal non-reducing β-d-Glc residues in oligosaccharides (or polysaccharides) or glucoconjugates. In plants, β-d-glucosidases are involved in various functions, including lignification (Dharmawardhana et al., 1995), regulation of the biological activity of cytokinins (Brzobohatý et al., 1993; Falk and Rask, 1995; Haberer and Kieber, 2002), control of the biosynthesis of indole-3-acetic acid (Ljung et al., 2001; Persans et al., 2001), and chemical defense against pathogens and herbivores (Niemeyer, 1988; Sicker et al., 2000; Zagrobelny et al., 2004). Many secondary products in plants occur as glucoconjugates with one or two Glc units attached to a hydroxy or thiol group. Hydrolysis of the glucosidic linkage in secondary metabolites, such as cyanogenic-, flavonoid-, and hydroxamic acid-glucosides, can drastically alter the biological activity, chemical stability, and water solubility of the molecule. The β-d-glucosidases implicated in the hydrolysis of plant secondary metabolites are members of the family GH1 glycoside hydrolases. The classification system of the glycoside hydrolases is available on the CAZy database at http://afmb.cnrs-mrs.fr/CAZY/.
Although β-d-glucosidases possess broad substrate specificity with respect to the aglycone moiety, the preferred aglycone structures vary with each glucosidase, reflecting their wide variety of physiological roles. Indeed, some β-d-glucosidases exhibit strict aglycone specificity. For example, the sorghum (Sorghum bicolor) glucosidase (dhurrinase 1, SbDhr1) acts specifically on its natural substrate, dhurrin. Dhurrin inhibits the activity of the maize (Zea mays) homolog, ZmGlu1, whose amino acid sequence shares about 70% identity with SbDhr1 (Hösel et al., 1987; Cicek and Esen, 1998). Recently, several research groups have investigated aglycone recognition mechanisms by SbDhr1 and ZmGlu1 using a combination of site-directed mutagenesis and x-ray crystallography (Czjzek et al., 2000, 2001; Verdoucq et al., 2003, 2004).
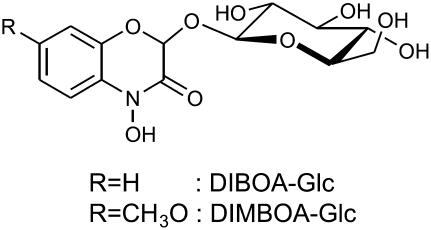
In previous studies, we purified β-d-glucosidases from the seedlings of wheat (Triticum aestivum) and rye (Secale cereale; Sue et al., 2000a, 2000b). The seedlings accumulate O-β-d-glucosides of hydroxamic acids (Hxs; 2,4-dihydroxy-1,4-benzoxazin-3-one, DIBOA, and its 7-methoxy derivative, DIMBOA; Fig. 1) as defensive compounds against pathogens and herbivores (Niemeyer, 1988). These compounds are thought to be stored in intact plants as glucosides within a different subcellular compartment from the glucosidase. Although the wheat and rye glucosidases hydrolyze DIBOA-Glc and DIMBOA-Glc, the preferred natural substrate for each enzyme is consistent with the predominant Hx found in each respective plant: DIMBOA-Glc in wheat and DIBOA-Glc in rye. These findings raise interesting questions concerning the evolution of this chemical defense mechanism. Recently, the cloning of a cDNA encoding the rye glucosidase (ScGlu) was reported by Nikus et al. (2003). The primary structure showed about 70% similarity to ZmGlu1 for which the preferred substrate is DIMBOA-Glc. Four hydrophobic amino acids (Trp-378, Phe-198, Phe-205, and Phe-466) of the maize glucosidase sandwich the aromatic aglycone moiety of the substrate (Czjzek et al., 2000, 2001; Zouhar et al., 2001) and are thought to be essential for the recognition of the aglycone moiety. Furthermore, Ala-467 was demonstrated to make contact with the methoxy group of DIMBOA-Glc. While Trp-378 and Phe-198 are conserved in rye at the corresponding positions, Phe-205, Phe-466, and Ala-467 are substituted by His, Gly, and Ser, respectively. Nikus et al. (2003) proposed that these substitutions allow the rye glucosidase to accept DIBOA-Glc as the preferred substrate. Because wheat is closely related to rye, the primary amino acid sequence of the wheat glucosidase is likely to resemble that of the rye enzyme. Indeed, the N-terminal sequences of the two enzymes are very similar to one other. Thus, the wheat glucosidase, with the preferred substrate DIMBOA-Glc, is an excellent target to investigate the structural factors that determine substrate preference between DIMBOA-Glc and DIBOA-Glc.
Figure 1.
Structures of the natural substrates in wheat. DIBOA-Glc, 2-O-β-d-Glucopyranosyl-4-hydroxy-1,4-benzoxazin-3-one; DIMBOA-Glc, 2-O-β-d-glucopyranosyl-4-hydroxy-7-methoxy-1,4-benzoxazin-3-one.
The β-d-glucosidases from both plants are thought to exist in an oligomeric form (probably tetrameric or hexameric; Sue et al., 2000a, 2000b). Specifically, the wheat glucosidase comprises 60- and 58-kD subunits and the rye enzyme comprises only 60-kD subunits, as determined by gel filtration and SDS-PAGE analysis. This is different from the maize homolog that is a homodimer of 60-kD subunits. Furthermore, the two subunits in the wheat enzyme are thought to make up several types of heterooligomers by assembling in different ratios, which is observed in a zymogram as multiple activity bands (Sue et al., 2000b). The N-terminal 12 amino acids of both subunits are identical, suggesting they are closely related isozymes. The zymogram of the maize glucosidase from a hybrid line has been shown to display three activity bands caused by two subunits derived from an allele (Stuber et al., 1977). The situation in wheat is more complicated because it has a hexaploid genome and oligomeric glucosidases. Furthermore, the origin of the subunits in the wheat enzyme, their substrate specificity, the actual quaternary structure, and the influence of oligomerization on activity are not yet known. The crystal structures of four family GH1 glucosidases from plants have been solved (Barrett et al., 1995; Burmeister et al., 1997; Czjzek et al., 2001; Verdoucq et al., 2004), and biochemical analysis has shown these enzymes function as dimers. Some bacterial enzymes have shown tetrameric or octameric quaternary structure in the crystal structure (Aguilar et al., 1997; Sanz-Aparicio et al., 1998; Chi et al., 1999; Hakulinen et al., 2000). However, the amino acid sequence of rye glucosidase, which forms an oligomer, shows a higher level of similarity to the plant enzymes than to the bacterial enzymes. Additionally, the monomer-monomer interaction and orientation of the bacterial glucosidases are different from those of the plant enzymes ZmGlu1 and SbDhr1. Thus, it is of interest to elucidate the quaternary structure of wheat glucosidase and investigate the structure-activity relationships of this enzyme. In this study, we cloned three cDNAs encoding the wheat β-glucosidase monomers (TaGlu1a–TaGlu1c) and characterized the hexameric enzymes of wheat and rye. In addition, we crystallized one of the wheat β-d-glucosidases (N-His-tagged TaGlu1b) as a hexamer and investigated the aglycone binding site using site-directed mutagenesis.
RESULTS
Primary Structure of the Wheat β-d-Glucosidase
Three cDNAs encoding wheat β-d-glucosidases were isolated by screening a cDNA library prepared from 48-h-old wheat shoots (cv Chinese Spring [CS]) and designated Taglu1a, Taglu1b, and Taglu1c (supplemental text named as “Cloning of the wheat glucosidases”). Taglu1a, Taglu1b, and Taglu1c comprised open reading frames of 1,710-, 1,710-, and 1,713-bp encoding polypeptides of 569, 569, and 570 amino acids, respectively (Supplemental Fig. 1). The deduced amino acid sequence of TaGlu1a shows 91%, 95%, and 95% identity and 95%, 98%, and 97% similarity to ScGlu, TaGlu1b, and TaGlu1c, respectively. Each TaGlu1 included the N-terminal sequence of the mature protein as confirmed by sequence analysis of the natural wheat glucosidases (Sue et al., 2000b). The predictive programs ChloroP v.1.1 (Emanuelsson et al., 1999) and TargetP v.1.01 (Emanuelsson et al., 2000) indicate that TaGlu1 may possess a signal peptide for a plastid (Supplemental Fig. 1) similar to the known monocot β-d-glucosidases from maize, rye, sorghum, and oats (Avena sativa).
