Abstract
Pteris vittata sporophytes hyperaccumulate arsenic to 1% to 2% of their dry weight. Like the sporophyte, the gametophyte was found to reduce arsenate [As(V)] to arsenite [As(III)] and store arsenic as free As(III). Here, we report the isolation of an arsenate reductase gene (PvACR2) from gametophytes that can suppress the arsenate sensitivity and arsenic hyperaccumulation phenotypes of yeast (Saccharomyces cerevisiae) lacking the arsenate reductase gene ScACR2. Recombinant PvACR2 protein has in vitro arsenate reductase activity similar to ScACR2. While PvACR2 and ScACR2 have sequence similarities to the CDC25 protein tyrosine phosphatases, they lack phosphatase activity. In contrast, Arath;CDC25, an Arabidopsis (Arabidopsis thaliana) homolog of PvACR2 was found to have both arsenate reductase and phosphatase activities. To our knowledge, PvACR2 is the first reported plant arsenate reductase that lacks phosphatase activity. CDC25 protein tyrosine phosphatases and arsenate reductases have a conserved HCX5R motif that defines the active site. PvACR2 is unique in that the arginine of this motif, previously shown to be essential for phosphatase and reductase activity, is replaced with a serine. Steady-state levels of PvACR2 expression in gametophytes were found to be similar in the absence and presence of arsenate, while total arsenate reductase activity in P. vittata gametophytes was found to be constitutive and unaffected by arsenate, consistent with other known metal hyperaccumulation mechanisms in plants. The unusual active site of PvACR2 and the arsenate reductase activities of cell-free extracts correlate with the ability of P. vittata to hyperaccumulate arsenite, suggesting that PvACR2 may play an important role in this process.
Arsenic is a naturally occurring, metalloid element that is potentially toxic to most organisms. Arsenic is a known human carcinogen (Hughes, 2002; Shi et al., 2004), and while there is great interest in the study of arsenic uptake and accumulation by plants for remediating arsenic polluted soils and waters (Salt et al., 1998), the metabolism of arsenic in plants is poorly understood. This process is especially important to human and environmental toxicology because plants, including food crops such as rice (Oryza sativa; Meharg, 2004; Laparra et al., 2005), are known to convert the arsenate that is taken up by the plant to the more toxic arsenite (for review, see Carter et al., 2003). In addition to environmental concerns, arsenic has important medicinal value. Arsenic trioxide has recently emerged as the chemotherapeutic agent of choice for the treatment of acute promyelocytic leukemia (Sanz et al., 2005) and also holds promise for the treatment of ovarian cancer (Kong et al., 2005). Whether arsenic is considered a detriment or a cure, understanding the mechanisms responsible for arsenic metabolism, toxicity, and resistance or tolerance can be of benefit for the prevention as well as effective treatment of human diseases.
The genus Pteris (Pteridaceae) is remarkable in that it has several species, including Pteris vittata, that hyperaccumulate arsenic (Visoottiviseth et al., 2002; Meharg, 2003; Raab et al., 2004). When grown in areas with elevated levels of arsenic, more than 1% of the dry weight of a P. vittata frond is arsenic (Tongbin et al., 2002; Wang et al., 2002). Previous studies have shown that this plant efficiently takes up arsenate As(V) from the soil and rapidly transports it to the shoot in the xylem mainly as As(V) (Kertulis et al., 2005), where it arrives in the petiole and midrib of the frond as arsenate (Hokura et al., 2006) and is finally stored in the fronds as free arsenite As(III), as determined by x-ray absorption spectroscopy (XAS; Lombi et al., 2002; Webb et al., 2003; Ze-Chun et al., 2004) and high-pressure liquid chromatography-inductively coupled plasma mass spectrometry (HPLC-ICP-MS; Wang et al., 2002; Zhao et al., 2003). Such a model suggests that arsenate in the frond is reduced to arsenite for storage. However, this model is not supported by Duan et al. (2005), who were unable to detect any arsenate reductase activity, measured as arsenate-dependant oxidation of NADPH in a crude frond extract.
Like other homosporous ferns, each P. vittata sporophyte produces and releases abundant haploid spores that germinate and develop as autotrophic haploid gametophytes. Each gametophyte consists mostly of a small (approximately 2 mm) single layer of cells. These free-living gametophytes tolerate up to 5 mm arsenate without showing symptoms of arsenic toxicity and hyperaccumulate up to 2% of their dry weight as arsenic (Gumaelius et al., 2004). Their simple morphology, rapid growth under highly controlled environments, and ability to hyperaccumulate arsenic make the gametophyte of P. vittata an experimentally tractable system for studying arsenic tolerance and hyperaccumulation in this fern (Gumaelius et al., 2004).
Arsenic nonaccumulating plants, such as Brassica juncea and Arabidopsis (Arabidopsis thaliana), also reduce of arsenate to arsenite. However, the arsenite that accumulates is coordinated to thiolate ligands and remains sequestered in the root (Pickering et al., 2000a; Dhankher et al., 2002). This suggests that while all plants may reduce arsenate to arsenite, there are significant differences between P. vittata and arsenic nonhyperaccumulating plants in how they store, transport, and ultimately tolerate or detoxify arsenic. While the physiology of arsenic hyperaccumulation in P. vittata sporophytes has been studied extensively, no genes that define the molecular mechanisms underlying this process have been identified.
One approach to identify P. vittata genes required for arsenic tolerance and hyperaccumulation is to make use of the yeast (Saccharomyces cerevisiae) where the molecular mechanisms of arsenic tolerance have been well studied (Bobrowicz et al., 1997; Ghosh et al., 1999; Mukhopadhyay et al., 2000; Wysocki et al., 2003, 2004; Haugen et al., 2004). The yeast arsenic resistance locus encodes a cluster of three arsenic resistance, or ARC, genes (ACR1, ACR2, and ACR3) that are required for arsenic tolerance (Bobrowicz et al., 1997). The yeast ACR2 gene (referred to here as ScACR2) encodes an arsenic-specific arsenate reductase (Mukhopadhyay and Rosen, 1998; Mukhopadhyay et al., 2000). Deletion of ScACR2 in yeast results in an arsenate-sensitive phenotype (Mukhopadhyay and Rosen, 1998). In this study, we describe the biochemical characterization of an arsenate reductase from P. vittata that was isolated by complementing a ScACR2 deletion mutant with a cDNA library generated from P. vittata gametophytes. The biochemical properties of the P. vittata arsenate reductase are described and compared to ScACR2 and the related protein Arath;CDC25 from Arabidopsis.
RESULTS
Chemical Speciation of Arsenic in P. vittata
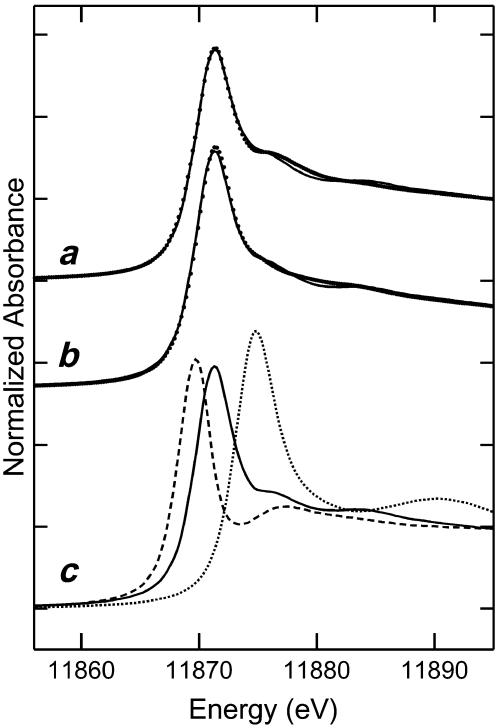
While P. vittata gametophytes hyperaccumulate arsenic, it is not known what form of arsenic is stored in the gametophyte. To address this, XAS was applied to bulked, intact, flash-frozen P. vittata gametophytes grown in the presence of 8 mm arsenate. These results show that gametophytes store 95% ± 3% of the accumulated arsenic as arsenite (AsIII), with none of the arsenite coordinated by thiol ligands (Fig. 1). The remaining 5% ± 3% of the accumulated arsenic remains as arsenate (AsV). Similar results were also obtained from gametophytes grown in 1.3 and 4 mm arsenate (data not shown). A similar analysis of P. vittata sporophyte frond tissue (Fig. 1) shows that 92% ± 3% of arsenic is stored as arsenite, with 6% ± 2% as arsenate and 2% ± 1% as an As(III) thiolate complex.
