Abstract
Upon localized attack by necrotizing pathogens, plants gradually develop increased resistance against subsequent infections at the whole-plant level, a phenomenon known as systemic acquired resistance (SAR). To identify genes involved in the establishment of SAR, we pursued a strategy that combined gene expression information from microarray data with pathological characterization of selected Arabidopsis (Arabidopsis thaliana) T-DNA insertion lines. A gene that is up-regulated in Arabidopsis leaves inoculated with avirulent or virulent strains of the bacterial pathogen Pseudomonas syringae pv maculicola (Psm) showed homology to flavin-dependent monooxygenases (FMO) and was designated as FMO1. An Arabidopsis knockout line of FMO1 proved to be fully impaired in the establishment of SAR triggered by avirulent (Psm avrRpm1) or virulent (Psm) bacteria. Loss of SAR in the fmo1 mutants was accompanied by the inability to initiate systemic accumulation of salicylic acid (SA) and systemic expression of diverse defense-related genes. In contrast, responses at the site of pathogen attack, including increases in the levels of the defense signals SA and jasmonic acid, camalexin accumulation, and expression of various defense genes, were induced in a similar manner in both fmo1 mutant and wild-type plants. Consistently, the fmo1 mutation did not significantly affect local disease resistance toward virulent or avirulent bacteria in naive plants. Induction of FMO1 expression at the site of pathogen inoculation is independent of SA signaling, but attenuated in the Arabidopsis eds1 and pad4 defense mutants. Importantly, FMO1 expression is also systemically induced upon localized P. syringae infection. This systemic up-regulation is missing in the SAR-defective SA pathway mutants sid2 and npr1, as well as in the defense mutant ndr1, indicating a close correlation between systemic FMO1 expression and SAR establishment. Our findings suggest that the presence of the FMO1 gene product in systemic tissue is critical for the development of SAR, possibly by synthesis of a metabolite required for the transduction or amplification of a signal during the early phases of SAR establishment in systemic leaves.
Plants generally possess multiple layers of defense to restrict the growth of potentially pathogenic microorganisms. Preformed mechanical or chemical barriers constitute an effective first line of defense against nonadapted or nonhost pathogens (Thordal-Christensen, 2003). Host pathogens that are able to overcome this first barrier provoke a whole set of inducible reactions. In specific or gene-for-gene resistance, plants rely on the presence of resistance gene products, which recognize matching avirulence factors from the pathogen to induce a multitude of protective responses (Dangl and Jones, 2001). Avirulent pathogens thus trigger rapid production of reactive oxygen species (ROS), accumulation of the defense signals salicylic acid (SA) and/or jasmonic acid (JA), increased expression of various defense-related genes, production of phytoalexins, and hypersensitive death of challenged cells (Kuć, 1995; Lamb and Dixon, 1997). Some of these responses also occur, albeit delayed, after infection with virulent pathogens, which manage to escape resistance protein recognition. Induced defenses thus limit the extent of pathogen spread not only in incompatible interactions to ensure specific resistance, but also in compatible interactions to centrally contribute to basal resistance (Parker et al., 1996).
Plant defense responses are initiated not only locally at the site of pathogen attack, but also in tissue distant from the site of infection (Cameron et al., 1994). These systemic resistance responses are generally subdivided into two broad categories, systemic acquired resistance (SAR) and induced systemic resistance. SAR develops in response to a pathogen that causes a necrotic lesion either as a consequence of a hypersensitive response (HR) or as a result of disease symptom development in the course of a compatible interaction (Hammerschmidt, 1999). Plants exhibiting SAR are generally resistant to a broad range of different pathogens. Establishment of SAR is dependent on the SA pathway and associated with both systemic increase of SA levels and systemic expression of pathogenesis-related (PR) genes (Ryals et al., 1996). By contrast, induced systemic resistance, a response to colonization of plant roots by certain rhizosphere bacteria, is dependent on JA and ethylene signaling (Pieterse et al., 2002).
The molecular mechanisms underlying SAR are under intensive study. The capability of plants to accumulate SA is known to be indispensable for SAR, as Arabidopsis (Arabidopsis thaliana) SA biosynthesis mutants SA induction deficient 1 and 2 (sid1 and sid2) and transgenic plants expressing the SA-degrading enzyme NahG are SAR defective (Gaffney et al., 1993; Nawrath and Métraux, 1999; Wildermuth et al., 2001; Nawrath et al., 2002). SA activates the SAR regulatory protein nonexpressor of PR genes (NPR1) through redox changes, which in turn drives systemic expression of antimicrobial PR proteins and facilitates their secretion by up-regulating protein secretory pathway genes (Mou et al., 2003; Wang et al., 2005). Several recent studies indicate that components of entirely distinct biochemical origin are necessary to realize SAR. Lipid metabolism turned out to play a central role in SAR signaling, as SAR is specifically compromised in Arabidopsis defective in induced resistance 1 (dir1) and suppressor of fatty acid desaturase deficiency 1 (sfd1), which bear mutations in a lipid transfer protein and a dihydroxyacetone phosphate reductase, respectively (Maldonado et al., 2002; Nandi et al., 2004). In addition, a peptide signal system mediated by the Asp protease constitutive disease resistance 1 (CDR1) appears to be essential for SAR long-distance signaling in Arabidopsis (Xia et al., 2004), and thiamine (vitamin B1) is capable of inducing SAR in a SA-dependent manner (Ahn et al., 2005). Moreover, ROS mediate a systemic signaling network that contributes to SAR (Alvarez et al., 1998). The complexity of systemic resistance regulation is further reflected by the fact that SAR establishment is subject to environmental and developmental plasticity. For instance, initiation of SAR has been demonstrated to occur in a light-dependent manner and the mechanisms of realizing SAR differ under variable light regimes (Zeier et al., 2004). Furthermore, leaf age influences the capability of initiating and executing SAR (Zeier, 2005).
Molecular events triggered by the primary infection process play a key role in SAR initiation. To identify uncharacterized genes involved in SAR establishment, we have selected Arabidopsis candidate genes up-regulated by SAR-inducing pathogens at the inoculation site, as indicated in microarray experiments publicly available from the Nottingham Arabidopsis Stock Centre (NASC) and from The Arabidopsis Information Resource (TAIR). T-DNA knockout lines corresponding to candidate genes were subsequently checked for an impaired SAR phenotype. This strategy revealed that the Arabidopsis flavin-dependent monooxygenase 1 (FMO1) gene is essential for the establishment of SAR and systemic defense responses provoked both by an avirulent (Psm avrRpm1) and a virulent (Psm) strain of Pseudomonas syringae. By contrast, FMO1 did not critically influence defense responses at the site of pathogen attack during these interactions.