Transcript Profiles of the Genes Encoding TaGlu1 Isozymes in Young Wheat
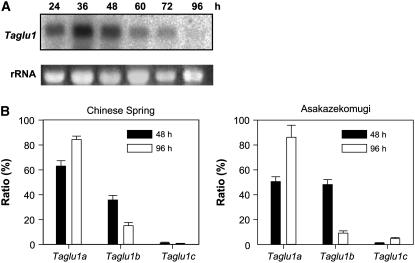
In our previous work, we reported that the glucosidase activity changes transiently, peaking 36 to 48 h after imbibition (Sue et al., 2000b). Northern-blot analysis showed that the genes are expressed at a high level 36 to 48 h after imbibition and that expression level then gradually decreases as the plant grows (Fig. 2A). This pattern correlated well with that of glucosidase activity. However, northern-blot analysis could not discriminate between each of the three Taglu1 genes because of the high level of sequence homology. Therefore, the expression of each glucosidase gene was analyzed by quantitative PCR using primers specific to each gene. In this experiment, RNA was also prepared from another bread wheat cultivar, Asakazekomugi (Ak), in addition to CS, to examine whether the expression pattern of each gene is conserved among the two cultivars.
Figure 2.
Expression level of Taglu1 mRNA. A, Northern-blot analysis. Taglu1, probed with Taglu1a; rRNA, ribosomal RNA stained with ethidium bromide. B, Quantification of Taglu1a to Taglu1c mRNA in 48- and 96-h-old wheat shoots (CS and Ak). The data of each growth stage are described as the ratio of each gene to the sum of Taglu1a to Taglu1c.
In CS, Taglu1a was most highly expressed (67%) in 48-h-old wheat, with Taglu1b expressed at about one-half this level (32%; Fig. 2B). The ratio was slightly different in a 96-h-old plant where Taglu1a expression increased to 85% and Taglu1b decreased to 14%, though the total amount of glucosidase gene expression declined as shown by the northern analysis. In both growth stages, the expression levels of Taglu1c were much less (0.4% and 0.1% in 48- and 96-h-old plants, respectively) than those of other glucosidase genes. These data were supported by the results of the cDNA library screening; 37 clones of Taglu1a, 11 clones of Taglu1b, and only one clone of Taglu1c were obtained. In contrast to the results of CS, Taglu1a and Taglu1b (Fig. 2B) were shown to be expressed in almost equal amounts in 48-h-old Ak (Taglu1a, 51%; Taglu1b, 48%). As the plant grows, however, the expression of Taglu1a and Taglu1b changed to a percentage comparable to that in CS. Similarly to CS, Taglu1c was expressed at a low level in both 48- and 96-h-old AK plants (1.3% and 4.7%, respectively).
Oligomeric Structures of the Wheat and Rye Glucosidases
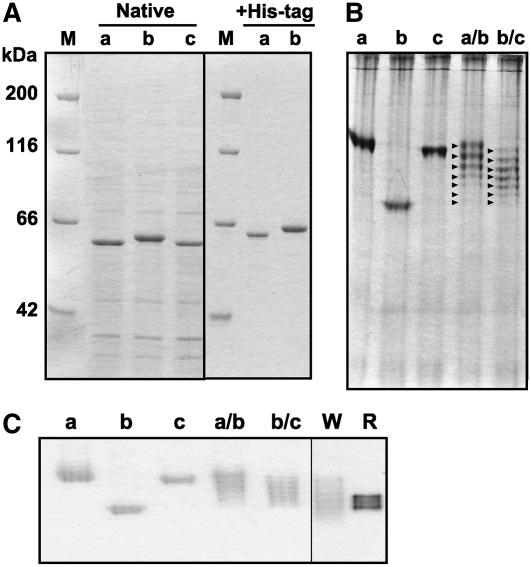
The diversity in the theoretical molecular mass of mature TaGlu1a to TaGlu1c (TaGlu1a, 59,155 D; TaGlu1b, 59,099 D; TaGlu1c, 59,245 D) is insufficient to explain the 2-kD disparity between the two subunits observed by SDS-PAGE of the natural wheat glucosidase (Sue et al., 2000b). Thus, we engineered the three wheat glucosidase genes for heterologous expression in Escherichia coli and examined the SDS-PAGE profiles of the respective recombinant proteins. The wheat glucosidases corresponding to the mature enzymes were overexpressed with and without an N-terminal His-tag in the BL21 CodonPlus(DE3)-RIL strain. While the native (without His-tag) TaGlu1a and TaGlu1c had similar mobility by SDS-PAGE, they evidently both migrated faster than TaGlu1b (Fig. 3A). Their molecular sizes on the gel were estimated as 58 kD (TaGlu1a and TaGlu1c) and 60 kD (TaGlu1b) that correspond to the two bands of the natural glucosidases purified from wheat shoots (Sue et al., 2000b). The same difference in mobility was evident with the N-His-tagged glucosidases (Fig. 3A). The precise Mr of N-His-tagged TaGlu1a and TaGlu1b was determined by mass spectrometry to make sure that they were not digested during extraction. TaGlu1a had a Mr of 64,141.25 ± 3.25 (theoretical Mr: 64,143), while TaGlu1b had a Mr of 64,085.41 ± 3.70 (theoretical Mr: 64,085). In contrast to the SDS-PAGE results, TaGlu1b showed increased mobility on a native-PAGE gel over the other two isozyme (Fig. 3, B and C).
Figure 3.
SDS- and native-PAGE of the wheat and rye glucosidases. A, The glucosidase monomers were analyzed by SDS-PAGE. For the native (without His-tag) glucosidases, the crude E. coli cell extracts were directly subjected to SDS-PAGE. The His-tagged glucosidases were electrophoresed after purification by affinity chromatography on a nickel-charged column. B, The cell extracts were subjected to native-PAGE (on an 8% separating gel for 4 h) and stained with Coomassie Brilliant Blue. The arrowheads indicate the seven types of glucosidase hexamers expressed in coexpression lines. C, The bands with β-glucosidase activity were detected by activity staining. The crude extracts of E. coli cells and 48-h-old shoots were separated under nondenaturing conditions. In each segment, M, a, b, c, a/b, b/c, W, and R indicate marker proteins, TaGlu1a, TaGlu1b, TaGlu1c, coexpressed TaGlu1a and TaGlu1b, coexpressed TaGlu1b and TaGlu1c, wheat shoots, and rye shoots, respectively.
The glucosidases overexpressed in E. coli were verified as constituting active homooligomers because the activity-stainable bands were detected on a native-PAGE gel at a similar position to the naturally occurring wheat glucosidase. However, the mobility of TaGlu1b and TaGlu1a (or TaGlu1c) was different (Fig. 3, B and C). To examine if the recombinant glucosidases had an ability to form heterooligomers, we coexpressed TaGlu1b and TaGlu1a (or TaGlu1c) in E. coli. Each glucosidase isozyme was expressed as the native form (without an N-terminal His-tag). As shown in Figure 3, B and C, both coexpression lines (Glu-1a/b and Glu-1b/c) exhibited seven bands when the cell extracts were subjected to native-PAGE. The uppermost and lowermost bands had the same electrophoretic mobility as the homooligomers. The results suggested that the wheat glucosidase monomers can form homo- and heterohexamers because formation of seven types of oligomers from two kinds of subunits can only be achieved when the subunits aggregate in a ratio from 0:6 to 6:0. The rye glucosidase had a similar mobility on a native-PAGE gel to the wheat glucosidase (Fig. 3C). Together with the gel filtration results (Fig. 4B; Sue et al., 2000a), the rye enzyme appeared to form a hexamer.
Figure 4.