Figure 1.
Arsenic K-near-edge XAS. Points represent data from frond tissue of a sporophyte grown in arsenic-contaminated soil (a), gametophytes grown in 8 mm arsenate-containing media (b), and As K-edge XAS of the arsenic model compounds used for fitting (c). The solid line represents the best fit to the spectrum from a linear combination of As K-edge XAS for aqueous standards of arsenate, arsenite, and As(III)-tris-glutathione. The fits represent 6% ± 2%, 92% ± 3%, and 2% ± 1% for the sporophyte frond, and 5% ± 3%, 95% ± 3%, and 0% for the gametophyte of arsenate, arsenite, and As(III)-tris-glutathione, respectively. Arsenate (dotted line), arsenite (solid line), and As(III)-tris-glutathione (dashed line) were included as model compounds.
Phenotype Suppression Cloning of P. vittata Arsenate Reductase
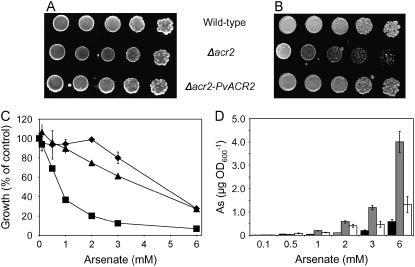
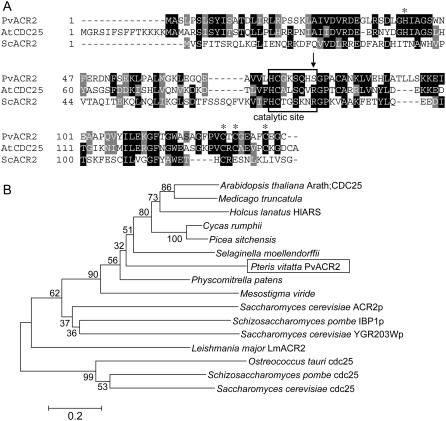
To identify an arsenate reductase gene in P. vittata, we transformed the arsenate-sensitive yeast strain RM1 (Δacr2) with a cDNA expression library generated from mRNA isolated from 1 mm arsenate-grown gametophytes. Yeast colonies that were able to grow in the presence of 10 mm arsenate, a concentration of arsenic that is lethal to RM1 yeast, were selected. One positive colony was characterized in detail. This colony displayed arsenate resistance comparable to wild type based upon growth on both solid and liquid media containing arsenate (Fig. 2, A–C) and accumulated levels of arsenic similar to wild-type yeast (Fig. 2D). The protein predicted from the nucleotide sequence of the transforming P. vittata cDNA contains 134 amino acids, four amino acids longer than ScACR2, and shares 25% identity (47% similarity) to ScACR2 (Fig. 3A). Based upon the suppression of the arsenic-related phenotypes of RM1 and its amino acid sequence similarity to ScACR2 (Fig. 3A), we named the P. vittata gene PvACR2.
Figure 2.
A, Wild-type yeast (W303), Δacr2 (RM1), and Δacr2 (RM1) expressing PvACR2 grown on solidified medium with no added arsenate. B, The same strains grown on media with 10 mm arsenate. C, Growth of wild type (diamonds), Δacr2 (squares), and Δacr2 expressing PvACR2 (triangles) in liquid culture at various concentrations of arsenate, with growth measured by A600. D, ICP-MS measurement of total arsenic in wild type (black bar), Δacr2 (gray bar), and Δacr2 expressing PvACR2 (white bar); data represents the mean (n = 3) ± se.
Figure 3.
A, Alignment of the predicted amino acid sequence of PvACR2, ScACR2 (GenBank accession no. NP_015526), and Arath;CDC25 (GenBank accession no. NP_568119); asterisks indicate conserved Zn-binding residues; the active site is boxed; novel Ser residue marked by an arrow. B, Neighbor-joining dendrogram showing the relationship among the C-terminal phosphatase domain of cdc25 phosphatases and single-domain homologs, which include arsenate reductases and phosphatases. Numbers at nodes show the bootstrap values obtained for the 1,000 replicate analysis. Bar, Kimura-2 parameter distance.
Sequence Analysis of PvACR2
A BLAST (Altschul et al., 1990) search of the genome of Arabidopsis, a nonarsenic accumulator, revealed that Arabidopsis has one gene, Arath;CDC25, that is closely related to PvACR2. Arath;CDC25 was initially characterized as a phosphatase (Landrieu et al., 2004a, 2004b; Sorrell et al., 2005) and more recently has been suggested to play a role in arsenate reduction (Bleeker et al., 2006; Dhankher et al., 2006). Arath;CDC25 shares 42% sequence identity (60% similarity) with PvACR2 and 26% identity (45% similarity) to ScACR2 (Fig. 3A). Plant, yeast, and protista CDC25-like proteins are all related to the C-terminal phosphatase domain of CDC25 phosphatases (Fig. 3B). All of the plant proteins form a distinct clade that has the conserved residues thought to bind zinc (Zn) in Arath;CDC25 (Fig. 3A; Landrieu et al., 2004a, 2004b), suggesting that Zn may be an important cofactor of plant but not yeast or protista CDC25/ACR2-like enzymes. The yeast CDC25-like proteins also form a distinct clade that contains a phosphatase (IBP1p; Snaith et al., 2003), an arsenate reductase (ACR2), and YGR203Wp, a protein shown not to be involved in arsenate resistance in yeast and whose function is unknown (Mukhopadhyay et al., 2000).
Arsenate Reductase Activity of PvACR2 and Arath;CDC25
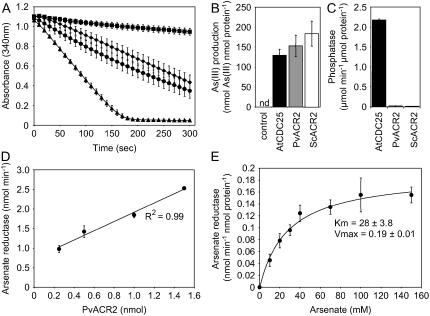
To determine the enzymatic activity of the PvACR2 and Arath;CDC25 proteins they were overproduced in Escherichia coli and purified. PvACR2 was expressed in E. coli from a T7 promoter as an N-terminal fusion with a thioredoxin, S-tag, and six-His tag. Arath;CDC25 was expressed in E. coli from a T7 promoter as an N-terminal fusion with a six-His tag (Landrieu et al., 2004a). Soluble proteins were purified using a Co2+ affinity resin and the entire N-terminal tag removed from PvACR2 using enteriokinase. ScACR2 protein was also overproduced in E. coli and purified as previously described by Mukhopadhyay et al. (2000) and used as a positive control. Arsenate reductase activity was assayed following the method developed for ScACR2 (Mukhopadhyay et al., 2000). In this assay, arsenate reduction is coupled to NADP (NADPH) oxidation via the reduction of oxidized glutathione by glutathione reductase and with the resulting glutathione (GSH) serving as the electron donor for arsenate reduction. Rates of NADPH oxidation were measured as loss of optical density at 340 nm and were found to be minimal in the absence of purified ScACR2, PvACR2, or Arath;CDC25 protein, while reaction rates increased dramatically in the presence of these proteins (Fig. 4A). Enzyme assays were also performed in the absence of arsenate, GSH, or glutaredoxin (GRX), and all were found to be required for arsenate reductase activity (data not shown). To confirm the results of the coupled assay, we modified an HPLC-ICP-MS method (Wangkarn and Pergantis, 2000) to allow the direct measurement of arsenite production in the same reaction mixture used in the coupled assay (Fig. 4B). In the presence of arsenate, GSH, and GRX, but in the absence of either ScACR2, PvACR2, or Arath;CDC25, no detectable arsenite was produced in 45 min (Fig. 4B). The addition of purified ScACR2, PvACR2, or Arath;CDC25 to the reaction mixture catalyzed the accumulation of arsenite (Fig. 4B), with the rate of arsenite accumulation mirroring that found using the coupled assay, with ScACR2 > PvACR2 > Arath;CDC25 (Fig. 4, A and B). The results of both assays clearly demonstrate that PvACR2 and Arath;CDC25 have arsenate reductase activity that requires both GSH and GRX for the production of arsenite, as does ScACR2 (Mukhopadhyay et al., 2000).
