Abstract
Recent studies have shown that cyclin-dependent kinase (CDK) inhibitors can have a tremendous impact on cell cycle progression in plants. In animals, CDK inhibitors are tightly regulated, especially by posttranslational mechanisms of which control of nuclear access and regulation of protein turnover are particularly important. Here we address the posttranslational regulation of INHIBITOR/INTERACTOR OF CDK 1 (ICK1)/KIP RELATED PROTEIN 1 (KRP1), an Arabidopsis (Arabidopsis thaliana) CDK inhibitor. We show that ICK1/KRP1 exerts its function in the nucleus and its presence in the nucleus is controlled by multiple nuclear localization signals as well as by nuclear export. In addition, we show that ICK1/KRP1 localizes to different subnuclear domains, i.e. in the nucleoplasm and to the chromocenters, hinting at specific actions within the nuclear compartment. Localization to the chromocenters is mediated by an N-terminal domain, in addition we find that this domain may be involved in cyclin binding. Further we demonstrate that ICK1/KRP1 is an unstable protein and degraded by the 26S proteasome in the nucleus. This degradation is mediated by at least two domains indicating the presence of at least two different pathways impinging on ICK1/KRP1 protein stability.
Eukaryotic cells encompass many different subcellular environments, and subcellular targeting is an important mechanism for regulating protein function in plants and animals (Merkle, 2003; Meier, 2005; Pemberton and Paschal, 2005). A paradigm for the importance of intracellular localization is the control of DNA replication and mitosis through the cell cycle by cyclin-dependent kinases (CDKs) in conjunction with their cyclin cofactors (Pines, 1999). CDK activity needs to be directed to many different targets during a cell cycle. For instance, to initiate a new round of DNA synthesis, one of the substrates of CDK-cyclin complexes are the prereplication complexes located at the origins of replication. During prophase CDK-cyclin complexes are targeted to chromatin to initiate chromosome condensation and later in mitosis CDK-cyclin activity is required at the spindle poles.
However, the localization of CDK activity is not the only control mechanism at work. The access of regulatory factors to CDKs or to CDK targets is also tightly controlled and represents an additional mechanism to regulate cell cycle progression as seen in the controlled import of the human CDK inhibitor p27Kip1 into the nucleus. Upon growth factor binding to membrane-associated receptors such as insulin, one branch of the triggered signaling cascade results in the activation of the Ser/Thr kinase Akt1/PKBα (Liang and Slingerland, 2003; Osaki et al., 2004). Among other targets, Akt1/PKBα phosphorylates residue Thr-157 in the nuclear localization sequence (NLS) of p27Kip1, creating a binding site for a 14-3-3 protein. 14-3-3 binding competes with the interaction of p27Kip1 with Importin α, and thus, sequesters phosphorylated p27Kip1 in the cytoplasm where it cannot inhibit CDK activity (Fujita et al., 2002, 2003; Sekimoto et al., 2004).
Moreover, the degradation of p27Kip1 is specific to particular cell compartments. One pathway acts in the nucleus and involves the phosphorylation of Thr-187 by CDK2-cyclin E. Phosphorylated p27Kip1 is recognized and subsequently marked for degradation by a Skp2-containing multisubunit Skp1-Cdc53/cullin-F-box (SCF) E3 ubiquitin-protein ligase complex (Carrano et al., 1999; Tsvetkov et al., 1999; Nakayama et al., 2001; Hengst, 2004). Another pathway of p27Kip1 degradation is independent of Thr-187 phosphorylation and takes place in the cytoplasm mediated by the E3 ubiquitin ligase Kip-ubiquitination-promoting complex (KPC; Kamura et al., 2004).
CDK inhibitors have also been identified in a number of plant species. All of them share a short sequence motif with p27Kip1 in their CDK- and cyclin-binding site and are therefore named KIP RELATED PROTEINS (KRP) or INHIBITOR/INTERACTOR OF CDK (ICK; Verkest et al., 2005b). ICK/KRPs are bona fide CDK inhibitors since in kinase assays performed with plant extracts or with in vitro translated proteins ICK/KRPs block A- and B-type CDK activity (Zhou et al., 2003; Coelho et al., 2005; Verkest et al., 2005a; Bisbis et al., 2006; Nakai et al., 2006; Pettko-Szandtner et al., 2006). Cell cycle control in plants appears to be very sensitive to ICK/KRP levels, and recent studies provide evidence that different cell cycle programs were executed depending on the ICK/KRP dosage. In plants strongly overexpressing ICK1/KRP1 or ICK2/KRP2, entry into both S and M phases was blocked, whereas in weakly overexpressing lines the entry into mitosis was preferentially inhibited (Verkest et al., 2005a; Weinl et al., 2005).