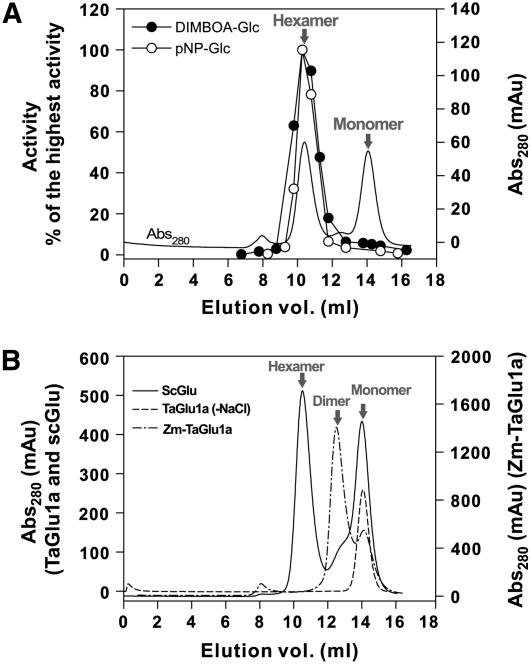
Gel filtration of TaGlu1a, ScGlu, and Zm-Glu1a. A, The enzyme solution eluted from the nickel column was further purified by gel filtration using a Superdex 200 column. Each fraction volume was 0.5 mL. The eluted protein was monitored by A280. The two protein peaks correspond to hexamer and monomer. B, Solid line, Gel filtration of ScGlu was performed after affinity chromatography. Two peaks of hexamer and monomer were observed. Dashed line, TaGlu1a (wild type) purified by affinity chromatography followed by gel filtration was dialyzed against HEPES buffer without NaCl and subjected to gel filtration analysis. Only a monomeric protein was detected. Dash-dot line, Zm-TaGlu1a purified by affinity chromatography was further purified by gel filtration. While the hexamer peak was not detected, the dimer and monomer peaks were observed.
N-His-tagged TaGlu1a expressed in E. coli was purified by metal chelation chromatography to almost complete homogeneity as judged by SDS-PAGE analysis (Fig. 3A). However, two major protein peaks were detected on the chromatogram of gel filtration corresponding to the hexamer and monomer, and only the hexamer peak showed activity with the natural (DIMBOA-Glc) and artificial (pNP-Glc) substrate (Fig. 4A). During the gel filtration experiments, minor peaks corresponding to the dimer and tetramer were occasionally observed (data not shown). Thus, the wheat glucosidase also can exist as a dimer and a tetramer. However, the fractions containing these oligomers, as well as the monomer, showed only slight activity with DIMBOA-Glc. The activity was so low that we could not eliminate the possibility that the activity was derived from minor contamination with the active hexamer. When the purified hexameric glucosidase was dialyzed against 50 mm HEPES without NaCl, the monomer readily formed (Fig. 4B), resulting in loss of activity.
Among the isozymes, TaGlu1b showed the highest activity with DIBOA-Glc and DIMBOA-Glc (the kcat/Km values 149 and 4,138 s−1/mm, respectively), while TaGlu1a, the major isozyme in 48-h-old CS, showed the lowest activity (4.5-fold lower than TaGlu1b). The lower activity of TaGlu1a may be caused by instability of the active hexamer, as suggested by gel filtration where the amount of hexameric TaGlu1a was lower than that of TaGlu1b or TaGlu1c (data not shown).
Crystal Structure of the Wheat β-d-Glucosidase and the Role of Its N-Terminal Region in Hexamer Formation
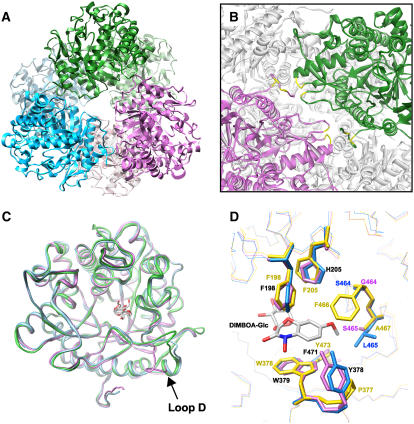
The structure of TaGlu1b in complex with DIMBOA was determined at 1.8-Å resolution from a crystal that was soaked in the DIMBOA solution. The final refinement statistics are shown in Table I. The overall structure of TaGlu1b was almost the same as those of known β-d-glucosidases, which have classical (β/α)8 barrel folds. The β-strands and α-helices within each β/α repeat were connected by loops at the top barrels. One disulfide bond was observed between Cys-210 and Cys-216, which is conserved among β-glucosidases in sorghum and maize. Although 11 residues at the N terminus and 18 at the C terminus of the mature enzyme were not included in the structure due to the lack of electron density, both termini were modeled three residues longer than those of sorghum or maize structures. The program PROCHECK (Laskowski et al., 1993) was applied to validate the structure; 374 residues were in most favored, 48 in additional allowed, and two (Ala-78 and Trp-463) were in generously allowed regions. When we superposed the TaGlu1b structure with those from sorghum (PDB code 1V03) and maize (PDB code 1E4N), they mostly fitted well, except that slightly different conformations were observed in the region from Val-413 to Pro-422 (loop D; Fig. 5C), probably due to low sequence similarity (Supplemental Fig. 1). The electron density of the amino acid chain was clearly observed, whereas that of the aglycone was not defined. This may have been due to interference caused by several glycerol molecules bound in the active site. The binding of a glycerol molecule at the active site was reported for the maize glucosidase Zm-p60.1 (Zouhar et al., 2001). A sulfate ion derived from LiSO4 in the crystallization buffer was also observed, which was fixed by Ser-366 and Asp-271, and Arg-434 of an adjacent subunit as well. Although the asymmetric unit contained one monomer of the enzyme, the symmetrical operation produced a hexamer conformation (Fig. 5A), where the dimer is obtained by the crystallographic 2-fold symmetry operation of the monomer and the hexamer by the 3-fold symmetry operation of the dimer. Both symmetrical operation axes are located perpendicular to each other.
Table I.
Refinement statistics
R factor = Σ‖Fobs| − Σ|Fcalc‖/Σ|Fobs|. Rfree was calculated with the 5% of reflections set aside randomly throughout the refinement.
| TaGlu1b | |
|---|---|
| Space group | P4132 |
| Cell dimensions a, b, c (Å) | 194.6, 194.6, 194.6 |
| Resolution (Å) | 50-1.8 |
| No. reflections | 2,667,849 |
| Unique reflections | 217,826 |
| Redundancy | 12.2 |
| Completeness (%) | 98.6 |
| Rwork/Rfree | 19.0/20.4 |
| No. atoms | |
| Water | 620 |
| Luzzati ESD (obs) | 0.19 |
| Luzzati Sigma A (obs) | 0.13 |
| Luzzati ESD (Rfree) | 0.21 |
| Luzzati Sigma A (Rfree) | 0.13 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 1.3 |
Figure 5.
Tertiary and quaternary structures of β-d-glucosidases. A, A ribbon diagram representation of the functional TaGlu1b hexamer. The dimers generated by crystallographic 2-fold symmetry operation are displayed as the same color. B, Side view of A (close-up view of dimer-dimer interface). The N-terminal regions of the monomer are positioned at the dimer-dimer interface. The four residues participating in direct hydrogen bonds to the monomer in the adjacent dimer are shown in yellow. These intermolecular hydrogen bonds are formed between T16 (side chain) and S366 (back bone), K17 (back bone) and Q270 (side chain), K19 (side chain) and S272 (side chain), and Q22 (side chain) and D271 (back bone). C, Superimposition of the back bones of TaGlu1b (magenta), ZmGlu1-E191D (PDB code 1E56; green), and SbDhr1 (PDB code 1V03; cyan). The structures of DIBOA-Glc and dhurrin in the substrate binding pockets of ZmGlu1 mutant and SbDhr1, respectively, are shown. D, Close-up view of the aglycone binding site. The crystal structures of TaGlu1b and ZmGlu1-E191D (PDB code 1E56) and the modeled structure of ScGlu are superimposed. A natural substrate, DIMBOA-Glc, bound to the maize enzyme is also shown. Blue, TaGlu1b; magenta, ScGlu; yellow, ZmGlu1-E191D. Identical residues found in TaGlu1b and ScGlu are shown in black. All of the molecular graphics images were produced using the UCSF Chimera package (Pettersen et al., 2004) from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institutes of Health P41 RR–01081).