Figure 4.
A, Time course of arsenate reductase activity, estimated as nmol NADPH oxidized; control (squares) complete assay mix with GRX2 but no reductase (squares), Arath;CDC25 (diamonds), P. vittata PvACR2 (circles), and yeast ScACR2 (triangles) in the presence of 40 mm arsenate in the assay mix. Data represents the mean (n = 3) ± se. B, Arsenate reductase activity measured as arsenite production over 45 min using LC-ICP-MS, with control (complete assay mix without enzyme), recombinant Arath;CDC25 (black), PvACR2 (gray), and ScACR2 (white) in the presence of 40 mm arsenate in the assay mix; nd, Not detected. Data represents the mean (n = 3) ± se. C, Phosphatase activity measured as hydrolysis of p-nitrophenol phosphate by Arath;CDC25, PvACR2, and ScACR2 recombinant protein. Data represents the mean (n = 3) ± se. D, Arsenate reductase activity (loss NADPH) with increasing concentrations of recombinant PvACR, at 40 mm arsenate in the assay mix. Data represents the mean (n = 3) ± se. E, Arsenate reductase activity of recombinant PvACR with increasing concentrations of arsenate in the assay mix. Data represents the mean (n = 3) ± se. Kinetic constants (±sd) determined using a nonlinear regression to the data, using the Marquardt-Levenberg algorithm.
Kinetic Properties of PvACR2 Catalyzed Arsenate Reduction
The initial rate of arsenate reduction as a function of PvACR2 concentration was determined and shown to increase linearly with increasing concentrations of purified PvACR2 protein (Fig. 4D). The rate of arsenate reduction as a function of arsenate concentration was also determined (Fig. 4E). The data was well fitted by the standard Michaelis-Menten model (V = Vmax × [{S}/{S} + KM]), and kinetic constants (±sd) determined using a nonlinear regression to the data using the Marquardt-Levenberg algorithm (Fig. 4E). The PvACR2 KM for arsenate is 28 ± 8 mm with a Vmax = 0.19 ± 0.02 nmol min−1 nmol protein−1. Preliminary evidence also suggests that the arsenate reductase of PvACR2, like ScACR2, is inhibited by arsenite (data not shown).
Phosphatase Activity of PvACR2 and Arath;CDC25
Because PvACR2 and Arath;CDC25 are also similar in sequence to phosphatases, purified PvACR2, ScACR2, and Arath;CDC25 were assayed for phosphatase activity in vitro at pH 6.5 and 7.5. PvACR2 and ScACR2 had very low levels of phosphatase activity (Fig. 4C) consistent with that previously reported for ScACR2 (Mukhopadhyay et al., 2000). However, Arath;CDC25 had high levels of phosphatase activity consistent with its previous identification as a phosphatase (Landrieu et al., 2004a, 2004b).
Steady-State Expression Levels of PvACR2 and Arsenate Reductase Activity in P. vittata Gametophytes
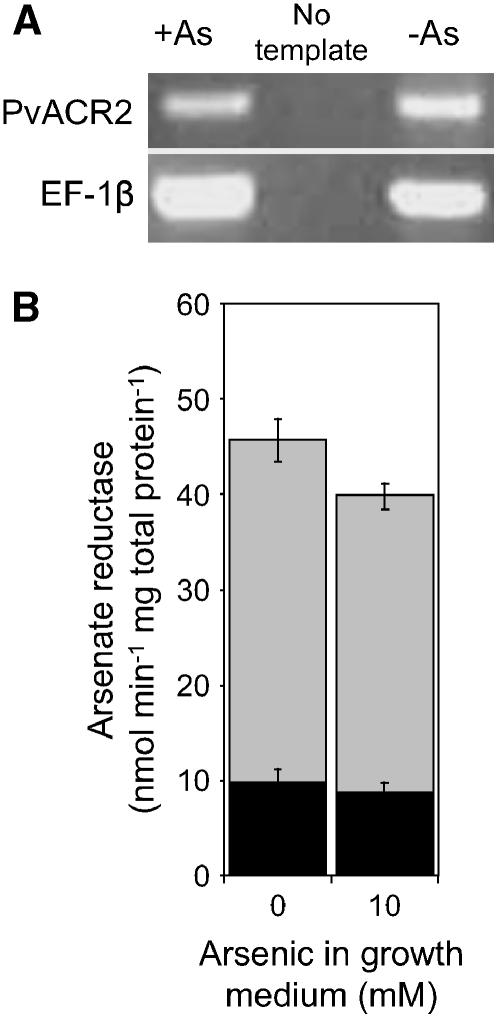
Steady-state levels of PvACR2 mRNA were determined by reverse transcription (RT)-PCR and quantitative (q)RT-PCR. As shown in Figure 5A, PCR products amplified using PvACR2-specific primers could be detected in approximately equal quantities in gametophyte samples grown in the absence or presence of 10 mm arsenate. PvACR2 mRNA levels were also established to be unaffected by arsenate exposure based on qRT-PCR, with little difference in expression in gametophytes grown in the presence of 0 and 10 mm arsenate (ΔΔCT = 0.4 ± 2.6). Similar results were also obtained with gametophytes grown in 0 and 1 mm arsenate (data not shown). To demonstrate that arsenate reductase activity occurs in P. vittata gametophytes in vivo, desalted cell free extracts of gametophytes were assayed for arsenate activity using the coupled assay previously described. Levels of NADPH oxidation increased upon addition of arsenate to gametophyte cell-free extracts (Fig. 5B). However, in the absence of GSH and GRX, NADPH oxidation was not detected, demonstrating that arsenate reduction in cell-free extracts is dependent upon both GSH and GRX. Gametophyte extracts were also found to reduce arsenate almost equally well independent of the amount of arsenate in their growth medium (Fig. 5B).
Figure 5.
A, Relative levels of PvACR2 mRNA in gametophytes grown in the presence or absence of arsenate, as determined by RT-PCR. The gene-specific primers used are indicated on the left, and the source of cDNA template indicated at the top of the figure; EF-1b-specific primers were used as an internal control. B, Arsenate reductase activity in P. vittata gametophyte cell-free extracts. Black bars represent rates of NADPH oxidation in the absence of arsenate in the assay, gray bars represent rates with 100 mm arsenate included in the assay. Data represents the mean of four replicate assays (±se).
DISCUSSION
Mechanisms for arsenic uptake and detoxification have been best studied in the yeast and the bacteria E. coli and Bacillus subtilis (for review, see Rosen, 2002). In these microbes, arsenate (a phosphate analog) is taken up by phosphate transporters (Willsky and Malamy, 1980; Bun-ya et al., 1996; Yompakdee et al., 1996), reduced to arsenite by arsenate reductases (Liu et al., 2002), and extruded from the cell by a variety of arsenite transporters (for review, see Rosen, 2002). In yeast, glutathione-conjugated arsenite may also be transported into the vacuole (Ghosh et al., 1999). Plants are known to reduce arsenate and store arsenite-thiol complexes in their vacuoles (Pickering et al., 2000a). Interestingly, recent evidence suggests that the CDC25-like protein Arath;CDC25, which has been previously shown to have phosphate activity (Landrieu et al., 2004a, 2004b; Sorrell et al., 2005), contributes to the arsenate reduction capacity of the arsenic nonaccumulator Arabidopsis (Duan et al., 2005; Bleeker et al., 2006; Dhankher et al., 2006). A T-DNA insertion mutation in the Arath;CDC25 sequence produced a complete loss of arsenate reductase activity in both shoots and roots of 3-week-old hydroponically grown plants treated with 300 μm arsenate for 9 d (Duan et al., 2005). In an independent study, roots of the same mutant line were also assayed for arsenate reductase activity by Bleeker et al. (2006). In this study, arsenate reductase activity was unchanged in crude extracts of roots from plants not exposed to arsenate and was reduced by 36% in plants exposed to arsenate. This study also established that purified recombinant Arath:CDC25 protein (termed AtASR) has arsenate reductase activity in vitro and concludes that Arath;CDC25 accounts for only that proportion of the arsenate reductase activity that is inducible by arsenate, which represents 36% of the total arsenate reductase activity in Arabidopsis roots. The basis for the difference between the Duan et al. (2005) and Bleeker et al. (2006) results remains to be resolved. The capacity of Arath;CDC25 to reduce arsenate was further established by the fact that Arath;CDC25 is capable of suppressing the arsenate sensitivity of E. coli lacking its native arsenate reductase (Dhankher et al., 2006). Interestingly, T-DNA insertional mutants of Arath;CDC25 accumulate 5-fold less total arsenic in shoots than wild-type plants over a range of arsenate concentrations in the nutrient solution (Bleeker et al., 2006). However, in a separate study, suppression of Arath;CDC25 mRNA (termed AtACR2) using an RNAi construct produced plants with a 7-fold increase in shoot arsenic accumulation (Dhankher et al., 2006). The discrepancy between these studies makes their interpretation difficult in the context of the physiological role of Arath;CDC25. However, they do clearly demonstrate that this dual function phosphatase/arsenate reductase plays a significant role in the physiological processes that affect arsenate metabolism in Arabidopsis.