RESULTS
FMO1 Is Expressed in Response to Virulent and Avirulent P. syringae
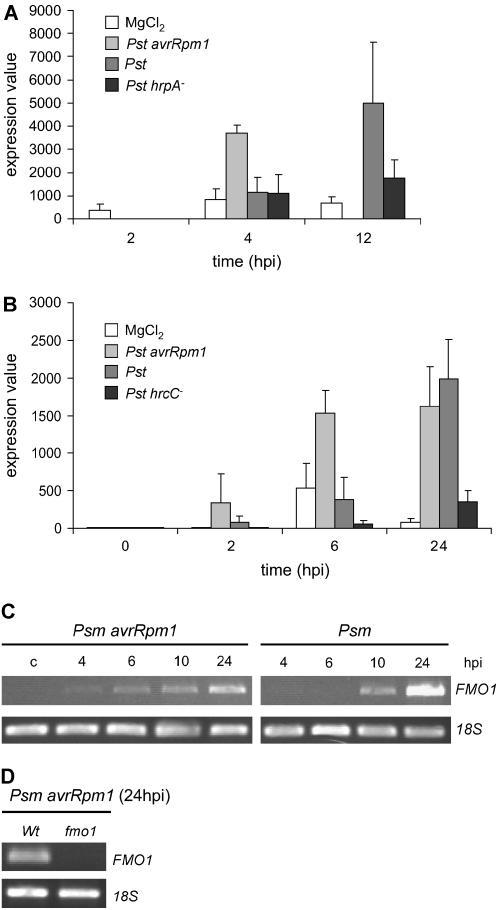
Gene expression profiling from two independent microarray datasets indicated that expression of the Arabidopsis FMO1 gene (At1g19250) is increased 12 h post leaf infection of Arabidopsis ecotype Columbia (Col-0) with virulent P. syringae pv tomato DC3000 (Pst; Fig. 1, A and B). Inoculation with the isogenic avirulent Pst avrRpm1 strain, which is recognized by Col-0 through the RPM1 resistance protein and consequently elicits an HR (Bisgrove et al., 1994), leads to earlier induction of FMO1 expression, starting from about 4 h postinfection (hpi; Fig. 1, A and B). Up-regulation of FMO1 by Pst is largely dependent on a functional bacterial type III secretion system because the type III secretion-defective Pst hrpA− or Pst hrcC− strains only weakly induce its expression (Fig. 1, A and B). To experimentally verify the microarray data, we inoculated Col-0 with Psm ES4326, another virulent pathogen that induces similar defense responses as Pst (Dong et al., 1991). Reverse transcription (RT)-PCR analysis revealed that FMO1 transcripts started to increase at about 6 hpi with avirulent Psm avrRpm1 and at 10 hpi with virulent Psm, confirming that FMO1 expression is triggered by host bacteria and that avirulent strains provoke an earlier transcription when compared with virulent strains (Fig. 1C).
Figure 1.
A and B, Expression levels of FMO1 (At1g19250) in Arabidopsis leaves challenged with Pst according to microarray analyses. Means (±sd) of Affymetrix expression values originating from three independent replicates are given. The data were normalized according to the Affymetrix MAS 5.0 scaling protocol. A, Expression data from the NASC array NASCARRAYS-59: impact of type III effectors on plant defense responses. B, Expression data from TAIR (TAIR-ME00331: response to virulent, avirulent, type III secretion system-deficient and nonhost bacteria). C, RT-PCR analysis of FMO1 expression triggered by Psm (virulent strain) and Psm avrRpm1 (avirulent strain). Numbers indicate hpi. Control leaves (c) were infiltrated with 10 mm MgCl2 for 24 h. 18S rRNA was amplified to standardize the transcript levels of each sample. D, Expression of FMO1 in leaves of wild-type and fmo1 mutant plants (T-DNA insertion line SALK_026163) 24 h after inoculation with Psm avrRpm1.
Experiments investigating the kinetics of SAR establishment in the Arabidopsis-Psm interaction indicated that pathogen-treated primary leaves start to initiate SAR at least 1 d postinoculation. Moreover, the avirulent strain activated SAR faster than the virulent strain (data not shown). This tendency correlated with the expression pattern of FMO1 in inoculated leaves (Fig. 1) and we thus postulated that FMO1 might play a role during SAR induction in primary leaves. A SALK insertion line (SALK_026163) harboring a T-DNA insertion in exon 4 of the FMO1 gene in the Col-0 background was obtained from the NASC to examine whether FMO1 contributes to SAR establishment. In contrast to Col-0 plants, fmo1 mutant plants failed to express FMO1 after inoculation with Psm (Fig. 1D), demonstrating the knockout of FMO1.
SAR Is Compromised in fmo1 Mutants
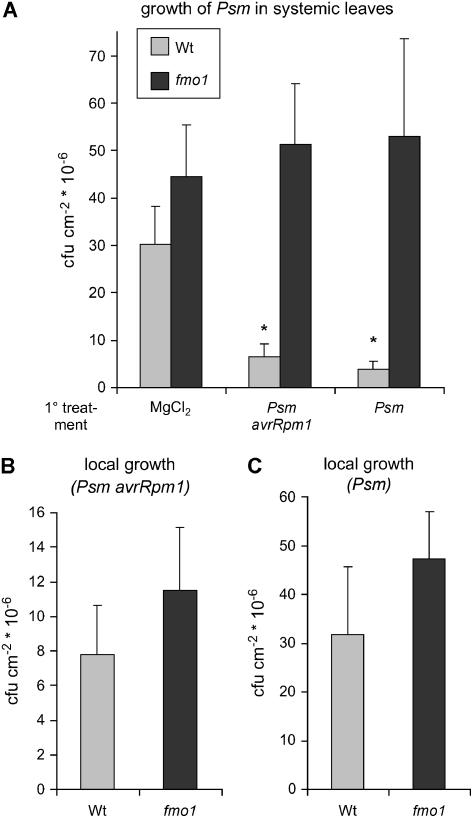
To investigate the biological induction of SAR, leaves of a given plant were treated with Psm avrRpm1 or Psm in a primary inoculation (designated as primary leaves) and 2 d later a secondary or challenge infection with virulent Psm was performed in rosette leaves located just above the primary leaves (systemic leaves). Bacterial growth was scored in systemic leaves 3 d later. After treatment of primary leaves with a control solution of MgCl2, growth of Psm during the challenge infection was vigorous in both wild-type and fmo1 mutant plants (Fig. 2A). When primary leaves of wild-type plants were preinoculated with Psm avrRpm1 or Psm, we observed a significant reduction of bacterial growth in the subsequent challenge infection in systemic leaves, demonstrating the establishment of SAR in both cases. In marked contrast, SAR did not develop in fmo1 mutant plants because growth of Psm in systemic leaves proved to be equally pronounced in plants pretreated in primary leaves with MgCl2, Psm avrRpm1, or Psm (Fig. 2A).