If we regard the hexamer as three molecules of dimer (equivalent to the maize and sorghum dimers), the N-terminal region of TaGlu1b is located at the interface between the adjacent dimers. With respect to this region, the subunits are linked by four direct hydrogen bonds (Fig. 5B). To examine the role of this region in the formation of hexameric structure, the N-terminal 25 residues of the mature TaGlu1a and TaGlu1b were replaced with the corresponding residues of ZmGlu1 (Supplemental Fig. 1), considering that this enzyme is known to exist as a dimer. The chimeric glucosidases (Zm-TaGlu1a and Zm-TaGlu1b) completely lost their ability to form a hexamer, which was confirmed by gel filtration chromatography (Fig. 4B). Instead of being hexameric, the dimeric structure was the major component on the chromatogram, suggesting the crucial role of the N-terminal sequence in maintaining the dimer-dimer association. The fractions containing dimeric Zm-TaGlu1a or Zm-TaGlu1b exhibited little activity toward DIMBOA-Glc (data not shown).
Site-Directed Mutagenesis of the Substrate Binding Pocket
The recombinant enzyme with an N-terminal His-tag displayed an activity comparable to the naturally occurring glucosidase; the Vmax value of the natural wheat glucosidase for DIMBOA-Glc was 4,100 nkat/mg protein (Sue et al., 2000b) and that of the recombinant TaGlu1a was 5,200 nkat/mg protein. We therefore used the N-His-tagged enzyme for site-directed mutagenesis of residues at the substrate binding pocket. The amino acid residues involved in the substrate binding pocket are absolutely conserved among TaGlu1a to TaGlu1c (Supplemental Fig. 1), deduced from the structural and the sequence alignments with ZmGlu1-E191D mutant in complex with DIMBOA-Glc (Czjzek et al., 2000). The data of enzyme activity are shown in Tables II and III.
Table II.
Kinetic parameters of the TaGlu1s and TaGlu1a mutants
The relative efficiency toward each substrate is the percent ratio of the kcat/Km values to that of TaGlu1a. n.d., Not detected. –, Not determined.
| DIBOA-Glc
|
DIMBOA-Glc
|
pNP-Glc
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Relative Efficiency | Km | kcat | kcat/Km | Relative Efficiency | Km | kcat | kcat/Km | Relative Efficiency | |
| mm | s−1 | s−1/mm | % | mm | s−1 | s−1/mm | % | mm | s−1 | s−1/mm | % | |
| TaGlu1a | 1.40 | 48.8 | 34.6 | 100 | 0.36 | 338 | 939 | 100 | 1.85 | 99.2 | 53.6 | 100 |
| TaGlu1b | 1.44 | 214 | 149 | 429 | 0.29 | 1,201 | 4,138 | 441 | 2.16 | 128.2 | 59.3 | 111 |
| TaGlu1c | 1.05 | 137 | 131 | 377 | 0.39 | 773 | 1979 | 211 | 1.75 | 235.9 | 135 | 252 |
| E191A | n.d. | n.d. | – | 0 | n.d. | n.d. | – | 0 | n.d. | n.d. | – | 0 |
| F198A | 27.5 | 6.7 | 0.24 | 0.7 | 35.7 | 110 | 3.1 | 0.3 | 9.28 | 10.7 | 1.2 | 2.2 |
| Y378A | 6.05 | 26.4 | 4.36 | 12.6 | 1.19 | 124 | 104 | 11.4 | 1.33 | 25.1 | 18.9 | 35.3 |
| Y378F | 0.41 | 4.7 | 11.5 | 33.2 | 0.13 | 85.1 | 655 | 69.8 | 1.23 | 46.1 | 37.5 | 70.0 |
| E407A | n.d. | n.d. | – | 0 | n.d. | n.d. | – | 0 | n.d. | n.d. | – | 0 |
| S464F | 2.89 | 84.0 | 29.1 | 84.1 | 1.42 | 73.3 | 51.6 | 5.5 | 2.31 | 31.2 | 13.5 | 25.2 |
| F471Y | 4.21 | 83.5 | 19.8 | 57.2 | 0.70 | 192 | 274 | 29.2 | 2.93 | 54.6 | 18.6 | 34.7 |
Table III.
Kinetic parameters of ScGlu and its mutants
The relative efficiency toward each substrate is the % ratio of the kcat/Km values to that of ScGlu. n.d., Not detected.
| DIBOA-Glc
|
DIMBOA-Glc
|
pNP-Glc
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Relative Efficiency | Km | kcat | kcat/Km | Relative Efficiency | Km | kcat | kcat/Km | Relative Efficiency | |
| mm | s−1 | s−1/mm | % | mm | s−1 | s−1/mm | % | mm | s−1 | s−1/mm | % | |
| ScGlu | 0.80 | 118 | 148 | 100 | 1.30 | 158 | 122 | 100 | 1.78 | 22.2 | 12.5 | 100 |
| E191A | n.d. | n.d. | – | 0 | n.d. | n.d. | – | 0 | n.d. | n.d. | – | 0 |
| F198A | 44.1 | 233 | 5.28 | 3.6 | 39.3 | 194 | 4.94 | 4.1 | 3.05 | 8.3 | 2.72 | 21.8 |
| Y378A | 7.99 | 1,098 | 137 | 92.6 | 1.42 | 826 | 582 | 477 | 2.02 | 102 | 50.5 | 404 |
| Y378F | 0.52 | 12.0 | 23.1 | 15.6 | 1.07 | 195 | 182 | 149 | 1.34 | 63.0 | 47.0 | 376 |
| E407A | n.d. | n.d. | – | 0 | n.d. | n.d. | – | 0 | n.d. | n.d. | – | 0 |
| G464F | 1.60 | 54.6 | 34.1 | 23.0 | 5.12 | 121 | 23.6 | 19.3 | 1.70 | 25.5 | 15.0 | 120 |
| G464S | 1.02 | 86.9 | 85.2 | 57.6 | 0.94 | 229 | 244 | 200 | 2.89 | 124 | 42.9 | 343 |
| S465L | 0.83 | 46.6 | 56.1 | 37.9 | 0.76 | 225 | 296 | 243 | 2.03 | 75.6 | 37.2 | 298 |
| G464S/S465L | 1.55 | 83.5 | 53.9 | 36.4 | 0.59 | 288 | 488 | 400 | 1.38 | 75.4 | 54.6 | 437 |
| F471Y | 2.03 | 243 | 120 | 81.1 | 1.04 | 321 | 309 | 253 | 0.92 | 46.3 | 50.3 | 402 |
The primary structures of the aglycone binding sites of TaGlu1a and ScGlu diverge from each other at the residues Ser-464 and Leu-465 in TaGlu1a and Gly-464 and Ser-465 in ScGlu. While the mutations G464S and S465L of ScGlu decreased the relative efficiency for DIBOA-Glc by 42% and 62%, respectively, they increased the efficiency for DIMBOA-Glc by 100% and 143%, respectively. The effects were enhanced by introduction of the double mutation G464S/S465L. Introduction of Phe at position 464 of both enzymes resulted in decreased efficiency for the natural substrates. The influence was most obvious in TaGlu1a with DIMBOA-Glc (relative efficiency 5.5%). The kcat/Km values of ScGlu-F198A for DIMBOA-Glc and DIBOA-Glc decreased by a factor of 24- to 28-fold as compared with those of wild-type ScGlu. However, the decrease in the kcat/Km value of this mutant was solely due to an increased Km, whereas the kcat value actually increased. TaGlu1a-F198A mutants showed some activity (kcat) toward the natural substrates, although the Km values increased dramatically. The Y378A mutation of ScGlu enhanced kcat/Km values for DIMBOA-Glc and pNP-Glc by 300% to 380%, whereas that of TaGlu1a was lowered to 11% to 35% for all substrates tested. Although the catalytic efficiency (kcat/Km) of the ScGlu mutant for DIBOA-Glc was comparable to that of wild type, both the Km and kcat were about 10-fold greater. Replacing Phe-471 in TaGlu1a with Tyr decreased the catalytic efficiency for all the substrates tested. However, the same mutation on ScGlu increased the kcat for the three substrates by about 100% and decreased Km for DIMBOA-Glc and pNP-Glc. TaGlu1 and ScGlu contain the well-conserved TFNEP and ITNEG motifs at the catalytic center of family GH1 glucosidases, and replacement of either of the two Glu residues in the motifs (Glu-191 and Glu-407, respectively) with Ala resulted in a complete loss of enzyme activity.