The arsenic hyperaccumulating fern P. vittata is an unprecedented system for the study of arsenic metabolism and the evolution of arsenic tolerance and resistance mechanisms in plants and other multicellular organisms. Here, we show that when grown in the presence of arsenate, the simple, haploid gametophytes of P. vittata convert 90% to 95% of accumulated arsenic to free arsenite, with only a very minor portion of the arsenic being accumulated as an As(III) thiolate complex. Similar results are observed for the diploid sporophyte plant, consistent with previous observations of this and the related species Pteris cretica (Tongbin et al., 2002; Meharg, 2003). Because the gametophyte is morphologically simple and is able to reduce and store arsenic in a manner similar to the sporophyte plant, we have used the gametophyte as a source of material to examine whether P. vittata metabolizes arsenic by a mechanism similar to that of yeast. Here, we focus on the identification and functional characterization of a P. vittata gene similar to the yeast arsenate reductase gene ScACR2.
Suppression-screening of yeast strain RM1 harboring a deletion of the ScACR2 gene resulted in the isolation of a cDNA from P. vittata gametophytes, which suppresses the arsenate sensitivity phenotype of this yeast mutant. The protein deduced from the cDNA sequence (PvACR2) is similar in size and sequence to ScACR2 and represents the first plant gene identified that almost completely complements the loss of the ScACR2 gene in yeast. The hyperaccumulation of arsenic in RM1, a phenotype not reported previously, is also complemented by the PvACR2 gene. This additional phenotype of RM1 is not unexpected since an arsenate-exporting protein that extrudes arsenate from the cell has not been detected in yeast, which is known to rely on ACR3 to efflux arsenite. That PvACR2 also suppresses the arsenic hyperaccumulation phenotype in Δacr2 provides additional support that PvACR2 is functionally similar to the ScACR2 gene of yeast.
With the exception of PvACR2, ScACR2 and other CDC25-like proteins have in common a HCX5R amino acid motif that also defines the active-site loop of ScACR2 (Streuli et al., 1990; Zhang et al., 1994; Hoff et al., 1999; Jackson and Denu, 2001). While ScACR2, PvACR2, and Arath;CDC25 all have the conserved His and Cys residues within their predicted catalytic site, only PvACR2 lacks the conserved Arg residue. In yeast, substitution of this residue with Ala abolishes the arsenate reductase activity of ScACR2, suggesting that this Arg is essential for its activity (Mukhopadhyay and Rosen, 2001). The occurrence of Ser instead of Arg within the putative active site of PvACR2 suggests that either PvACR2 is not an arsenate reductase or that the amino acid sequence of the active site can be more flexible than previously thought.
To directly establish that PvACR2 can function as an arsenate reductase, we measured its capacity to catalyze the reduction of arsenate to arsenite using both an indirect coupled assay, first established to measure the arsenate reductase activity of ScACR2, and the direct measurement of arsenite. Both assays confirmed that PvACR2 acts as an arsenate reductase that requires GRX for activity and GSH as the primary electron donor. PvACR2 has a KM for arsenate of 28 ± 8 mm, similar to that reported for ScACR2, which has a KM for arsenate of 35 mm (Mukhopadhyay et al., 2000). PvACR2 has a Vmax = 0.19 ± 0.02 nmol min−1 nmol protein−1, which is approximately 30-fold lower than that previously established for ScACR2 (Mukhopadhyay et al., 2000). It is important to note that the reported Vmax value for ScACR2 is based upon the coupled reaction using the appropriate yeast GRX1 enzyme and a reaction temperature of 37°C (Mukhopadhyay et al., 2000). Because the in vivo coupling mechanism for PvACR2 is unknown, we used the E. coli GRX 2 (EcGRX2) enzyme in this study and performed the coupled enzyme reactions at room temperature. Similarly, the discrepancy between the reported KM value of 2.3 mm for an arsenate reductase activity in crude extracts from P. vittata sporophyte roots (Duan et al., 2005) could also be attributed to the use of the EcGRX2 in our in vitro assay. The difference in the Vmax values for ScACR2 and PvACR2 may also be attributed to the choice of GRXs used in each reaction. Given that Arabidopsis has 30 GRX genes (Lemaire, 2004) and a genomic sequence is not available for P. vittata, it is difficult to determine which GRX would be most appropriate for these reactions. Nevertheless, our observations of the enzymatic properties of PvACR2 demonstrate that the unique configuration of the active site of PvACR2 does allow it to reduce arsenate, although the mechanism may differ from that of ScACR2.
We also observed that the closely related Arath;CDC25 protein has a similar capacity to reduce arsenate to arsenite in a GSH- and GRX-dependent manner as PvACR2. Although Arath;CDC25 has been characterized as a phosphatase (Landrieu et al., 2004a, 2004b; this work), it clearly also has the in vitro capacity to function as an arsenate reductase. Our observation that Arath;CDC25 has an arsenate reductase activity in vitro agrees with Bleeker et al. (2006). However, we are puzzled as to the mechanism of the arsenate reductase activity of Arath;CDC25 observed by Bleeker and coworkers, as their arsenate reductase assays were performed in the absence of GRX. To date, all characterized arsenate reductases have a requirement for GRX or thioredoxin, which serve as electron donors (Rosen, 2002).
PvACR2 and Arath;CDC25 are similar in sequence to both arsenate reductases and phosphatases (Fig. 3B) so it is unclear if they function as arsenate reductases, phosphatases, or both. Arath;CDC25 has been shown to function as a phosphatase (Landrieu et al., 2004a, 2004b; this work), although more recent evidence has shown that Arath;CDC25 may act as an arsenate reductase in vivo (Bleeker et al., 2006; Dhankher et al., 2006). Like ScACR2 (Mukhopadhyay et al., 2003), we found that PvACR2 has very low phosphatase activity, whereas Arath;CDC25 has strong phosphatase activity. Previous phylogenetic studies have used the observation that Arath;CDC25 is more closely related to the yeast ScACR2 than to CDC25 phosphatases as support for the model that Arath;CDC25 functions as an arsenate reductase rather than as a phosphatase (Dhankher et al., 2006). However, it is clear when such a phylogenetic study is extended by the addition of more CDC25-like protein sequences including the Schizosaccharomyces pombe IBP1p phosphatase (Fig. 3B), that Arath;CDC25 and PvACR2 are both equally related to arsenate reductase (ScACR2; Mukhopadhyay and Rosen, 1998; Mukhopadhyay et al., 2000), phosphatase (IBP1p; Snaith et al., 2003), and dual arsenate reductase/phosphatase sequences (LmACR2; Zhou et al., 2006). With our current limited understanding of arsenate reductase structure/function relationships such phylogenies have no predictive power for the assignment of function to biochemically uncharacterized CDC25-like proteins. However, based on our biochemical characterization of PvACR2, although it is more similar in amino acid sequence to Arath;CDC25 than ScACR2, its arsenate reductase activity and lack of phosphatase activity indicate that it functionally more closely resembles ScACR2 than Arath;CDC25. PvACR2 from P. vittata, therefore, represents the first functional ortholog of the yeast arsenate reductase (ScACR2) characterized from plants or other eukaryotes.