Figure 2.
A, Bacterial growth quantification of Psm in systemic leaves to assess SAR in wild-type and fmo1 mutant plants. Five-week-old Arabidopsis plants were pretreated with MgCl2, Psm avrRpm1, or Psm (OD = 0.02 for each pathogen) in three primary leaves (1° treatment), and 2 d later, three systemic leaves located directly above the primary leaves were inoculated with Psm (OD = 0.002). Bacterial growth in systemic leaves was assessed 3 d (3 dpi) after infection of systemic leaves. Bars represent mean values (±sd) of colony-forming units per square centimeter from seven parallel samples each consisting of three leaf discs. Asterisks denote pathogen treatments with statistically significant differences to the respective MgCl2 control (P < 0.001; Student's t test). Light bars, Wild-type plants; dark bars, fmo1 plants. B and C, Quantification of bacterial growth to assess local resistance. B, Growth of Psm avrRpm1 in leaves 3 d after inoculation with a bacterial suspension of OD = 0.005. C, Growth of Psm in leaves 3 d after inoculation (OD = 0.002). In both B and C, no statistical differences between the wild type and fmo1 existed (P > 0.05; Student's t test). In addition, to ensure the uniformity of the experiments, initial bacterial numbers (1 hpi) were quantified. No significant differences in bacterial numbers were detected at 1 hpi for comparable treatments in A, B, and C (data not shown).
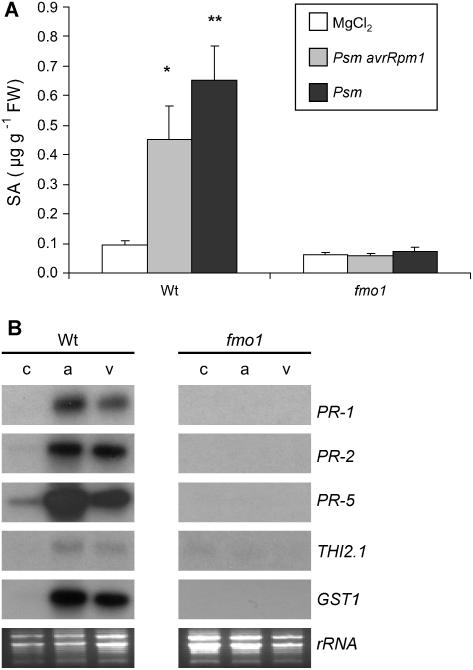
Systemic accumulation of SA and enhanced expression of defense genes in systemic leaves are characteristic features of SAR (Ryals et al., 1996). When primary leaves of wild-type plants were treated with Psm avrRpm1 or Psm, systemic leaves exhibited about 5- and 7-fold higher levels of free SA, respectively, compared to naive plants that were pretreated with MgCl2 solution only (Fig. 3A). Additionally, both avirulent and virulent bacteria triggered systemic expression of the SA-inducible defense gene PR-1 (Nawrath and Métraux, 1999), the jasmonate-dependent thionin gene THI2.1 (Epple et al., 1995), and the SA- and JA-independent defense genes PR-2 and PR-5 in the wild type (Fig. 3B). Moreover, the glutathione S-transferase gene GST1, a reliable marker for ROS production during plant-pathogen interactions (Levine et al., 1994; Alvarez et al., 1998), was systemically up-regulated in wild-type plants inoculated with Psm or Psm avrRpm1 (Fig. 3B). Unlike the wild type, fmo1 mutant plants exhibited neither elevated systemic SA levels after a local infection with Psm or Psm avrRpm1 nor increased systemic expression of any of the defense genes under examination (Fig. 3).
Figure 3.
Systemic defense responses in wild-type and fmo1 plants. Primary leaves of 5-week-old plants were treated as described in Figure 2A and untreated systemic leaves were harvested 2 d later for analysis. A, Systemic accumulation of SA. Bars represent mean values (±sd) of three independent samples. Each sample consisted of six leaves from two different plants. Asterisks denote pathogen treatments with statistically significant differences to the respective MgCl2 control (*, P < 0.02; **, P < 0.005; Student's t test). White bars, MgCl2 treatment; gray bars, Psm avrRpm1 inoculation; black bars, Psm inoculation. B, Systemic expression of defense-related genes assessed by northern-blot analysis (c, MgCl2 treatment; a, Psm avrRpm1 inoculation; v, Psm inoculation).
Local Resistance in fmo1 Mutants Is Similar to Wild-Type Plants
To examine whether the loss of SAR in fmo1 mutants was associated with compromised local disease resistance in the P. syringae-Arabidopsis interaction, we determined bacterial growth of Psm avrRpm1 and Psm in naive plants. Bacterial multiplication of both avirulent and virulent isolates was similar in wild-type and fmo1 mutants (Fig. 2, B and C). In some experiments, a slightly enhanced growth tendency of fmo1 mutant plants could be observed for Psm or Psm avrRpm1, but this tendency was not statistically significant. These results indicate that specific or basal disease resistance in the examined interactions is not compromised in fmo1.