DISCUSSION
Taglu1a and Taglu1b Genes Are Highly Expressed in Young Wheat Shoots
We isolated three genes encoding family GH1 glucosidases that are responsible for the hydrolysis of Hx-Glcs. The transcript profile of Taglu1 genes, as analyzed by northern hybridization, agreed with the transient occurrence of glucosidase activity and Hx-Glc occurrence in young plants (Sue et al., 2000b). However, the expression profiles of each gene after 48- and 96-h imbibition do not appear to be synchronized, suggesting that the expression of these three genes is controlled independently of each other. This is similar to the expression profiles of the three homoeologous genes of each of the five Hx biosynthetic genes, which vary between developmental stages of wheat seedlings (Nomura et al., 2005). The mechanism underlying the different expression profiles of the three Taglu1s as well as the biosynthetic genes has not yet been uncovered.
The ratio of each Taglu1 gene differs between two wheat cultivars at the same growth stage. At 48 h, CS expressed the Taglu1a gene at a higher ratio than Taglu1b, whereas Ak expressed both genes almost equally. In consideration of the result that TaGlu1a moves more slowly than TaGlu1b on a native-PAGE gel, the higher expression level of Taglu1a over Taglu1b in CS can explain the different zymogram patterns of the two cultivars; the intensity of cathodic bands was stronger than those of the anodic bands in CS, while the intermediate bands were of higher intensity for Ak (Sue et al., 2000b). The purified glucosidase from 48-h-old Ak showed seven bands on a native-PAGE gel. Because TaGlu1c was considered to be expressed at a much lower level than TaGlu1a and TaGlu1b in the plant, the natural glucosidase would mainly exist as homo- and heterooligomers of TaGlu1a and TaGlu1b.
The Wheat and Rye Glucosidases Function as a Hexamer
The recombinant TaGlu1b showed greater electrophoretic mobility than TaGlu1a and TaGlu1c on a native-PAGE gel. The mobility of proteins in native-PAGE is influenced by the shape, or holding, and charge of the proteins as well as their Mr. The results of ion-spray mass spectrometric analyses of recombinant TaGlu1a and TaGlu1b subunits excluded the possibility of proteolytic digestion during cell disruption. Furthermore, the overall holding of the TaGlu1b must be almost identical with the other isozymes because it shares high homology with TaGlu1a and TaGlu1c with respect to the primary structures and has similar enzyme activity and substrate specificity. Therefore, the greater mobility of TaGlu1b is presumably due to its more substantial net negative charge compared with TaGlu1a and TaGlu1c. This hypothesis is supported by the lower theoretical pI value of TaGlu1b (pI 5.2) in comparison with that of TaGlu1a and TaGlu1c (both with pI 5.8). Nevertheless, the anomalously low mobility of TaGlu1b under denaturing conditions cannot be predicted from the pI value. There may be additional factors that cause the mobility discrepancy by SDS-PAGE. The slight differences in the amino acid composition might result in different migration rates for the protein even under denaturing condition.
In the previous study, we demonstrated that the wheat glucosidase exhibits activity as an oligomer (Sue et al., 2000b), but the number of subunits constituting the oligomer remained unclear. These results from coexpression of two TaGlu1 isozymes (TaGlu1a and TaGlu1b) and the corresponding gel filtration analysis have established that TaGlu1 exhibits its activity only as a hexamer. Because wheat and rye glucosidases show similar gel filtration and electrophoretic profiles, it is reasonable to assume that the active form of ScGlu is also a hexamer. Although only three bands were detected in the sample of natural ScGlu on a native-PAGE gel (Fig. 3C), they must comprise a larger number of stacking bands because the rye glucosidase isolated from shoots separated into at least six protein peaks by anion-exchange chromatography (Sue et al., 2000a). Presumably, the rye used in our studies contains multiple isozymes whose electrophoretic mobility is very similar to each other. The maize glucosidase gene (Zmglu1) is known to be a highly polymorphic gene, and a zymogram using hybrid lines shows three bands of β-glucosidase (Stuber et al., 1977) representing two homodimers and one heterodimer consisting of two allozymes. Bread wheat is a hexaploid plant consisting of homoeologous A, B, and D genomes. Therefore, it is not surprising that the three almost identical Taglu1 genes were isolated from a bread wheat cultivar, CS. However, the chromosomal locations of the Taglu1a to Taglu1c genes are unknown at present.
Multimeric forms of some plant β-d-glucosidases have been reported previously. Among monocot glucosidases, the β-d-glucosidases from oat (avenacosidase), sorghum (SbDhr1), and maize (ZmGlu1) have about 75% similarity to TaGlu1 and ScGlu. Although the maize glucosidase can form a large complex mediated by an aggregating factor (Esen and Blanchard, 2000), it primarily exists as a dimer. SbDhr1 was demonstrated to exist as a tetramer in the plant (Hösel et al., 1987), but it does not lose activity even when it dissociates into a dimer. The avenacosidase is known to form 300- to 350-kD aggregates and multimers thereof (Gus-Mayer et al., 1994; Kim and Kim, 1998). Gus-Mayer et al. (1994) reported that the avenacosidase (As-p60) dissociates into a dimer by treatment with urea or freeze-thawing accompanied by a loss of activity, although the recombinant T7. Tag-As-Glu2 (an isozyme of As-p60) was shown to form an active dimer (Kim et al., 2000). Thus, TaGlu1 and ScGlu seem to be unique in that they require a hexameric structure to exhibit enzyme activity.
On replacing the N-terminal 25 residues with the corresponding region of ZmGlu1, the wheat glucosidase (Zm-TaGlu1a or Zm-TaGlu1b) monomer could not make up a hexamer but a dimer (Fig. 4B). The amino acid sequences of the N-terminal region are least conserved among the family GH1 β-d-glucosidases. This result suggests a novel function of the N-terminal region in maintaining the quaternary structure of the β-d-glucosidases. The dimer of Zm-TaGlu1 easily dissociated into monomers (data not shown), suggesting that the monomer of TaGlu1 interacts very weakly in comparison with the ZmGlu1 or SbDhr1 monomers. These characteristics differ from the monocot family GH1 β-d-glucosidases, where the dimeric forms represent the stable structural units from which the oligo- and multimers are composed. The crystal structure of the TaGlu1b monomer resembles the known structures of the maize and sorghum homologs (Fig. 5C). The two residues in ZmGlu1 (Arg-295 and Asp-342) that were shown to form the intermonomer salt bridges (Czjzek et al., 2001) are conserved in TaGlu1b (Arg-296 and Asp-343). The intramolecular disulfide bond responsible for maintaining the dimeric form of Zm-p60.1 (Zouhar et al., 2001) is also conserved in the wheat glucosidase. Furthermore, the other amino acids mapping to the monomer-monomer interface of ZmGlu1 show high similarity to the corresponding regions of the wheat and rye glucosidases. Thus, the reasons for the weak monomer-monomer interaction within the dimer are unclear at present.