We have established that in vitro PvACR2 acts as a specific GSH-dependent arsenate reductase; however, its role in vivo is still speculative. Noninvasive XAS of arsenate-grown P. vittata gametophytes shows that they accumulate arsenite, suggesting that they have the capacity to efficiently reduce arsenate to arsenite in vivo. An analysis of cell-free extracts of P. vittata gametophytes grown in the absence or presence of arsenate revealed that a biochemical capacity to reduce arsenate is present in gametophytes and that this activity is constitutive. The steady-state levels of PvACR2 expression also indicate that the gene is constitutively expressed as PvACR2 mRNA accumulates in both the absence and presence of arsenate. The constitutive activation of various biochemical processes known to be involved in metal hyperaccumulation has been observed in a number of metal hyperaccumulating plants. In the hyperaccumulators Thlaspi caerulescens (Cd/Ni/Zn), Thlaspi goesingense (Ni/Zn), and Arabidopsis halleri (Zn), for example, genes involved in Zn homeostasis, including Zn influx transporters in the ZRT1- and IRT1-like protein (ZIP) family, Zn efflux transporters in the P-type ATP-dependent metal transporter family, Nramp ion-transporter, and cation diffusion facilitator (CDF) families are constitutively expressed at relatively high levels (Pence et al., 2000; Assuncao et al., 2001; Persans et al., 2001; Becher et al., 2004; Freeman et al., 2004; Papoyan and Kochian, 2004; Weber et al., 2004). Furthermore, levels of the antioxidant glutathione are also constitutively elevated in various nickel hyperaccumulating Thlaspi species, driven by constitutive activation of the sulfur assimilation enzyme Ser acetyltransferase (Freeman et al., 2004). It is thought that these types of permanent biochemical adjustments reflect the fact that hyperaccumulation is not an inducible environmental stress response but rather, a constitutively expressed adaptive trait (Reeves and Baker, 1984).
While the requirement of the PvACR2 gene for arsenic tolerance or hyperaccumulation in P. vittata has not been established at this time, the results of this study suggest that PvACR2 is likely to play an important role in arsenic metabolism in this species. Its similarity to ScACR2 in structure and function, together with the differences between PvACR2 and Arath;CDC25 in phosphatase activity, also suggests that the evolution of arsenic tolerance in P. vittata may have involved mutations that affected the function of a protein-Tyr phosphatase. A recent study in yeast (Mukhopadhyay et al., 2003) demonstrated that changing only three amino acid residues of ScACR2 was sufficient to change its activity from an arsenate reductase to a protein-Tyr phosphatase. A functional and phylogenetic characterization of ACR2-like genes from other ferns and angiosperms is likely to shed light on the evolutionary relationships between Arath;CDC25 and PvACR2, and, more importantly, how the arsenic hyperaccumulating trait may have evolved in P. vittata. While the role of PvACR2 in arsenic hyperaccumulation is speculative, its identification provides a starting point for dissecting the molecular mechanisms that underlie arsenic tolerance and hyperaccumulation in this extraordinary plant.
MATERIALS AND METHODS
Arsenic K-Edge XAS
Field collected Pteris vittata sporophytes grown in arsenic-contaminated soil were used for XAS. Gametophytes were grown from spores in media (Gumaelius et al., 2004) containing 8 mm arsenate. Living P. vittata sporophytes and gametophytes were transported to the Stanford Synchrotron Radiation Laboratory for XAS analysis of bulk tissue samples following established procedures (Pickering et al., 2000b).
Bacterial Strains, Yeast Strains, and Plasmids
Strains and plasmids used in this study are described in Table I.
Table I.
Strains and plasmids used in this study
| Strain/Plasmid | Genotype | Reference or Source |
|---|---|---|
| Bacterial Strains | ||
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(araA-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| DH5α | supE44 ΔlacU169 (phi80 lacZ ΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Invitrogen |
| Rossetta (DE3)pLysS | F−ompT hsdSB(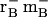 ) gal dcm lacY1 (DE3) pLysSRARE6, Cmr ) gal dcm lacY1 (DE3) pLysSRARE6, Cmr
|
Novagen |
| BL21(DE3)pLysS | F−ompT hsdSB( ) gal dcm (DE3) pLysS, Cmr ) gal dcm (DE3) pLysS, Cmr
|
Novagen |
| Yeast strains | ||
| W303-1B | Mat-αade2-1 his3-11, 15 leu2-3, 112 ura3-1 trp-1 | Mukhopadhyay et al. (2000) |
| RM1 | Mat-αade2-1 his3-11, 15 leu2-3, 112 ura3-1 trp-1 ACR2∷HIS3 | Mukhopadhyay et al. (2000) |
| Plasmids | ||
| pGEM-T | Multicopy E. coli cloning vector, Apr | Promega |
| pBAD-ScACR2 | pBad/myc-HIS A, E. coli expression vector, with ScACR2 inserted into the NcoI-HindIII site, Apr | Mukhopadhyay et al. (2000) |
| pET-Grx2 |
E. coli expression vector, pET28a with E. coli GrxB inserted into the NdeI-BamHI sites, 
|
Shi et al. (1999) |
| p424-GPD | Multicopy, shuttle vector, Apr, TRP, GPD | Mumberg et al. (1995) |
| pDNR-Lib | Donor vector used for library construction, Cmr | Clontech |
| p424-Sfi | A fragment of the multicloning site cut from pDNR-Lib with EcoRI and XhoI was inserted into the EcoRI-XhoI sites of p424-GPD, Apr | This study |
| p424-PvACR2 | Yeast expression vector, p424-Sfi with PvACR2 cDNA inserted into SfiI sites, Apr | This study |
| pET-32a | E. coli expression vector, Apr | Novagen |
| pET-PvACR2 | E. coli expression vector, pET32a with PvACR2 inserted into the NcoI-XhoI sites, Apr | This study |
| pET-Arath;CDC25 | E. coli expression vector, pET15b with Arath;CDC25 inserted into the NdeI-XhoI sites, Apr | Landrieu et al. (2004a, 2004b) |
Cloning of PvACR2
The yeast (Saccharomyces cerevisiae) expression library was constructed from RNA purified from P. vittata gametophytes grown in liquid culture (Gumaelius et al., 2004) containing 1 mm KH2AsO4 for 6 weeks, using the Creator SMART cDNA library construction kit (CLONTECH) following the manufacturer's instructions. The library was constructed in the yeast expression vector p424-Sfi containing a Trp selectable marker. The library was transformed into the yeast strain RM1 (Δacr2), and the resulting transformants were selected on minimal media without Trp containing 2% Glc and 10 mm Na2HAsO4. Plates were incubated for 5 to 7 d at 30°C. All colonies were restreaked onto fresh selection plates, and those with the best growth were selected, the plasmid isolated, amplified in Escherichia coli, and retransformed back into RM1 to confirm its ability to suppress the arsenate sensitivity of RM1. Isolated plasmid was also used for sequencing of the cDNA. The sequence of PvACR2 was confirmed by sequencing two other independent cDNA inserts isolated from the P. vittata cDNA library. The PvACR2 sequence was deposited into GenBank (accession no. DQ310370).
Complementation of Yeast
Liquid culture complementation experiments followed existing procedures (Mukhopadhyay et al., 2000). For plate assays, yeast strains Δacr2 p424, Δacr2 p424-PvACR2, and W303-B p424 (Mumberg et al., 1995) were grown at 30°C in minimal media without Trp supplemented with 2% Glc. Spots of 5-fold serial dilutions of the cultures (7 μL) were applied onto 1% agar plates containing minimal media without Trp supplemented with Na2HAsO4 as indicated. The plates were incubated at 30°C for 4 to 5 d.
Phylogenetic Analysis
Phylogenetic analyses were conducted using MEGA version 3 (Kumar et al., 2004). Protein sequences were aligned using ClustalW and the dendrogram constructed using distance methods of neighbor joining (Saitou and Nei, 1987), performed on a Kimura-2 parameter (pairwise distances; Kimura, 1980). Bootstraps were carried out with 1,000 replications. The GenBank accession numbers for the protein sequences or the nucleotide sequences from which protein sequence was deduced are: NP_568119 (Arath;CDC25), DW019041 (Medicago truncatula), AA411500 (HlARS), CB091734 (C. rumphii), DR483768 (P. sitchensis), DQ310370 (PvACR2), BJ947601(P. patens), DN263821 (M. viride), NP_015526 (ScACR2p),CAB46700 (IBP1p), Q06597(YGR203Wp), AA573185 (LmACR2), AAQ16122 (O. tauri cdc25), CAA90849 (S. pombe cdc25), and NP_013750 (yeast cdc25). The protein from S. moellendorffii was deduced from expressed sequence tag 1590812 obtained from http://selaginella.genomics.purdue.edu.