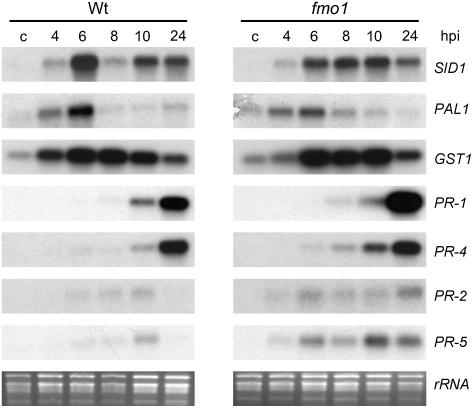
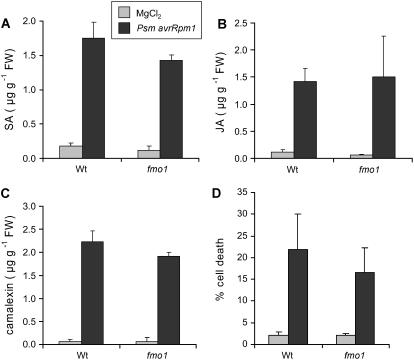
To further address this issue, we investigated typical defense responses that are induced by Psm avrRpm1 in Col-0 wild-type plants at inoculation sites. SA and JA are well-characterized signaling molecules accumulating during incompatible interactions. Up-regulation of the SA biosynthesis gene SID1 occurred in a similar manner in both wild-type and fmo1 plants starting 4 hpi (Fig. 5). Accordingly, local SA accumulation in fmo1 closely resembled SA elevation in wild-type plants at 10 hpi (Fig. 4A). Levels increased from about 0.15 μg g−1 fresh weight in control leaves to about 1.5 μg g−1 fresh weight in inoculated leaves. Likewise, Psm avrRpm1 induced accumulation of JA to a comparable extent in wild-type and fmo1 plants (Fig. 4B). Further downstream in these pathways, SA and JA trigger the expression of distinct sets of PR genes (Reymond and Farmer, 1998). Again, striking similarities were obvious in the expression patterns of the SA-inducible PR-1 gene and the JA-inducible PR-4 gene after Psm avrRpm1 inoculation (Fig. 5). Moreover, Psm avrRpm1-induced transcription of the SA- and JA-independent defense genes PR-2 and PR-5 was detected in both wild type and fmo1, yet to a somewhat higher extent in the mutant. These data indicate that, at the site of pathogen inoculation, FMO1 is neither required for the execution of SA- and JA-dependent defense pathways nor for the accomplishment of pathways independent of these defense signals.
Figure 5.
Local defense responses in wild-type and fmo1 plants. Expression of defense-related genes in leaves challenged with Psm avrRpm1 (OD = 0.005), as assessed by northern-blot analysis. Numbers indicate hpi. Control leaves (c) were treated with 10 mm MgCl2 (4 h).
Figure 4.
Local defense responses in wild-type and fmo1 plants. A to C, Accumulation of signaling and antimicrobial compounds in leaves challenged with Psm avrRpm1 (OD = 0.005). Control samples were treated with 10 mm MgCl2. All samples were collected 10 h post treatment. A, SA levels. B, JA content. C, Accumulation of the phytoalexin camalexin. Mean values (±sd) of three independent samples are given. No statistical differences between equally treated wild-type and fmo1 plants existed for each metabolite (P > 0.05; Student's t test). D, Quantification of microscopic HR lesions in leaves inoculated with Psm avrRpm1 that were stained with trypan blue 24 hpi. Bars represent mean values (±sd) of dead cells in infiltrated areas from at least seven independent leaf samples. Light bars, Areas infiltrated with 10 mm MgCl2; dark bars, Psm avrRpm1-infiltrated areas.
Increased production of secondary metabolites represents a further characteristic response to host pathogens. The indole derivative camalexin constitutes the major phytoalexin in Arabidopsis and accumulates in response to elicitor and pathogen treatment (Tsuji et al., 1992; Zhou et al., 1998). Camalexin was essentially absent in noninoculated leaves, but accumulated substantially in Psm avrRpm1-treated leaves already at 10 hpi (Fig. 4C). Again, no significant difference between wild type and fmo1 existed. Moreover, expression of Phe ammonia lyase (PAL), the key enzyme of phenylpropanoid biosynthesis, is up-regulated upon infection with avirulent Pseudomonas (Zeier et al., 2004). PAL1 transcripts were elevated at 4 to 6 hpi in both wild-type and fmo1 leaves, indicating that the phenylpropanoid pathway is initiated independently from FMO1.
The oxidative burst at the site of pathogen ingress and the subsequent hypersensitive cell death response represent hallmarks of incompatible plant-pathogen interactions (Lamb and Dixon, 1997). During the oxidative burst, ROS are produced that contribute to triggering the HR in infected cells and driving expression of protective genes in neighboring tissue (Levine et al., 1994). The expression of GST1 is triggered by ROS produced during the oxidative burst (Alvarez et al., 1998; Zeier et al., 2004). Enhanced GST1 expression was observed from 4 to 10 h after Psm avrRpm1 inoculation in wild-type leaves and a similar pattern was evident in fmo1 mutant leaves. FMO1 has recently been described as a marker gene for cell death pathways in plants (Olszak et al., 2006). To investigate whether FMO1 contributes to hypersensitive cell death lesion formation upon infection with Psm avrRpm1, we performed trypan blue-staining experiments with inoculated leaves (Zeier et al., 2004). At 24 hpi, wild-type plants exhibited a considerable amount of stained cells inside the pathogen-treated leaf area (Fig. 4D) and similar staining patterns were observed in inoculated fmo1 mutant leaves. Thus, FMO1 does not play a critical role in the regulation of the oxidative burst or the hypersensitive cell death response at the site of pathogen attack.
Local and Systemic Expression of FMO1 in Arabidopsis Defense Mutants
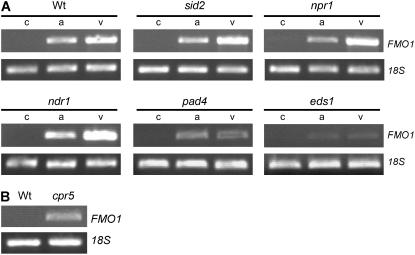
The SA-signaling pathway is essential for the full establishment of local and systemic disease resistance in the Arabidopsis-P. syringae interaction (Nawrath and Métraux, 1999). To examine whether expression of FMO1 is dependent on SA signaling and SA-related defense pathways, we checked the pathogen-induced up-regulation of FMO1 in different Arabidopsis defense mutants (Fig. 6). Psm avrRpm1 or Psm induced FMO1 expression to a similar extent in the SA biosynthesis mutant sid2, in the SA-insensitive mutant npr1, and in wild-type plants at the site of inoculation (Fig. 6A), demonstrating that local FMO1 expression does not require SA signaling. Additionally, FMO1 was strongly expressed in the no disease resistance 1 (ndr1) defense mutant (Century et al., 1995) upon P. syringae infection. In the defense-signaling mutants phytoalexin-deficient 4 (pad4; Zhou et al., 1998) and enhanced disease susceptibility 1 (eds1; Parker et al., 1996), pathogen-induced FMO1 expression was clearly attenuated or entirely suppressed, respectively. Moreover, the constitutive expression of PR 5 (cpr5) mutant that constitutively exhibits resistance in both an npr1-dependent and -independent manner (Bowling et al., 1997) exhibited marked constitutive FMO1 expression (Fig. 6B).
Figure 6.