F198A Mutation Reduces the Catalytic Efficiency of TaGlu1a and ScGlu
The crystal structure of a β-glucosidase responsible for hydrolysis of Hx-Glc was first solved for maize (Czjzek et al., 2000). The aglycone moiety of DIMBOA-Glc was shown to be stabilized by four aromatic side chains: Trp-378 on one side and three Phe residues (Phe-198, Phe-205, and Phe-466) on the other side (Czjzek et al., 2001; Zouhar et al., 2001). Although Trp-378 and Phe-198 are conserved among the five monocot β-d-glucosidases (glucosidases from wheat, rye, maize, sorghum, and oats), Phe-205 and Phe-466 are substituted by a His and Ser, respectively, in TaGlu1 and His and Gly, respectively, in ScGlu. These amino acid substitutions change the electrostatic and spatial environment of the aglycone binding pocket, although substrate recognition and binding must resemble that of ZmGlu1 because all three enzymes favor Hx-Glcs. Assuming the mechanism of substrate recognition is the same for TaGlu1a, ScGlu, and ZmGlu1, the sole aromatic residue (Phe-198) positioned opposite Trp-379 in TaGlu1a and ScGlu is likely to play a significant role in substrate binding. This is supported by the results of the F198A mutants, where the catalytic efficiency (kcat/Km) decreased by more than 95% except for the activity of ScGlu-F198A against pNP-Glc. However, the kcat of ScGlu for DIBOA-Glc and DIMBOA-Glc was enhanced by this mutation, and the kcat of TaGlu1a-F198A toward DIMBOA-Glc was about 30% that of the wild-type protein. These results are somewhat different from those for the maize glucosidase, where replacement of Phe-198 by smaller amino acids caused a more drastic reduction in kcat (Zouhar et al., 2001; Verdoucq et al., 2003). The crystal structure of the ZmGlu1 mutants revealed that the F198V mutation changes the orientation of the side chain of Phe-466, one of the Phe residues constituting the hydrophobic binding pocket, leading to complete loss of activity against pNP-Glc (Verdoucq et al., 2003). In the case of TaGlu1a and ScGlu, the Phe-466 is substituted by Ser and Gly, respectively. Thus, the mutation may alter the environment around the binding pocket to a lesser extent in the wheat and rye glucosidases. Additionally, the Tyr at the entrance to the aglycone binding sites in both glucosidases (Tyr-378) may compensate for the deletion of Phe-198.
Bulky Aromatic Side Chains at the Entrance of the Substrate Binding Site Play a Significant Role in TaGlu1a
One of the greatest differences around the aglycone binding pocket between the wheat and rye glucosidases and the maize glucosidase is Tyr-378 in TaGlu1 and ScGlu (equivalent to Pro-377 in ZmGlu1). The bulky side chain of Tyr-378 in TaGlu1a and ScGlu may obstruct access to the binding pocket by the substrate. Indeed, we initially assumed that replacing the Tyr with a small residue might increase the activity of the enzymes. Substitution of the Tyr with Ala caused the kcat/Km of ScGlu to increase for DIMBOA-Glc and pNP-Glc by a factor of 4- to 5-fold. However, the efficiency for all substrates was lower for the TaGlu1a-Y378A mutant, suggesting a different role for the Tyr residue in the wheat enzyme. The Ser-464 and Leu-465 residues in TaGlu1a, which are bulkier than the corresponding residues in ScGlu, may require a lid to maintain the substrate in a favorable position. The higher Km values of the Y378A mutants and the lower Km values in the Y378F mutants indicate the significance of an aromatic residue positioned at the entrance of the binding pockets. In the case of the maize glucosidase, the lack of an aromatic residue may still give high activity toward DIMBOA-Glc because the binding pockets, comprising four aromatic side chains, can still hold the aromatic aglycone moiety firmly. The lower kcat values of TaGlu1a- and ScGlu-Y378F mutants compared to those of wild-type proteins may indicate the hydroxy group is necessary for placing the glucosidic bond of the substrate at the optimal position for attack by the catalytic residues. However, a more hydrophobic environment seems to be more favorable for enzyme-substrate binding.
The Effects of the Mutations at Positions 464, 465, and 471
This result that the S465L mutation of ScGlu increased the efficiency toward DIMBOA-Glc seems to match with the results of ZmGlu1, where the maize counterpart of the residue is involved in the recognition of the methoxy group in DIMBOA-Glc (Czjzek et al., 2000). However, the single G464S mutation alone could raise the specificity to DIMBOA-Glc. Therefore, amino acid 464 as well as 465 plays an important role in distinguishing DIMBOA-Glc from DIBOA-Glc. The substitutions of Gly-464 and Ser-465 of ScGlu by the TaGlu1 counterparts made the substrate preference of ScGlu resemble that of TaGlu1s. However, the kcat/Km value toward DIMBOA-Glc was about 30 times larger than that toward DIBOA-Glc in TaGlu1a, although the value against DIMBOA-Glc of ScGlu-G464S/S465L mutant was only about 10 times larger than that against DIBOA-Glc. This would suggest that there are other unknown factors in TaGlu1a that contribute to distinguish DIMBOA-Glc from DIBOA-Glc.
In ZmGlu1, Tyr-473 was shown to form a hydrogen bond with Trp-378, and the mutation of Tyr-473 into Phe caused an increase in efficiency (Verdoucq et al., 2003). The authors suggested that the loss of the hydrogen bond increased the flexibility of Trp-378, resulting in an enhanced catalytic efficiency. Replacing the corresponding residue of TaGlu1a, Phe-471, into Tyr resulted in a decrease in efficiency, which agrees with the results of ZmGlu1. However, the same mutation in ScGlu enhanced the efficiency toward DIMBOA-Glc and pNP-Glc, while it decreased the efficiency toward DIBOA-Glc. It is notable that every ScGlu mutant that showed a higher efficiency toward DIMBOA-Glc than the wild type exhibited an increased value toward pNP-Glc but not toward DIBOA-Glc, although the detailed mechanisms are not known at present.
Because none of the mutations introduced in this study enhanced the efficiency of TaGlu1a, a more precise organization of the substrate binding site is likely to be required to recognize and hydrolyze the substrates in TaGlu1a by comparison with ScGlu. The broader substrate specificity of ScGlu may be due to the wider aglycone binding site compared with those of ZmGlu1 and TaGlu1a. Further structural analyses of TaGlu1 and ScGlu crystals will allow elucidation of the details of substrate recognition and organization of the active hexamers.
MATERIALS AND METHODS
Plant Materials
Two cultivars of bread wheat (Triticum aestivum), cv Ak and cv CS, and rye (Secale cereale) were grown at 25°C as described previously (Sue et al., 2000a, 2000b).
Northern-Blot Analysis and Quantitative-PCR
Total RNA was isolated from wheat shoots (Ak) using RNeasy Plant Mini kit (QIAGEN). The RNA (15 μg) was separated on a 1.2% agarose MOPS-formaldehyde gel and transferred to a nylon membrane, Hybond N+ (Amersham Biosciences). The membrane was probed with an [α-32P]-labeled cDNA fragment from the 3′-RACE (see supplemental text) as described by Church and Gilbert (1984). The hybridization and washing procedures were carried out at 65°C. The membrane was autoradiographed with Imaging Plate and analyzed by a bio-imaging analyzer, BAS2000 (Fujifilm).
The primers used for quantitative PCR are shown in Supplemental Table I. Total RNA was isolated from 48- and 96-h-old wheat shoots (Ak and CS), and cDNA was synthesized using 1 μg of total RNA and Omniscript reverse transcriptase (QIAGEN) by priming with oligo(dT) primer. Prior to performing real-time PCR, the sequences of the DNA fragments amplified with the above primer sets were confirmed to be identical to the corresponding cDNA sequences. The cDNA (2 μL of 1/50 dilution of the RT sample) was used for real-time PCR on a LightCycler with LightCycler FastStart DNA Master SYBR Green kit (Roche). Each sample was quantified at least three times with respect to standard DNA (ranging from 102 to 106 copies/reaction tube) quantified under the same conditions. An annealing temperature of 64°C was employed. The PCR buffer contained 3 mm MgCl2. All other PCR conditions were set according to the manufacturer's instructions. The purity of the DNA products was confirmed by agarose gel electrophoreses after every real-time PCR reaction.