Protein Expression and Purification
The PvACR2 reading frame was amplified by PCR with primers that added a NcoI and XhoI site to the 5′ and 3′ ends of the fragment, respectively. The PCR fragment was cloned into pGEM-T (Promega). The resulting plasmid was digested with NcoI and XhoI and the fragment ligated into the NcoI-XhoI site of pET32A (Novagen) in frame with the N-terminal thioredoxin, S-tag, and six-His tag, creating plasmid pET-PvACR2. For protein expression in E. coli, Rosseta pLysS (Novagen) cells transformed with pET-PvACR2 were grown at 37°C in Luria-Bertani medium containing 50 mg/mL ampicillin and 20 μm ZnSO4. At an A600 of 0.5, isopropylthio-β-galactoside was added to a final concentration of 0.4 mm, and the culture grown for 3 h at 30°C. Expression of EcGRX2, ScACR2, and Arath;CDC25 were as previously described (Shi et al., 1999; Mukhopadhyay et al., 2000; Landrieu et al., 2004a, 2004b). Total proteins were extracted according to Mukhopadhyay et al. (2000). Recombinant proteins were affinity purified using TALON cobalt resin (CLONTECH). The PvACR2 n-terminal thioredoxin and His tag were cleaved following the manufacturer's instructions (Novagen). Proteins were concentrated and extraction buffer exchanged to 50 mm MOPS, pH 6.5, 0.5 m NaCl, 20% glycerol, and 10 mm β-mercaptoethanol using Vivaspin columns with a 2,000 Mr cutoff. Arath;CDC25 recombinant protein was stored at 4°C and used promptly; other proteins were stored at −80°C. Protein concentrations were determined at 280 nm using the extinction coefficients 14,300 m−1 cm−1 for PvACR2; 11,170 m−1 cm−1 for Arath;CDC25; 14,300 m−1 cm−1 for ScACR2; and 21,860 m−1 cm−1 for EcGrx2, and calculated according to Gill and von Hippel (1989).
Arsenate Reductase Assay
Arsenate reductase activity was measured using a previously established coupled assay (Mukhopadhyay et al., 2000). The assay buffer consisted of 50 mm MES, 50 mm MOPS, pH 6.5, 300 mm NaCl, 0.1 mg/mL bovine serum albumin, 1 mm GSH, 250 μm NADPH, 50 nm glutathione reductase, and 1 μm EcGrx2. Purified protein and Na2HAsO4 were added as indicated. The assays were carried out in a final reaction volume of 100 mL at room temperature. Reductase activity was measured as a change in A340. The nmoles of NADPH oxidized during the assay were calculated using a molar extinction coefficient of 6,200 m−1 cm−1. Enzyme kinetic constants were calculated according to the Marquardt-Levenberg algorithm that uses a nonlinear, least-squares regression to find the minimum sum of squares (Brooks, 1992).
Arsenate Reductase Assays of Cell-Free Extracts
Cell-free extracts were obtained from 1-month-old shake-grown P. vittata gametophyte cultures grown in 0.5× Murashige and Skoog salts (Sigma M5524) plus 3.9 g/L MES, pH 6.5. Three replicates of cultures grown in 0 or 10 mm KH2AsO4 were assayed. Gametophytes were filtered onto a number 1 Whatman cellulose filter and washed with 2 L distilled, deionized water; 0.3 g of tissue was ground in liquid nitrogen for 15 min, resuspended in 10 mL degassed extraction buffer (0.3 m KCl, 50 mm MOPS/MES, 10 mm β-mercaptoethanol, 1% proteinase inhibitor cocktail [Sigma P9599], 0.33 g polyvinylpyridine, and 5% glycerol), and centrifuged at 1,857g for 15 min at 4°C. The supernatant was centrifuged for 35 min at 18,000g and the resulting supernatant centrifuged at 100,000g for 1 h. The supernatant was transferred to Vivaspin 2,000 Mr cutoff concentrators (VivaScience VS15RH92), concentrated to 500 μL, resuspended in 2.5 mL extraction buffer, and this procedure repeated twice. Total protein concentration was determined using the Coomassie Plus protein assay reagent (Pierce 23236) using a bovine serum albumin standard curve. Arsenate reductase assays were performed with 1.7 mg mL−1 total protein per assay, as previously described.
Phosphatase Assay
Purified PvACR2, ScACR2, and Arath;CDC25 (10 μm) were incubated for 15 min at 30°C with 100 mm p-nitrophenol phosphate (Sigma) phosphatase substrate in 50 mm MES, 50 mm MOPS, pH 6.5 or 7.5, and 0.3 m NaCl (Mukhopadhyay et al., 2003; Landrieu et al., 2004a, 2004b). The reaction was terminated by the addition of 200 mL of 1 m NaOH and the absorbance measured at 410 nm. Phosphatase activity was calculated using a molar extinction coefficient of 1.78 × 104 m−1 cm−1.
RT-PCR
P. vittata gametophyte cultures were grown in liquid media for 2 weeks in the presence of 0 or 10 mm KH2AsO4. Gametophytes were harvested, washed with distilled, deionized water, and ground in liquid nitrogen. Total RNA was extracted from approximately 100 mg of gametophytes using a RNeasy Plant Mini kit (Qiagen). First-strand cDNA was synthesized using the SuperScript III kit (Invitrogen). PCR primers were designed using Primer Express 2.0 software (ABI Biosystems). The PvACR2 primers used were 5′-GCATAGCGGACCTGCATGT-3′ (forward) and 5′-GCGGCTTCGATTTCTTTCTTG-3′ (reverse). P. vittata elongation factor-1b (EF-1b) was used as the internal control and the primers used were EF-1b forward 5′-GAAGCCTTGGGATGATGAAA-3′ and EF-1b reverse 5′-CCTCGATCAGGTTGTCCACT-3′. PCR conditions were 2 min at 94°C, 30 cycles of 20 s at 94°C, 20 s at 55°C, and 30 s at 72°C, followed by 5 min at 72°C.
Real-Time qRT-PCR
P. vittata gametophyte mRNA was extracted as described above. Primers were designed using Primer Express 2.0 software by ABI Biosystems. The PvACR2 primers used were PvACR2 forward 5′-GCATAGCGGACCTGCATGT-3′ and PvACR2 reverse 5′-GCGGCTTCGATTTCTTTCTTG-3′. Actin was used as the internal control and the primers used were Act forward 5′-CAACAGGTATCGTGCTCGACTCT-3′ and Act reverse 5′-GGCAATGCGTAACCCTCATAA-3′. The qRT-PCR was run on the ABI Prism 7000 with a dilute cDNA/SYBR green mix (1:2). The primer pair concentration was at 300/300 nm determined from optimization runs with the cDNAs of interest. The manual baseline was set at 9 and 25. The data reflects three technical replications of three different biological samples by qRT-PCR. The ΔCT values were averaged, and the ΔΔCT value calculated as the difference between the ΔCT values for gametophytes grown in the presence of 0 and 10 mm arsenate (ΔCT10 − ΔCT0).
ICP Quantification of Arsenic in Yeast
One milliliter of a 24-h yeast culture was filtered onto a 0.45-μm nitrocellulose filter (Whatman) and washed with 10 mL of ice-cold 1 mm EDTA, 20 mm sodium citrate buffer, pH 7.0, followed by a 10-mL ultrapure water wash. The filter plus yeast was digested with 1 mL of concentrated nitric acid (Omnitrace) for 4 h at 115°C, and diluted with 4 mL of ultrapure water. Samples were analyzed using a Perkin Elmer-Sciex DRC-E ICP-MS fitted with an APEX-Q high-sensitivity desolvation system (Elemental Scientific).
Arsenic Speciation Using HPLC-ICP-MS
Arsenic reductase assays were performed as described above and incubated for 45 min. Arsenate and arsenite levels were measured in 10-μL aliquots of 100,000:1 dilutions of the assay mixture. Analytical conditions were modified from Wangkarn and Pergantis (2000). Diluted samples were injected onto a Waters XTerra 5-μm C18 narrow-bore reverse-phase column (2.1 × 150 mm) and arsenic species eluted in a 12 mm tetrabutylammonium hydroxide, pH 6, mobile phase at a flow rate of 0.7 mL min−1. Arsenic was detected postcolumn with an in-line VG PQ3 EXCELL ICP-MS (VG Elemental) equipped with an APEX-Q high-sensitivity desolvation system (Elemental Scientific Instruments), coupled via a tuneable microsplitter. Arsenite was quantified using an arsenite calibration curve; arsenate present in the samples was used as an internal standard for normalization.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number DQ310370.