A, Expression of FMO1 at the site of pathogen inoculation in wild-type plants and Arabidopsis defense mutants (24 hpi) as assessed by RT-PCR analysis (c, MgCl2 treatment; a, Psm avrRpm1 inoculation; v, Psm inoculation; OD = 0.005 for each pathogen). B, Levels of FMO1 transcripts in untreated leaves of wild-type and cpr5 mutant plants.
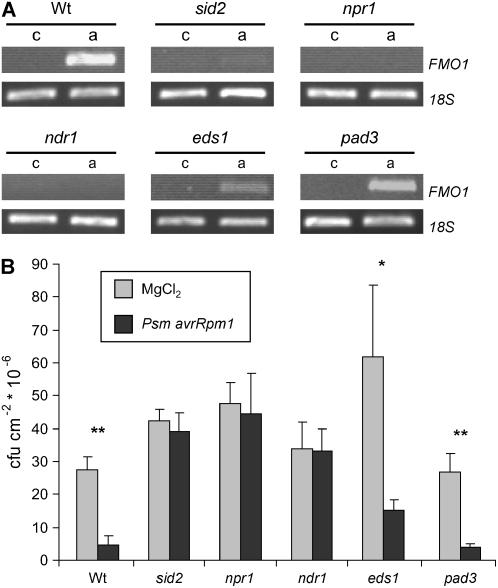
To further analyze the function of FMO1 during SAR, we tested whether FMO1 is systemically expressed upon local Psm avrRpm1 inoculation (Fig. 7). The pathogen-induced SAR response was associated with an up-regulation of FMO1 in systemic leaves of wild-type plants (Fig. 7, A and B). In SA-signaling mutants sid2 and npr1, however, SAR was fully compromised and FMO1 failed to be expressed systemically. The ndr1 mutant constitutes a further SAR-deficient mutant, and systemic expression of FMO1 was not enhanced upon Psm avrRpm1 infection. The eds1 mutation, by contrast, did not abolish Psm avrRpm1-triggered SAR, and systemic FMO1 up-regulation still took place in eds1, albeit to a lesser extent than in the wild type. Moreover, the camalexin-deficient mutant pad3 exhibited a wild-type-like SAR response and showed a systemic FMO1 expression pattern similar to the wild type. Thus, establishment of SAR closely correlated with systemic elevation of FMO1 transcript levels in the lines under investigation and, in contrast to the expression characteristics at the site of pathogen attack (Fig. 6A), systemic FMO1 expression was dependent on an intact SA-signaling pathway.
Figure 7.
Correlation of systemic FMO1 expression and SAR establishment. A, Systemic expression of FMO1 in wild-type plants and Arabidopsis defense mutants in response to Psm avrRpm1 as assessed by RT-PCR analysis (c, MgCl2 treatment; a, Psm avrRpm1 inoculation; OD = 0.02). For further details, see legend to Fig. 3. B, Growth quantification of Psm in systemic leaves (3 dpi) to assess SAR induced by Psm avrRpm1 in wild-type and Arabidopsis defense mutants. For further details, see legend to Fig. 2A. Asterisks denote lines with statistically significant differences between plants pretreated with MgCl2 and Psm avrRpm1 (*, P < 0.01; **, P < 0.001; Student's t test).
DISCUSSION
SAR can be activated in many plant species by necrotizing pathogens and, once established, it confers long-lasting resistance toward a broad spectrum of different pathogens (Durrant and Dong, 2004). SAR turns out to be under complex molecular regulation because several components of entirely distinct biochemical origin are necessary for its induction in Arabidopsis (Nawrath and Métraux, 1999; Maldonado et al., 2002; Nandi et al., 2004; Xia et al., 2004; Wang et al., 2005). We show here that FMO1, whose expression is induced by virulent and avirulent strains of P. syringae both at the site of pathogen ingress and in systemic tissue, constitutes a further component essential for the successful activation of SAR in Arabidopsis because fmo1 knockout mutants proved to be totally compromised in the activation of systemic defense responses and the establishment of SAR (Figs. 2A and 3).
Currently, a central role for FMO1 in plant disease resistance is emerging. In fact, the FMO1 gene has recently been recognized by distinct approaches to be involved in plant defense (Bartsch et al., 2006; Koch et al., 2006; Olszak et al., 2006). FMO1 was demonstrated to be up-regulated in Arabidopsis acd11, a mutant exhibiting constitutively activated SA-, PAD4-, and EDS1-dependent defenses and spontaneous HR lesions (Brodersen et al., 2002; Olszak et al., 2006). In addition, it was shown that FMO1 expression is enhanced in the runaway cell death lesion-simulating disease 1 (lsd1) mutant (Dietrich et al., 1997), but not in the constitutive defense-signaling mutants ctr1, cev1, mpk4, and cpr6 that do not develop spontaneous cell death (Olszak et al., 2006). Thus, FMO1 was suggested as a marker gene for certain forms of defense and cell death. In a screen for genes whose expression depends on EDS1 and PAD4, Bartsch et al. (2006) showed the requirement of functional FMO1 in the execution of basal resistance against a virulent isolate of the oomycete pathogen Hyaloperonospora parasitica and of specific resistance against H. parasitaca isolate Noco2 or P. syringae carrying the avrRps4 avirulence gene. Moreover, an activation-tagging approach identified an Arabidopsis line constitutively overexpressing FMO1, which is characterized by enhanced disease resistance against P. syringae (Koch et al., 2006).
Basal resistance is triggered by a multitude of relatively unspecific pathogen-associated molecular patterns to limit the growth of virulent pathogens to a certain extent (Nürnberger and Lipka, 2005). By contrast, resistance gene-mediated resistance is based on specific recognition events and two subclasses of resistance proteins with distinct signaling requirements are generally distinguished, depending on the presence of either an N-terminal coiled-coil domain (CC-NB-LRR) or a domain with similarity to the Drosophila Toll and mammalian interleukin-1 receptors (TIR-NB-LRR; Aarts et al., 1998). We have demonstrated that, at the site of pathogen inoculation, basal resistance against virulent Psm and specific resistance against Psm avrRpm1 are not compromised in fmo1 mutants (Fig. 2, B and C). Moreover, various characteristic defense responses locally triggered by Psm avrRpm1, including oxidative burst, accumulation of SA, JA, and camalexin, and expression of defense genes as well as the hypersensitive cell death response, are not significantly altered in fmo1 (Figs. 4 and 5). In line with these findings, Bartsch et al. (2006) showed that resistance to Pst avrRpm1 is not affected in fmo1 mutants, using the same T-DNA insertion line (SALK_026163). Because the AvrRpm1 avirulence protein is recognized by RPM1, a CC-NB-LRR-type resistance protein, we conclude that basal resistance to P. syringae and specific resistance mediated by CC-NB-LRR receptors are largely FMO1 independent. By contrast, resistance to Pst avrRps4 has been reported to be attenuated in fmo1 and therefore FMO1 is required for specific resistance against P. syringae mediated by TIR-NB-LRR resistance proteins (Bartsch et al., 2006). The contribution of FMO1 to basal resistance against H. parasitica, specific resistance against Pst avrRps4, and SAR triggered by Psm reveals that overlapping molecular principles exist in distinct kinds of resistance within different pathosystems.