The posttranslational regulation of ICK/KRPs appears to be important since the accumulation of transcript does not match protein abundance, as seen for the Arabidopsis (Arabidopsis thaliana) ICK2/KRP2 and the maize (Zea mays) KRP2 (Coelho et al., 2005; Verkest et al., 2005a). However, in spite of their potential role during the plant cell cycle, not much is known about the posttranslational regulation of ICK/KRPs. Previously, it has been shown that ICK2/KRP2 is an unstable protein (Verkest et al., 2005a). Conversely, ICK1/KRP1109–191 appeared on western blots of whole plant extracts to be more stable than the full-length ICK1/KRP1 protein (Zhou et al., 2003; Weinl et al., 2005). ICK1/KRP1109–191 also appears to be a more potent inhibitor than the full-length ICK1/KRP1 protein. This was revealed by the observation that trichomes on plants misexpressing ICK1/KRP1109–-191 under the control of the GLABRA2 (GL2) promoter displayed a stronger phenotype with fewer branches than trichomes expressing the full-length ICK1/KRP1 protein (Schnittger et al., 2003). Concomitantly, a yellow fluorescent protein (YFP) fusion with ICK1/KRP1109–191 was detected not only in the nucleus but also in the cytoplasm of trichomes (Weinl et al., 2005). The same relationship between phenotypic strength, cellular localization, and protein abundance was obtained by Zhou and coworkers (Zhou et al., 2003). The combined data suggest an intricate relationship between ICK/KRP nuclear localization, protein abundance, and protein function.
Here, we set out to unravel this relationship and to investigate the posttranslational regulation of plant CDK inhibitors. Similar to animals, the subcellular localization and protein abundance of ICK1/KRP1 from Arabidopsis appear to be tightly controlled and our data show that the nuclear compartment takes a central role in ICK/KRP action and regulation.
RESULTS
ICK1/KRP1 Exerts Its Function in the Nucleus and Its Access to the Nucleus Is Controlled by Multiple Import Pathways
To understand how the subcellular localization and function of ICK1/KRP1 are regulated, we first searched for sequence motifs present in its open reading frame. Previously, putative NLS had been identified based on sequence comparisons only for ICK2/KRP2, KRP4, KRP5, and KRP7 (De Veylder et al., 2001).
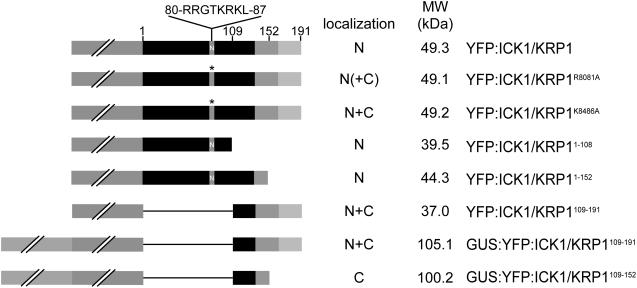
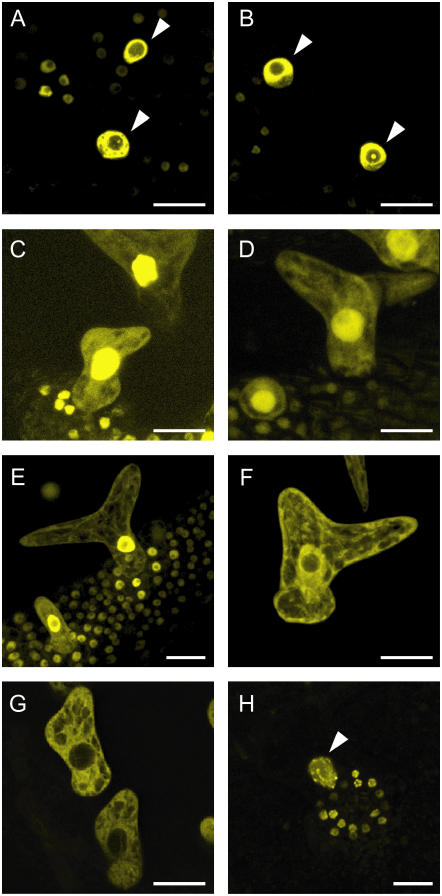
Searching the PredictNLS server (http://cubic.bioc.columbia.edu/predictNLS/; Cokol et al., 2000) we retrieved a putative NLS spanning residues 80 to 87 in ICK1/KRP1 (Table I). To test the functionality of this predicted NLS, two pairs of conserved Lys and/or Arg residues were replaced by Ala to create two different constructs with mutant NLS (Table I). Next, transgenic plants were generated expressing a fusion of the YFP with each of these two ICK1/KRP1 mutant proteins under the control of the trichome-directed GL2 promoter. We have previously shown that full-length ICK1/KRP1 fused to YFP gives rise to an exclusively nuclear fluorescent signal (Figs. 1 and 2A; Weinl et al., 2005).