Acknowledgments
D.E.S., J.A.B., D.R.E., and L.G. conceived the experiments. D.R.E. generated the data for Figures 2 (A–C), 3, and 4 (A, D, and E); L.G. generated the data for Figures 2D, 4 (B and C), and 5; E.I. performed the RT-PCR assays; and I.J.P. performed XAS analysis and interpretation. D.E.S., J.A.B., L.G., and D.R.E. wrote the article. We thank Barry Rosen and Rita Mukhopadhyay for helpful advice and supplying the yeast strains and the E. coli expression constructs for ScACR2 and EcGRX2, I. Landrieu for supplying the Arath;CDC25 E. coli expression construct, Brett Lahner for help with ICP-MS analysis, Graham George for help with XAS analysis, Hugh Harris for assistance with XAS data acquisition, and Om Parkash and Richard Meager for useful discussions.
This work was supported by the U.S. Department of Energy (grant no. DE–FG02–03ER63622), by the Indiana 21st Century Research and Technology Fund, by a Canada Research Chair award (to I.J.P.), and by the Natural Sciences and Engineering Research Council, Canada. Work at the Stanford Synchrotron Radiation Laboratory is funded by the U.S. Department of Energy and by the National Institutes of Health.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jo Ann Banks (banksj@purdue.edu).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.084079.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Assuncao AG, Da Coata Martins P, De Folter S, Vooijs R, Schat H, Aarts MG (2001) Elevated expression of metal transporter genes in three accessions of the metal hyperaccumulator Thlaspi caerulescens. Plant Cell Environ 24: 217–226 [Google Scholar]
- Becher M, Talke IN, Krall L, Kramer U (2004) Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J 37: 251–268 [DOI] [PubMed] [Google Scholar]
- Bleeker PM, Hakvoort HW, Bliek M, Souer E, Schat H (2006) Enhanced arsenate reduction by a CDC25-like tyrosine phosphatase explains increased phytochelatin accumulation in arsenate-tolerant Holcus lanatus. Plant J 45: 917–929 [DOI] [PubMed] [Google Scholar]
- Bobrowicz P, Wysocki R, Owsianik G, Goffeau A, Ulaszewski S (1997) Isolation of three contiguous genes, ACR1, ACR2 and ACR3, involved in resistance to arsenic compounds in the yeast Saccharomyces cerevisiae. Yeast 13: 819–828 [DOI] [PubMed] [Google Scholar]
- Brooks SPJ (1992) A simple computer program for the analysis of enzyme kinetics. Biotechniques 13: 906–911 [PubMed] [Google Scholar]
- Bun-ya M, Shikata K, Nakade S, Yompakdee C, Harashima S, Oshima Y (1996) Two new genes, PHO86 and PHO87, involved in inorganic phosphate uptake in Saccharomyces cerevisiae. Curr Genet 29: 344–351 [PubMed] [Google Scholar]
- Carter DE, Aposhian HV, Gandolfi AJ (2003) The metabolism of inorganic arsenic oxides, gallium arsenide, and arsine: a toxicochemical review. Toxicol Appl Pharmacol 193: 309–334 [DOI] [PubMed] [Google Scholar]
- Dhankher OP, Li Y, Rosen BP, Shi J, Salt D, Senecoff JF, Sashti NA, Meagher RB (2002) Engineering tolerance and hyperaccumulation of arsenic in plants by combining arsenate reductase and gamma-glutamylcysteine synthetase expression. Nat Biotechnol 20: 1140–1145 [DOI] [PubMed] [Google Scholar]
- Dhankher OP, Rosen BP, McKinney EC, Meagher RB (2006) Hyperaccumulation of arsenic in the shoots of Arabidopsis silenced for arsenate reductase, ACR2. Proc Natl Acad Sci USA 103: 5413–5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan G-L, Zhu Y-G, Tong Y-P, Cai C, Kneer R (2005) Characterization of arsenate reductase in the extract of root and fronds of chinese brake fern, an arsenic hyperaccumulator. Plant Physiol 138: 461–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JL, Persans MW, Nieman K, Albrecht C, Peer W, Pickering IJ, Salt DE (2004) Increased glutathione biosynthesis plays a role in nickel tolerance in Thlaspi nickel hyperaccumulators. Plant Cell 16: 2176–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Shen J, Rosen BP (1999) Pathways of As(III) detoxification in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 96: 5001–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182: 319–326 [DOI] [PubMed] [Google Scholar]
- Gumaelius L, Lahner B, Salt D, Banks JA (2004) Arsenic hyperaccumulation in gametophytes of Pteris vittata: a new model system for analysis of arsenic hyperaccumulation. Plant Physiol 136: 3198–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen AC, Kelley R, Collins JB, Tucker CJ, Deng C, Afshari CA, Brown JM, Ideker T, Houten V (2004) Integrating phenotypic and expression profiles to map arsenic-response networks. Genome Biol 5: R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff RH, Wu L, Zhou B, Zhang ZY, Hengge AC (1999) Does positive charge at the active sites of phosphatases cause a change in mechanism? The effect of the conserved arginine on the transition state for phosphoryl transfer in the protein-tyrosine phosphatase from Yersinia. J Am Chem Soc 121: 9514–9521 [Google Scholar]
- Hokura A, Omuma R, Terada Y, Kitajima N, Abe T, Saito H, Yoshida S, Nakai I (2006) Arsenic distribution and speciation in an arsenic hyperaccumulator fern by x-ray spectroscopy utilizing a synchrotron radiation source. J Anal At Spectrom 21: 321–328 [Google Scholar]
- Hughes MF (2002) Arsenic toxicity and potential mechanisms of action. Toxicol Lett 133: 1–16 [DOI] [PubMed] [Google Scholar]
- Jackson MD, Denu JM (2001) Molecular reactions of protein phosphatases: insights from structure and chemistry. Chem Rev 101: 2313–2340 [DOI] [PubMed] [Google Scholar]
- Kertulis GM, Ma LQ, MacDonald GE, Chen R, Winefordner JD, Cai Y (2005) Arsenic speciation and transport in Pteris vittata L. and the effects on phosphorus in the xylem sap. Environ Exp Bot 54: 239–247 [Google Scholar]
- Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120 [DOI] [PubMed] [Google Scholar]
- Kong B, Huang S, Wang W, Ma D, Qu X, Jiang J, Yang X, Zhang Y, Wang B, Cui B, et al (2005) Arsenic trioxide induces apoptosis in cisplatin-sensitive and -resistant ovarian cancer cell lines. Int J Gynecol Cancer 15: 872–877 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Landrieu I, da Costa M, de Veylder L, Dewitte F, Vandepoele K, Hassan S, Wieruszeski J-M, Faure JD, Montagu MV, Inze D, et al (2004. a) A small CDC25 dual-specificity tyrosine-phosphatase isoform in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 13380–13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrieu I, Hassan S, Sauty M, Dewitte F, Wieruszeski J-M, Inze D, De Veylder L, Lippens G (2004. b) Characterization of the Arabidopsis thaliana Arath;CDC25 dual-specificity tyrosine phosphatase. Biochem Biophys Res Commun 322: 734–739 [DOI] [PubMed] [Google Scholar]
- Laparra JM, Velez D, Barbera R, Farre R, Montoro R (2005) Bioavailability of inorganic arsenic in cooked rice: practical aspects for human health risk assessments. J Agric Food Chem 53: 8829–8833 [DOI] [PubMed] [Google Scholar]
- Lemaire SD (2004) The glutaredoxin family in oxygenic photosynthetic organisms. Photosynth Res 79: 305–318 [DOI] [PubMed] [Google Scholar]
- Liu Z, Shen J, Carbrey JM, Mukhopadhyay R, Agre P, Rosen BP (2002) Arsenite transport by mammalian aquaglyceroporins AQP7 and AQP9. Proc Natl Acad Sci USA 99: 6053–6058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombi E, Zhao F-J, Fuhrman M, Ma L, McGrath SP (2002) Arsenic distribution and speciation in the fronds of the hyperaccumulator Pteris vittata. New Phytol 156: 195–203 [DOI] [PubMed] [Google Scholar]
- Meharg AA (2003) Variation in arsenic accumulation-hyperaccumulation in ferns and their allies. New Phytol 157: 25–31 [DOI] [PubMed] [Google Scholar]