Our finding that FMO1 represents a critical component of SAR in Arabidopsis is further underlined by recent work demonstrating that constitutive overexpression of FMO1 leads to enhanced disease resistance toward P. syringae (Koch et al., 2006). FMO1 might function in the induction of SAR in inoculated tissue, in the propagation of a mobile signal to distant tissue, in the perception of this long-distance signal in systemic tissue, or in the potentiation of defense responses in systemic tissue. Induced expression of FMO1 in inoculated leaves is attenuated in eds1 and pad4 mutants, confirming that FMO1 contributes to the EDS1/PAD4 pathway in local defense signaling (Bartsch et al., 2006). In contrast, FMO1 expression is not affected in sid2, npr1, and ndr1 mutants, demonstrating that local FMO1 induction is independent of the SA-signaling pathway and NDR1-mediated signaling (Fig. 6). However, in contrast to the wild type, these three mutants fail to express FMO1 systemically (Fig. 7A), and this is associated with a loss of SAR (Fig. 7B). In the eds1 mutant, pathogen-induced FMO1 expression is abolished at the site of inoculation, yet still observable in systemic tissue, and a significant SAR response is established in eds1. Thus, the failure to systemically rather than locally up-regulate FMO1 transcription correlates with the development of SAR in all investigated lines. Additionally, fmo1 mutants, despite exhibiting unaltered local defenses, are totally compromised in any of the examined systemic responses (Fig. 3). These include systemic SA accumulation, systemic expression of SA-dependent and -independent PR genes, as well as up-regulation of GST1, a reliable marker for ROS generation (Levine et al., 1994). Moreover, FMO1 transcripts are up-regulated in the absence of a pathogen in defense of the mutant cpr5, which exhibits constitutive disease resistance (Fig. 6B; Bowling et al., 1997). Taking these findings together, we propose a model in which the presence of FMO1 in systemic tissue is critical for the realization of SAR. A metabolite generated by FMO1 might be necessary during the early phase of SAR establishment in systemic leaves, presumably for the transduction or amplification of a long-distance signal originating from primary leaves. Feedback loops, including SA and ROS, exist to amplify plant defense responses (Shirasu et al., 1997) and oxidative microbursts in systemic tissue have been shown to mediate a reiterative signal network during SAR (Alvarez et al., 1998). Moreover, superoxide has been demonstrated to induce FMO1 expression (Olszak et al., 2006). We thus propose that FMO1 contributes to a signal amplification loop involving ROS, SA, NPR1, and NDR1 that is required to potentiate SAR responses in systemic tissue.
Although SA represents a central and necessary signaling component for the establishment of SAR, there are controversial data as to whether it functions as a mobile signal that moves from infected leaves to systemic tissue. 18O2 feeding experiments in tobacco mosaic virus-infected tobacco (Nicotiana tabacum) demonstrate that about 60% to 70% of the SA detected in systemic leaves originates from inoculated tissue, with the remainder resulting from de novo synthesis (Shulaev et al., 1995). Similarly, 14C-labeling experiments in cucumber (Cucumis sativus) plants inoculated with tobacco necrosis virus showed that SA accumulation in systemic leaves results both from transport and from de novo synthesis (Mölders et al., 1996). Although SA transport from inoculated to systemic tissue is feasible in these species, SA does not necessarily represent the SAR long-distance signal. In cucumber, removal of pathogen-treated leaves led to systemic resistance induction before a rise in SA levels was detectable in petiole exudates of inoculated leaves (Rasmussen et al., 1991). Moreover, grafting experiments using transgenic tobacco expressing the salicylate hydroxylase NahG indicate that SA is not the long-distance signal during SAR, but it is required for signal transduction in systemic tissue (Vernooij et al., 1994). Considering Arabidopsis, Kiefer and Slusarenko (2003), by applying 14C-SA to rosette leaves, have demonstrated that exogenous SA is able to move from source to sink tissue. On the other hand, we have shown here that Pseudomonas-infected fmo1 mutant plants locally accumulate wild-type levels of SA, whereas no SA accumulation occurs systemically (Figs. 3A and 4A). A similar trend is observed in the SAR-defective mutants ndr1 and npr1 (Fig. 7B; T.E. Mishina and J. Zeier, unpublished data). This indicates that systemic accumulation of SA that is normally observed during biologically induced SAR in Arabidopsis is not due to transport of SA produced at the site of infection, but is largely caused by de novo synthesis in systemic tissue in which the above proposed feedback loop, including FMO1 and SA, might operate.
Mammalian FMO either contribute to oxidative xenobiotic metabolism or catalyze the oxygenation of endogenous metabolites, i.e. biogenic amines (Krueger and Williams, 2005). Besides FMO1, the only plant FMO genes characterized so far represent Arabidopsis YUCCA and its petunia (Petunia hybrida) ortholog FLOOZY, which are involved in auxin biosynthesis (Zhao et al., 2001; Tobena-Santamaria et al., 2002). YUCCA has been demonstrated to catalyze the hydroxylation of the amino group in tryptamine. A challenging future task represents the identification of the putative metabolite generated by FMO1 and the clarification of its role in disease resistance.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) L. Heynh. plants were grown in a mixture of soil (Fruhstorfer Pflanzenerde), vermiculite, and sand (9:1:1) in a controlled environment chamber (J-66LQ4; Percival) with a 9 h day (photon flux density 70 μmol m−2 s−1)/15 h night cycle and 70% relative humidity. Growth temperatures were set to 22°C during the day and 18°C during the night.
Arabidopsis ecotype Col-0 was used as the wild type in all experiments. The fmo1 line represents the Salk T-DNA insertion line SALK_026163 in the Col-0 background. Homozygous insertion mutants were identified by PCR, using a gene-specific and a T-DNA-specific primer (Alonso et al., 2003), and used for experiments. Further, the following Arabidopsis defense mutants were used in this study: sid2-1 (Nawrath and Métraux, 1999), npr1-2 (NASC ID no., N3801), ndr1 (Century et al., 1995), pad3-1 (Glazebrook and Ausubel, 1994), pad4-1 (Glazebrook et al., 1997), eds1-2 (Aarts et al., 1998), and cpr5-2 (Bowling et al., 1997).