Table I.
Predicted NLS in ICK1/KRP1 and here generated mutant variants
[KR], K or R; [KR]{2,3}, between 2 and 3 Ks or Rs; x, any amino acid. PredictNLS server is at http://cubic.bioc.columbia.edu/predictNLS/. Amino acid sequence present in ICK1/KRP1 that shows similarity to the predicted NLS sequence is given in bold face. Amino acid residues that have been mutated to interfere with the NLS are underlined.
| Object | Amino Acid Sequence |
|---|---|
| Consensus 1 | [KR]{2,3}xxKR[KR][QLM] |
| Consensus 2 | RRxxKRK |
| ICK1/KRP1 wild type | 75-ETSTYRRGTKRKLFENLREEE-95 |
| R80/81A | 75-ETSTYAAGTKRKLFENLREEE-95 |
| K84/86A | 75-ETSTYRRGTARALFENLREEE-95 |
Figure 1.
Overview of ICK1/KRP1 constructs analyzed in planta. ICK1/KRP1 was fused to the C terminus of YFP or to a GUS-YFP fusion (both reporters represented by a gray box with two slashes). The position of the NLS from amino acids R80 to L87 is marked with N and the amino acid sequence is given on top. The mutated NLS is marked by an asterisk. The cyclin- and CDK-binding sites at the C terminus of ICK1/KRP1 are highlighted by a medium and light gray box, respectively. The subcellular localization of the fusion constructs is given as nuclear (N) and/or cytoplasmic (C), parentheses indicate weak localization.
Figure 2.
Subcellular localization of ICK1/KRP1 in trichomes (A–H). Confocal laser-scanning micrographs of Arabidopsis trichomes on young rosette leaves. A, In plants expressing YFP:ICK1/KRP1 under control of the GL2 promoter the YFP signal is only detectable in the nucleus. B, Most plants expressing the NLS mutant version YFP:ICK1/KRP1R80/81A show only nuclear fluorescence. C, In a few transgenic lines a weak YFP signal can be also detected in the cytoplasm by increasing the gain setting of the detector. D, All transgenic lines expressing YFP:ICK1/KRP1K84/86A show a YFP signal in the cytoplasm as well as in the nucleus. E and F, A nuclear and cytoplasmic localization of YFP fluorescence can be seen in plants expressing ProGL2:YFP:ICK1/KRP1109–191 and ProGL2:GUS:YFP:ICK1/KRP1109–191, respectively. G, In plants expressing ProGL2:GUS:YFP:ICK1/KRP1109–152 the YFP signal is excluded from the nucleus. H, Misexpression of YFP:ICK1/KRP11–108 results in an exclusively nuclear localization; note the patchy appearance of the YFP signal in the nuclei of trichomes and the surrounding socket cells. Arrowheads in A, B, and H indicate trichome nuclei. Scale bars in A to H represent 20 μm.
The two mutations placed in the putative NLS affected to varying degrees the nuclear localization of ICK1/KRP1. YFP fusion proteins with the mutated ICK1/KRP1R80/81A predominantly localized in the nucleus although some fluorescence could be detected in the cytoplasm in a number of trichomes in a few transgenic lines, indicating that nuclear transport was only slightly affected (Figs. 1 and 2, B and C). In contrast, fusions with the KRP1K84/86A occurred in both the cytoplasm and the nucleus in all transgenic lines analyzed, demonstrating the presence of a functional NLS between the amino acid residues K80 and L87 (Fig. 2D; Table I).
Since a major fraction of the fusion protein still localized to the nucleus in plants expressing YFP:ICK1/KRP1K84/86A, we searched the ICK1/KRP1 sequence for another motif that could be responsible for nuclear import. We have previously shown that YFP:ICK1/KRP1109–191 and also a β-glucuronidase (GUS):YFP:ICK1/KRP1109–191 fusion protein with a mass of about 105 kD were still able to enter the nucleus (Figs. 1 and 2, E and F; Weinl et al., 2005). Since the threshold for passive entry into the nucleus is about 50 kD (Gorlich and Kutay, 1999; Merkle, 2003), one conclusion from this experiment is that, besides the NLS at positions 80 to 87, a second NLS is present in the C-terminal part of ICK1/KRP1, i.e. between residues 109 and 191. To further characterize this NLS, we generated plants expressing the ICK1/KRP1 residues 109 to 152 combined with a GUS:YFP fusion (Fig. 1). This fusion protein was solely detectable in the cytoplasm (Fig. 2G), suggesting that a second signal resides between residues 152 and 191, where the cyclin- and the CDK-interaction domains of ICK1/KRP1 are also situated.