- Meharg AA (2004) Arsenic in rice: understanding a new disaster for South-East Asia. Trends Plant Sci 9: 415–417 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R, Rosen BP (1998) Saccharomyces cerevisiae ACR2 gene encodes an arsenate reductase. FEMS Microbiol Lett 168: 127–136 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R, Rosen BP (2001) The phosphatase C(X)5R motif is required for catalytic activity of the Saccharomyces cerevisiae Acr2p arsenate reductase. J Biol Chem 276: 34738–34742 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R, Shi J, Rosen BP (2000) Purification and characterization of ACR2p, the Saccharomyces cerevisiae arsenate reductase. J Biol Chem 275: 21149–21157 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R, Zhou Y, Rosen BP (2003) Directed evolution of a yeast arsenate reductase into a protein-tyrosine phosphatase. J Biol Chem 278: 24476–24480 [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Papoyan A, Kochian LV (2004) Identification of Thlaspi caerulescens genes that may be involved in heavy metal hyperaccumulation and tolerance: characterization of a novel heavy metal transporting ATPase. Plant Physiol 136: 3814–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pence NS, Larsen PB, Ebbs SD, Letham DL, Lasat MM, Garvin DF, Eide D, Kochian LV (2000) The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens. Proc Natl Acad Sci USA 97: 4956–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persans MW, Nieman K, Salt DE (2001) Functional activity and role of cation-efflux family members in Ni hyperaccumulation in Thlaspi goesingense. Proc Natl Acad Sci USA 98: 9995–10000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering IJ, Prince RC, George MJ, Smith RD, George GN, Salt DE (2000. a) Reduction and coordination of arsenic in Indian mustard. Plant Physiol 122: 1171–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering IJ, Prince RC, Salt DE, George GN (2000. b) Quantitative, chemically specific imaging of selenium transformation in plants. Proc Natl Acad Sci USA 97: 10717–10722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab A, Feldman J, Meharg AA (2004) The nature of arsenic-phytochelatin complexes in Holcus lanatus and Pteris cretica. Plant Physiol 134: 1113–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves RD, Baker AJM (1984) Studies on metal uptake by plants from serpentine and non-serpentine populations of Thlaspi goesingense Ha'la'csy (Cruciferae). New Phytol 98: 191–204 [DOI] [PubMed] [Google Scholar]
- Rosen BP (2002) Biochemistry of arsenic detoxification. FEBS Lett 529: 86–92 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Salmeen A, Barford D (2005) Functions and mechanisms of redox regulation of cysteine-based phosphatases. Antioxid Redox Signal 7: 560–575 [DOI] [PubMed] [Google Scholar]
- Salt DE, Smith RD, Raskin I (1998) Phytoremediation. Annu Rev Plant Physiol Plant Mol Biol 49: 643–668 [DOI] [PubMed] [Google Scholar]
- Sanz MA, Fenaux P, Lo Coco F (2005) Arsenic trioxide in the treatment of acute promyelocytic leukemia: a review of current evidence. Haematologica 90: 1231–1235 [PubMed] [Google Scholar]
- Shi H, Shi X, Liu K-J (2004) Oxidative mechanisms of arsenic toxicity and carcinogenesis. Mol Cell Biochem 255: 67–78 [DOI] [PubMed] [Google Scholar]
- Shi J, Vlamis-Gardikas A, Aslund F, Holmgren A, Rosen BP (1999) Reactivity of glutaredoxins 1, 2, and 3 from Escherichia coli shows that glutaredoxin2 is the primary hydrogen donor to ArsC-catalyzed arsenate reduction. J Biol Chem 274: 36039–36042 [DOI] [PubMed] [Google Scholar]
- Snaith HA, Marlett J, Forsburg SL (2003) Ibp1p, a novel Cdc25-related phosphatase, suppresses Schizosaccharomyces pombe hsk1 (cdc7). Curr Genet 44: 38–48 [DOI] [PubMed] [Google Scholar]
- Sorrell DA, Chrimes D, Dickinson JR, Rogers HJ, Francis D (2005) The Arabidopsis CDC25 induces a short cell length when overexpressed in fission yeast: evidence for cell cycle functions. New Phytol 165: 425–428 [DOI] [PubMed] [Google Scholar]
- Streuli M, Krueger NX, Thai T, Tang M, Saito H (1990) Distinct functional roles of the two intracellular phosphatase like domains of the receptor-linked protein tyrosine phosphatases LCA and LAR. EMBO J 9: 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tongbin C, Chaoyang W, Zechun H, Qifei H, Quanguo L, Zilian F (2002) Arsenic hyperaccumulator Pteris vittata L. and its arsenic accumulation. Chin Sci Bull 47: 902–905 [Google Scholar]
- Visoottiviseth P, Francesconi K, Sridokchan W (2002) The potential of Thai indigenous plant species for the phytoremediation of arsenic contaminated land. Environ Pollut 118: 453–461 [DOI] [PubMed] [Google Scholar]
- Wang J, Zhao F-J, Meharg AA, Raab A, Feldman J, McGrath SP (2002) Mechanisms of arsenic hyperaccumulation in Pteris vittata: uptake kinetics, interactions with phosphate, and arsenic speciation. Plant Physiol 130: 1552–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangkarn S, Pergantis SA (2000) High-speed separation of arsenic compounds using narrow-bore high performance liquid chromatography on-line with inductively coupled plasma mass spectrometry. J Anal At Spectrom 15: 627–633 [Google Scholar]
- Webb SM, Gaillard JF, Ma L, Tu C (2003) XAS speciation of arsenic in a hyperaccumulating fern. Environ Sci Technol 37: 754–760 [DOI] [PubMed] [Google Scholar]
- Weber M, Harada E, Vess C, Roepenack-Lahaye E, Clemens S (2004) Comparative microarray analysis of Arabidopsis thaliana and Arabidopsis halleri roots identifies nicotianamine synthase, a ZIP transporter and other genes as potential metal hyperaccumulation factors. Plant J 37: 269–281 [DOI] [PubMed] [Google Scholar]
- Willsky GR, Malamy MH (1980) Characterization of two genetically separable inorganic phosphate transport systems in Escherichia coli. J Bacteriol 144: 356–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki R, Clemens S, Augustyniak D, Golik P, Maciaszczyk E, Tamas MJ, Dziadkowiec D (2003) Metalloid tolerance based on phytochelatins is not functionally equivalent to the arsenite transporter Acr3p. Biochem Biophys Res Commun 304: 293–300 [DOI] [PubMed] [Google Scholar]
- Wysocki R, Fortier P-K, Maciaszczyk E, Thorsen M, Leduc A, Odhagen A, Owsianik G, Ulaszewski S, Ramotor D, Tamas MJ (2004) Transcriptional activation of metalloid tolerance genes in Saccharomyces cerevisiae requires the AP-1-like proteins Yap1p and Yap8p. Mol Biol Cell 15: 2049–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yompakdee C, Bun-ya M, Shikata K, Ogawa N, Harashima S, Oshima Y (1996) A putative new membrane protein, Pho86p, in the inorganic phosphate uptake system of Saccharomyces cerevisiae. Gene 171: 41–47 [DOI] [PubMed] [Google Scholar]
- Ze-Chun H, Tong-Bin C, Lei M, Tian-Dou H (2004) Direct determination of arsenic species in arsenic hyperaccumulator Pteris vittata by EXAFS. Acta Bot Sin 46: 46–50 [Google Scholar]
- Zhang ZY, Wang Y, Wu L, Fauman EB, Stuckey JA, Schubert HL, Saper MA, Dixon JE (1994) The Cys(X)5Arg catalytic motif in phosphoester hydrolysis. Biochemistry 33: 15266–15270 [DOI] [PubMed] [Google Scholar]
- Zhao FJ, Wang JR, Barker JHA, Schat H, Bleeker PM, McGrath SP (2003) The role of phytochelatins in arsenic tolerance in the hyperaccumulator Pteris vittata. New Phytol 159: 403–410 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Bhattacharjee H, Mukhopadhyay R (2006) Bifunctional role of the leishmanial antimonate reductase LmACR2 as a protein tyrosine phosphatase. Mol Biochem Parasitol doi/10.1016/j.molbiopara.2006.03.009 [DOI] [PubMed]