Growth of Plant Pathogens and Infection
Pseudomonas syringae pv maculicola ES4326 lacking (Psm) or carrying (Psm avrRpm1) the avrRpm1 avirulence gene were grown at 28°C in King's B medium containing the appropriate antibiotics (Zeier et al., 2004). Overnight log phase cultures were washed three times with 10 mm MgCl2 and diluted to a final optical density (OD) concentration of 0.02, 0.005, or 0.002. The bacterial suspensions were infiltrated from the abaxial side into a sample leaf using a 1-mL syringe without a needle. Control inoculations were performed with 10 mm MgCl2. Bacterial growth was assessed by homogenizing discs originating from infiltrated areas of three different leaves in 1 mL 10 mm MgCl2, plating appropriate dilutions on King's B medium, and counting colony numbers after incubating the plates at 28°C for 2 d.
All pathogen experiments depicted in the figures were repeated at least twice with similar results.
Characterization of Systemic Resistance Responses
Plants were first infiltrated into three lower leaves with a suspension of Psm or Psm avrRpm1 (OD = 0.02), or with 10 mm MgCl2 as a control. Two days after the primary inoculation, nontreated upper leaves were harvested for SA determination and gene expression analysis or plants were inoculated on three upper leaves with virulent Psm (OD = 0.002). Growth of Psm in upper leaves was scored 3 d later.
Quantification of Microscopic HR Lesions
The extent of microscopic HR lesion formation was assessed by trypan blue staining, light microscopy, and quantification of stained cells as described by Zeier et al. (2004).
Northern-Blot Analysis
Total RNA was isolated from frozen leaves using peqGOLD RNAPure reagent (peqLab) following the manufacturer's instructions. For each sample, two leaves from different plants of the same treatment were used. One microgram of total RNA was loaded on formaldehyde-agarose gels, separated by electrophoresis, and blotted on nylon membranes (Hybond-N; Amersham). RNA-blot hybridization was performed with specific 32P-labeled DNA probes generated by PCR using appropriate oligonucleotide primers. The probes represented the following Arabidopsis genes: SID1 (Arabidopsis annotation At4g39030), PAL1 (At2g37040), GST1 (At1g02930), PR-1 (At2g14610), PR-2 (At3g57260), PR-4 (At3g04720), PR-5 (At1g75040), and THI2.1 (At1g72260).
RT-PCR Analysis
One microgram of extracted RNA was treated with DNase I (Fermentas) for 30 min at 37°C to remove genomic DNA, the DNase inactivated by incubation at 70°C for 5 min in the presence of 2.5 mm EDTA, and cDNA synthesized in a final reaction volume of 20 μL at 42°C for 1 h using random primer mix, ribonuclease inhibitor (RNaseOUT; Invitrogen), and reverse transcriptase (SuperScript II; Invitrogen). After another enzyme inactivation step for 15 min at 70°C, the cDNA mixture was diluted in water (1:10) and 3 to 10 μL of the final dilution used in a 30-μL RT-PCR reaction (3 μL for 18S rRNA, 10 μL for FMO1). The following primers were used for the amplification of cDNA derived from 18S rRNA and FMO1 mRNA, respectively: 5′-AAACGGCTACCACATCCAAG-3′ (18S-forward), 5′-ACCCATCCCAAGGTTCAACT-3′ (18S-reverse), 5′-CTTCTACTCTCCTCAGTGGCAAA-3′ (FMO1-forward), and 5′-CTAATGTCGT-CCCATCTTCAAAC-3′ (FMO1-reverse). The PCR reaction was performed as follows: 95°C for 10 min, 25 (18S) or 30 (FMO1) cycles of 92°C for 60 s, 60°C for 90 s, 72°C for 90 s, and a final extension step at 72°C for 5 min. Ten microliters of each PCR reaction were visualized by agarose gel electrophoresis with ethidium bromide staining.
Gas Chromatographic Determination of SA, JA, and Camalexin
The determination of SA, JA, and camalexin levels in leaves was performed by a modified vapor-phase extraction method (Schmelz et al., 2004). Briefly, 150 mg of frozen leaf tissue were homogenized with 600 μL of extraction buffer (water:1-propanole:HCl = 1:2:0.005). After addition of internal standards (D4-SA, dihydrojasmonic acid, and indolepropionic acid; 100 ng each) and 1 mL of methylene chloride, the mixture was shaken thoroughly and centrifuged at 14,000 rpm for phase separation. The lower, organic phase was then removed, dried over Na2SO4, and treated with 2 μL of 2 m trimethylsilyldiazomethane in hexane (Sigma-Aldrich) for 5 min at room temperature to convert carboxylic acids into their corresponding methyl esters. After stopping the methylation reaction with 2 m acetic acid in hexane, the sample was subjected to a vapor-phase extraction procedure using a volatile collector trap packed with Super-Q absorbent (VCT-1/4X3-SPQ; Analytical Research Systems). The final evaporation temperature was set to 200°C, and samples were eluted from the collector trap with 1 mL methylene chloride. Finally, the sample volume was reduced to 50 μL in a stream of nitrogen, and the sample was subjected to gas chromatography-mass spectrometry analysis. The sample mixture (2 μL) was separated on a gas chromatograph (GC 6890 N; Agilent Technologies) equipped with a fused silica capillary column (DB-1; Fisons), and combined with a 5975 mass spectrometric detector (Agilent Technologies). For quantitative determination of metabolites, peaks originating from selected ion chromatograms were integrated. The area of a substance peak was related to the peak area of the corresponding internal standard (SA/D4-SA; JA/dihydrojasmonic acid, camalexin/indolepropionic acid). Experimentally determined correction factors for each substance/standard pair were considered.
Acknowledgments
We thank Volker Lipka and Juriaan Ton for the donation of Arabidopsis mutant seeds, and Michael Rostás, Markus Riederer, and Volker Lipka for proofreading the manuscript. We are grateful to M. de Torres Zabala, M. Grant, B. Kemmerling, and T. Nürnberger for publicly sharing their microarray data.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 567).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jürgen Zeier (zeier@botanik.uni-wuerzburg.de).
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.081257.