Expression of the β-d-Glucosidases in Escherichia coli
The full-length cDNAs (Taglu1a–Taglu1c) were used as template for PCR to prepare DNA fragments flanked by NcoI and XhoI recognition sites at the 5′ and 3′ ends, respectively. The sequences of the primers used in the PCR are shown in Supplemental Table I. Scglu was amplified by RT-PCR using total RNA prepared from 48-h-old rye shoots (cv Haru-ichiban) with the primers given in Supplemental Table I. The primer for Scglu were designed based on the sequence of the rye glucosidase (GenBank accession no. AF293849) reported by Nikus et al. (2003). The reaction was carried out utilizing KOD polymerase (TOYOBO) with denaturing at 98°C, annealing at 58°C, and polymerization at 74°C. The amplified DNA fragments corresponding to the mature glucosidases were digested by NcoI and XhoI and then cloned into pET21d or pET30a. By introducing the DNA fragments into the NcoI and XhoI sites of pET21d and pET30a, we obtained plasmids for native (without His-tag) and N-terminal His-tagged glucosidases, respectively. The ligation products were transformed into BL21 CondonPlus(DE3)-RIL (Stratagene) competent cells. For coexpression of the glucosidase genes, Taglu1a (or Taglu1c) was cloned into pET21d and Taglu1b into pCDFDuet-1 (Novagen). Both plasmids were then introduced into the BL21-CodonPlus(DE3)-RIL strain. The E. coli was cultured in 50 mL of Luria-Bertani broth supplemented with appropriate antibiotics at 37°C with shaking until the OD600 reached approximately 0.5. Heterologous gene expression was then induced by adding 1 mm isopropylthio-β-galactoside followed by an overnight culture at 20°C. The cells were pelleted by centrifugation (1,500g for 15 min) and then resuspended in 5 mL of 50 mm HEPES, pH 7.2, containing a protease inhibitor cocktail (Sigma). The cells were disrupted by sonication on ice (several 20-s pulses at a power setting of 100 W). The soluble protein fraction was recovered by collecting the supernatant after centrifugation at 15,000g for 15 min.
The recombinant His-tagged protein was purified by metal chelation chromatography. The soluble fraction was applied to a HiTrap Chelating HP column (Amersham Biosciences) charged with Ni2+ and equilibrated in 0.02 m phosphate buffer, pH 7.4, containing 0.5 m NaCl and 10 mm imidazole. After washing the column with the same buffer containing 0.5 m NaCl and 60 mm imidazole, the glucosidase was eluted by increasing the concentration of imidazole to 300 mm. The eluate was concentrated by ultracentrifugation and then subjected to gel filtration chromatography on Superdex 200 (Amersham Biosciences) equilibrated with 50 mm HEPES and 150 mm NaCl, pH 7.2. To estimate the molecular mass on the gel filtration column, the following proteins were used as standards: ferritin (440 kD), human IgG (160 kD), transferrin (81 kD), ovalbumin (43 kD), and myoglobin (17.6 kD).
Electrophoresis and Activity Staining
The protein profile was analyzed by SDS-PAGE and native-PAGE using an 8% gel as described previously (Sue et al., 2000b). The bands corresponding to β-glucosidase were detected on a native-PAGE gel using a chromogenic substrate, 6-bromo-2-naphthyl-β-d-glucopyranoside, as described previously (Sue et al., 2000b).
Mass Analyses of TaGlu1a and TaGlu1b
The purified N-His-tagged TaGlu1a and TaGlu1b were concentrated by ultrafiltration, followed by dilution with MilliQ water to reduce the salt concentration in the buffer. After several rounds of concentration and dilution, acetonitrile and formic acid were added to give a final concentration of 20% and 0.1%, respectively. The protein concentration was adjusted to 1 mg/mL. The molecular masses of the glucosidases were measured using a Perkin-Elmer-Sciex API-165 (ion-spray voltage 5 kV, orifice voltage 30 V, nebulizer gas N2, curtain gas N2). The theoretical Mr and pI were determined by the Compute pI/Mw program on the ExPASy server (http://kr.expasy.org/tools/).
Structure Determination
The recombinant β-glucosidase was expressed, purified, and crystallized as described previously (Sue et al., 2005). To obtain the complex of TaGlu1b and its substrate aglycone, DIMBOA, crystals were soaked in the crystallization buffer with 0.5 mm DIMBOA and 30% glycerol as a cryoprotectant for 15 min and then cooled in a nitrogen stream at 100 K. The diffraction data set was collected on beamline BL-6A at Photon Factory and processed by the program HKL2000 (Otwinowski and Minor, 1997). The initial model was obtained by the molecular replacement method using the program MOLREP (Vagin and Teplyakov, 1997) and the β-glucosidase molecule from sorghum (Sorghum bicolor; PDB code; 1v03; Verdoucq et al., 2004) as the search model. The crystal belonged to the space group P4132 and contains one monomer in an asymmetric unit. The iterative refinement was performed using the programs CNS (Brunger et al., 1998) and XtalView (McRee, 1992).
Site-Directed Mutagenesis of His-Tagged TaGlu1a and ScGlu
Mutated DNA fragments for expression of TaGlu1a and ScGlu mutants were prepared by PCR-mediated overlap extension. The sequences of the mutagenic PCR primers used in this study are shown in Supplemental Table I. The DNA fragments for Zm-TaGlu1a and Zm-TaGlu1b were amplified by PCR using the primers shown in the Supplemental Table I. The amplified fragments were digested by NcoI and XhoI, introduced into pET30a, expressed in E. coli, and purified by affinity chromatography followed by gel filtration chromatography as described above.
Substrate Preparation and Enzyme Assay
DIBOA-Glc and DIMBOA-Glc were isolated from shoots of 48-h-old rye and maize (Zea mays), respectively, according to methods described previously (Sue et al., 2000b).
Enzyme activity was measured in 100 mm citrate-200 mm phosphate buffer, pH 5.5, at 30°C. The product was quantified by HPLC (eluent, 30% [v/v] methanol containing 0.1% [v/v] acetic acid; column, Inertsil ODS-3 [4.6 × 150 mm; GL Sciences]; temperature, 40°C). The amount of protein and reaction time were carefully chosen so that the product did not exceed 5% to 10% of the residual substrate. Kinetic parameters were determined from several independent experiments. The preliminary Km and kcat values were determined from reaction rates at various concentrations of substrates ranging from 0.02 mm to 10 mm. Precise kinetic parameters were subsequently obtained by varying the substrate concentration from 20% to 200% of the preliminary Km value. Km and kcat values were calculated by fitting the data to the Michaelis-Menten equation using SigmaPlot 2000 (SYSTAT Software). The protein concentration was measured by the method of Bradford (Bradford, 1976) using bovine serum albumin as a standard.
Modeling of ScGlu
The three-dimensional model of ScGlu was calculated by homology modeling with MODELLER6v2 program (Sali and Blundell, 1993; Marti-Renom et al., 2000) using the structures of TaGlu1b and ZmGlu1 (PDB code 1E56). First, the primary structures of ScGlu, TaGlu1b, and ZmGlu1 were aligned using ClustalW, and the result was checked based on the actual (TaGlu1b and ZmGlu1) or predicted (ScGlu) secondary structures. Then, five models were calculated by MODELLER6v2 using the above sequence alignment. Since all structures obtained were almost identical to each other, one model was employed as a predicted structure of ScGlu. The model was evaluated by PROCHECK. Although one residue (Lys-251) was in a disallowed region, it was located on the surface of the protein and far from the substrate binding pocket.
Sequence data from this article can be found in the DDBJ/EMBL/GenBank data libraries under the accession numbers AB100035 (Taglu1a), AB236422 (Taglu1b), and AB236423 (Taglu1c). The atomic coordinates of TaGlu1b were deposited in the Protein Data Bank under the code 2DGA.
Supplementary Material
Acknowledgments
We thank Dr. Atsushi Ishihara (Division of Applied Life Sciences, Kyoto University) for the mass analyses of the wheat glucosidases.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Masayuki Sue (sue@nodai.ac.jp).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.077693.