References
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signalling pathways in Arabidopsis. Proc Natl Acad Sci USA 95: 10306–10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn I-P, Kim S, Lee Y-H (2005) Vitamin B1 functions as an activator of plant disease resistance. Plant Physiol 138: 1505–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Alvarez ME, Pennell RI, Meijer P-J, Ishikawa A, Dixon RA, Lamb C (1998) Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92: 773–784 [DOI] [PubMed] [Google Scholar]
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE (2006) Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the nudix hydrolase NUDT7. Plant Cell 18: 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove SR, Simonich MT, Smith NM, Sattler A, Innes RW (1994) A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell 6: 927–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Petersen M, Pike HN, Olszak B, Skov-Petersen S, Oedum N, Joergensen LB, Brown RE, Mundy J (2002) Knockout of Arabidopsis ACCELERATED-CELL-DEATH11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev 16: 490–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron RK, Dixon RA, Lamb CJ (1994) Biologically induced systemic acquired resistance in Arabidopsis thaliana. Plant J 5: 715–725 [Google Scholar]
- Century KS, Holub EB, Staskawicz BJ (1995) NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc Natl Acad Sci USA 92: 6597–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Dietrich RA, Richberg MH, Schmidt R, Dean C, Dangl JL (1997) A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 88: 685–694 [DOI] [PubMed] [Google Scholar]
- Dong X, Mindrinos M, Davis KR, Ausubel FM (1991) Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell 3: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42: 185–209 [DOI] [PubMed] [Google Scholar]
- Epple P, Apel K, Bohlmann H (1995) An Arabidopsis thaliana thionin gene is inducible via a signal transduction pathway different from that for pathogenesis-related proteins. Plant Physiol 109: 813–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754–756 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Ausubel FM (1994) Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc Natl Acad Sci USA 91: 8955–8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J, Zook M, Mert F, Kagan I, Rogers EE, Crute IR, Holub EB, Hammerschmidt R, Ausubel FM (1997) Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146: 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt R (1999) Induced disease resistance: how do induced plants stop pathogens? Physiol Mol Plant Pathol 55: 77–84 [Google Scholar]
- Kiefer IW, Slusarenko AJ (2003) The pattern of systemic acquired resistance induction within the Arabidopsis rosette in relation to the pattern of translocation. Plant Physiol 132: 840–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M, Vorwerk S, Masur C, Sharifi-Sirchi G, Olivieri N, Schlaich N (2006) A role for a flavin-containing monooxygenase in pathogen resistance in Arabidopsis. Plant J (in press) [DOI] [PubMed]
- Krueger SK, Williams DE (2005) Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol Ther 106: 357–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuć J (1995) Phytoalexins, stress metabolism, and disease resistance in plants. Annu Rev Phytopathol 33: 275–297 [DOI] [PubMed] [Google Scholar]
- Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon RA, Lamb C (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive response. Cell 79: 583–593 [DOI] [PubMed] [Google Scholar]
- Maldonado AM, Doerner P, Dixon RA, Lamb C, Cameron RC (2002) A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419: 399–403 [DOI] [PubMed] [Google Scholar]
- Mölders W, Buchala A, Métraux J-P (1996) Transport of salicylic acid in tobacco necrosis virus-infected cucumber plants. Plant Physiol 112: 787–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–944 [DOI] [PubMed] [Google Scholar]
- Nandi A, Welti R, Shah J (2004) The Arabidopsis thaliana dihydroxyacetone phosphate reductase gene SUPPRESSOR OF FATTY ACID DESATURASE DEFICIENCY 1 is required for glycerolipid metabolism and for the activation of systemic acquired resistance. Plant Cell 16: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Heck S, Parinthawong N, Métraux JP (2002) EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE-transporter family. Plant Cell 14: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Métraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger T, Lipka V (2005) Non-host resistance in plants: new insights into an old phenomenon. Mol Plant Pathol 6: 335–345 [DOI] [PubMed] [Google Scholar]
- Olszak B, Malinovsky FG, Brodersen P, Grell M, Giese H, Petersen M, Mundy J (2006) A putative flavin-containing mono-oxygenase as a marker for certain defense and cell death pathways. Plant Sci 170: 614–623 [Google Scholar]
- Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8: 2033–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, Van Wees SCM, Ton J, Van Pelt JA, Van Loon LC (2002) Signalling in rhizobacteria-induced systemic resistance in Arabidopsis thaliana. Plant Biol 4: 535–544 [Google Scholar]
- Rasmussen JB, Hammerschmidt R, Zook MN (1991) Systemic induction of salicylic acid accumulation in cucumber after inoculation with Pseudomonas syringae pv syringae. Plant Physiol 97: 1342–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Farmer EE (1998) Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol 1: 404–411 [DOI] [PubMed] [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8: 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Engelberth J, Tumlinson JH, Block A, Alborn HT (2004) The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J 39: 790–808 [DOI] [PubMed] [Google Scholar]
- Shirasu K, Nakajima H, Rajasekhar VK, Dixon RA, Lamb C (1997) Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defence mechanisms. Plant Cell 9: 261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V, Leon J, Raskin I (1995) Is salicylic acid a translocated signal of systemic acquired resistance in tobacco? Plant Cell 7: 1691–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H (2003) Fresh insights into processes of nonhost resistance. Curr Opin Plant Biol 6: 351–357 [DOI] [PubMed] [Google Scholar]
- Tobena-Santamaria R, Bliek M, Ljung K, Sandberg G, Mol JNM, Souer E, Koes R (2002) FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev 16: 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji J, Jackson EP, Gage DA, Hammerschmidt R, Somerville SC (1992) Phytoalexin accumulation in Arabidopsis thaliana during the hypersensitive reaction to Pseudomonas syringae pv syringae. Plant Physiol 98: 1304–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernooij B, Friedrich L, Morse A, Reist R, Kolditzjawhar R, Ward E, Uknes S, Kessmann H, Ryals J (1994) Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell 6: 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, Dong X (2005) Induction of protein secretory pathway is required for systemic acquired resistance. Science 308: 1036–1040 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Xia Y, Suzuki H, Borevitz J, Blount J, Guo Z, Patel K, Dixon RA, Lamb C (2004) An extracellular aspartic protease functions in Arabidopsis disease resistance signalling. EMBO J 23: 980–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeier J (2005) Age-dependent variations of local and systemic defense responses in Arabidopsis leaves towards an avirulent strain of Pseudomonas syringae. Physiol Mol Plant Pathol 66: 30–39 [Google Scholar]
- Zeier J, Pink B, Mueller MJ, Berger S (2004) Light conditions influence specific defence responses in incompatible plant-pathogen interactions: uncoupling systemic resistance from salicylic acid and PR-1 accumulation. Planta 219: 673–683 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J (1998) PAD4 functions upstream from salicylic acid to control defence responses in Arabidopsis. Plant Cell 10: 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]