References
- Aguilar CF, Sanderson I, Moracci M, Ciaramella M, Nucci R, Rossi M, Pearl LH (1997) Crystal structure of the β-glycosidase from the hyperthermophilic archeon Sulfolobus solfataricus: resilience as a key factor in thermostability. J Mol Biol 271: 789–802 [DOI] [PubMed] [Google Scholar]
- Barrett T, Suresh CG, Tolley SP, Dodson EJ, Hughes MA (1995) The crystal structure of a cyanogenic β-glucosidase from white clover, a family 1 glycosyl hydrolase. Structure 3: 951–960 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Brzobohatý B, Moore I, Kristoffersen P, Bako L, Campos N, Schell J, Palme K (1993) Release of active cytokinin by a β-glucosidase localized to the maize root meristem. Science 262: 1051–1054 [DOI] [PubMed] [Google Scholar]
- Burmeister WP, Cottaz S, Driguez H, Iori R, Palmieri S, Henrissat B (1997) The crystal structures of Sinapis alba myrosinase and a covalent glycosyl-enzyme intermediate provide insights into the substrate recognition and active-site machinery of an S-glycosidase. Structure 5: 663–675 [DOI] [PubMed] [Google Scholar]
- Chi YI, Martinez-Cruz LA, Jancarik J, Swanson RV, Robertson DE, Kim SH (1999) Crystal structure of the β-glycosidase from the hyperthermophile Thermosphaera aggregans: insights into its activity and thermostability. FEBS Lett 445: 375–383 [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicek M, Esen A (1998) Structure and expression of a dhurrinase (β-glucosidase) from sorghum. Plant Physiol 116: 1469–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czjzek M, Cicek M, Zamboni V, Bevan DR, Henrissat B, Esen A (2000) The mechanism of substrate (aglycone) specificity in β-glucosidases is revealed by crystal structures of mutant maize β-glucosidase-DIMBOA, -DIMBOAGlc, and -dhurrin complexes. Proc Natl Acad Sci USA 97: 13555–13560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czjzek M, Cicek M, Zamboni V, Burmeister WP, Bevan DR, Henrissat B, Esen A (2001) Crystal structure of a monocotyledon (maize ZMGlu1) β-glucosidase and a model of its complex with p-nitrophenyl β-D-thioglucoside. Biochem J 354: 37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmawardhana DP, Ellis BE, Carlson JE (1995) A β-glucosidase from lodgepole pine xylem specific for the lignin precursor coniferin. Plant Physiol 107: 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esen A, Blanchard DJ (2000) A specific β-glucosidase-aggregating factor is responsible for the β-glucosidase null phenotype in maize. Plant Physiol 122: 563–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk A, Rask L (1995) Expression of a zeatin-O-glucoside-degrading β-glucosidase in Brassica napus. Plant Physiol 108: 1369–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gus-Mayer S, Brunner H, Schneider-Poetsch HA, Rudiger W (1994) Avenacosidase from oat: purification, sequence analysis and biochemical characterization of a new member of the BGA family of β-glucosidases. Plant Mol Biol 26: 909–921 [DOI] [PubMed] [Google Scholar]
- Haberer G, Kieber JJ (2002) Cytokinins. New insights into a classic phytohormone. Plant Physiol 128: 354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakulinen N, Paavilainen S, Korpela T, Rouvinen J (2000) The crystal structure of β-glucosidase from Bacillus circulans sp. alkalophilus: ability to form long polymeric assemblies. J Struct Biol 129: 69–79 [DOI] [PubMed] [Google Scholar]
- Hösel W, Tober I, Eklund SH, Conn EE (1987) Characterization of β-glucosidases with high specificity for the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) moench seedlings. Arch Biochem Biophys 252: 152–162 [DOI] [PubMed] [Google Scholar]
- Kim YW, Kang KS, Kim SY, Kim IS (2000) Formation of fibrillar multimers of oat β-glucosidase isoenzymes is mediated by the As-Glu1 monomer. J Mol Biol 303: 831–842 [DOI] [PubMed] [Google Scholar]
- Kim YW, Kim IS (1998) Subunit composition and oligomer stability of oat β-glucosidase isozymes. Biochim Biophys Acta 1388: 457–464 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Ljung K, Ostin A, Lioussanne L, Sandberg G (2001) Developmental regulation of indole-3-acetic acid turnover in Scots pine seedlings. Plant Physiol 125: 464–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A (2000) Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct 29: 291–325 [DOI] [PubMed] [Google Scholar]
- McRee DE (1992) A visual protein crystallographic software system for X11/Xview. J Mol Graph 10: 44–46 [Google Scholar]
- Niemeyer H (1988) Hydroxamic acids (4-hydroxy-1,4-benzoxazin-3-ones), defence chemicals in the Gramineae. Phytochemistry 11: 3349–3358 [Google Scholar]
- Nikus J, Esen A, Jonsson LMV (2003) Cloning of a plastidic rye (Secale cereale) β-glucosidase cDNA and its expression in Escherichia coli. Physiol Plant 118: 337–345 [Google Scholar]
- Nomura T, Ishihara A, Yanagita RC, Endo TR, Iwamura H (2005) Three genomes differentially contribute to the biosynthesis of benzoxazinones in hexaploid wheat. Proc Natl Acad Sci USA 102: 16490–16495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Persans MW, Wang J, Schuler MA (2001) Characterization of maize cytochrome P450 monooxygenases induced in response to safeners and bacterial pathogens. Plant Physiol 125: 1126–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234: 779–815 [DOI] [PubMed] [Google Scholar]
- Sanz-Aparicio J, Hermoso JA, Martinez-Ripoll M, Lequerica JL, Polaina J (1998) Crystal structure of β-glucosidase A from Bacillus polymyxa: insights into the catalytic activity in family 1 glycosyl hydrolases. J Mol Biol 275: 491–502 [DOI] [PubMed] [Google Scholar]
- Sicker D, Frey M, Schulz M, Gierl A (2000) Role of natural benzoxazinones in the survival strategy of plants. Int Rev Cytol 198: 319–346 [DOI] [PubMed] [Google Scholar]
- Stuber CW, Goodman MM, Johnson FM (1977) Genetic control and racial variation of β-glucosidase isozymes in maize (Zea mays L.). Biochem Genet 15: 383–394 [DOI] [PubMed] [Google Scholar]
- Sue M, Ishihara A, Iwamura H (2000. a) Purification and characterization of a β-glucosidase from rye (Secale cereale L.) seedlings. Plant Sci 155: 67–74 [DOI] [PubMed] [Google Scholar]
- Sue M, Ishihara A, Iwamura H (2000. b) Purification and characterization of a hydroxamic acid glucoside β-glucosidase from wheat (Triticum aestivum L.) seedlings. Planta 210: 432–438 [DOI] [PubMed] [Google Scholar]
- Sue M, Yamazaki K, Kouyama J-i, Sasaki Y, Ohsawa K, Miyamoto T, Iwamura H, Yajima S (2005) Purification, crystallization and preliminary X-ray analysis of a hexameric β-glucosidase from wheat. Acta Crystallogr Sect F Struct Biol Cryst Commun 61: 864–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Crystallogr 30: 1022–1025 [Google Scholar]
- Verdoucq L, Czjzek M, Moriniere J, Bevan DR, Esen A (2003) Mutational and structural analysis of aglycone specificity in maize and sorghum β-glucosidases. J Biol Chem 278: 25055–25062 [DOI] [PubMed] [Google Scholar]
- Verdoucq L, Moriniere J, Bevan DR, Esen A, Vasella A, Henrissat B, Czjzek M (2004) Structural determinants of substrate specificity in family 1 β-glucosidases: novel insights from the crystal structure of sorghum dhurrinase-1, a plant β-glucosidase with strict specificity, in complex with its natural substrate. J Biol Chem 279: 31796–31803 [DOI] [PubMed] [Google Scholar]
- Zagrobelny M, Bak S, Rasmussen AV, Jorgensen B, Naumann CM, Moller BL (2004) Cyanogenic glucosides and plant-insect interactions. Phytochemistry 65: 293–306 [DOI] [PubMed] [Google Scholar]
- Zouhar J, Vévodová J, Marek J, Damborský J, Su XD, Brzobahatý BE (2001) Insights into the functional architecture of the catalytic center of a maize β-glucosidase Zm-p60.1. Plant Physiol 127: 973–985 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.