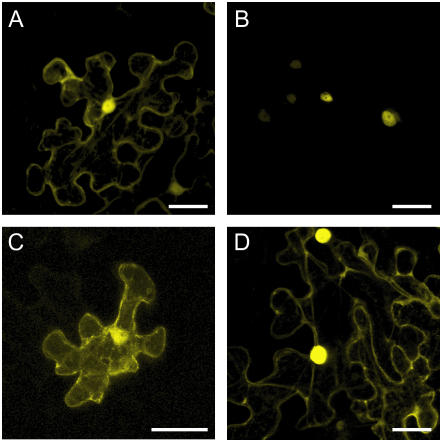
Since no NLS could be identified with various prediction tools in this part of the protein, one explanation could be that ICK1/KRP1 binds to a CDK-cyclin complex in the cytoplasm and travels piggyback with this complex into the nucleus. To explore such a cotransport mechanism further, we investigated in which cell compartment ICK1/KRP1 might interact with a CDK-cyclin complex by using the bimolecular fluorescence complementation (BiFC) technique (also called split YFP assay; Walter et al., 2004). As an interacting partner of ICK1/KRP1 we chose CDKA;1. CDKA;1 fused to a complete YFP is able to rescue the cdka;1 mutant phenotype (M. Nowack, N. Dissmeyer, and A. Schnittger, unpublished data). Consistent with previous studies on functional green fluorescent protein (GFP):CDKA;1 fusions, CDKA;1 fused to the complete YFP was found in both the nucleus and the cytoplasm (Fig. 3A; Weingartner et al., 2001). Interaction of CDKA;1 with ICK1/KRP1 in the BiFC assays was only detectable in nuclei, in agreement with the strong nuclear localization signal of ICK1/KRP1 identified above (Fig. 3B). However, the two mutant ICK/KRP versions, ICK1/KRP1109–191 and ICK1/KRP1K84/86A, were found to bind to CDKA;1 in both the cytoplasm and the nucleus, consistent with a cotransport of ICK1/KRP1 with a CDK-cyclin complex from the cytoplasm into the nucleus (Fig. 3, C and D).
Figure 3.
Subcellular interaction of ICK1/KRP1 with CDKA;1 (A–D). Confocal laser-scanning micrographs of the abaxial surface of Nicotiana benthamiana leaves. A, Pro35S:CDKA;1:YFP transient expression in a N. benthamiana leaf. The YFP signal can be detected in the cytoplasm and the nucleus. B, BiFC signal of CDKA;1 and the full-length ICK1/KRP1 fluorescence can be detected exclusively in the nucleus. C, BiFC signal of CDKA;1 and ICK1/KRP1109–191 can be detected in the cytoplasm and the nucleus. D, BiFC signal of CDKA;1 and ICK1/KRP1K84/86A fluorescence can be detected in the cytoplasm and the nucleus. Scale bars in A to D represent 40 μm.
Next we asked whether the different localization patterns of ICK1/KRP1 protein variants are dependent on the cell cycle program or the developmental background. Previously, a different response was observed upon ICK1/KRP1 expression in dividing cells of the stomata lineage and in endoreplicating trichomes (Weinl et al., 2005). Therefore, we generated transgenic plants expressing all generated ICK1/KRP1 mutants from the TOO MANY MOUTH (TMM) promoter that is active in cells of the stomata lineage of which most are still dividing (Supplemental Fig. 1A; Nadeau and Sack, 2002). Production of YFP:ICK1/KRP1R80/81A from the TMM promoter also resulted in predominantly nuclear localization of the fusion protein and production of YFP:ICK1/KRP1K84/86A again gave rise to both nuclear and cytoplasmic fluorescence. The YFP:ICK1/KRP1109–191 and GUS:YFP:ICK1/KRP1109–191 fusion proteins were also found in both the nucleus and the cytoplasm, whereas a GUS:YFP:ICK1/KRP1109–152 fusion protein driven from the TMM promoter was excluded from the nucleus (Supplemental Fig. 1, B–H). Similar results were also obtained by transient expression of the above mentioned fusion constructs in Arabidopsis leaves under the control of the ubiquitous 35S promoter (data not shown). Taken together, these results demonstrate that the nuclear localization of ICK1/KRP1 is independent of the cellular setting.
Finally, we addressed whether the altered subcellular localization also results in an altered protein function. Plants producing the GUS:YFP:ICK/KRP1109–152 fusion protein under the control of the GL2 or the TMM promoter had a trichome and leaf morphology identical to that of wild-type plants (data not shown). These data are consistent with previous experiments in which the expression of a mutant ICK1/KRP11–152 allele lacking the cyclin and the CDK interaction domain (amino acids 152–191) did not alter the typical trichome morphology of three to four branches (Schnittger et al., 2003). In contrast, plants expressing YFP:ICK1/KRP1R80/81A or YFP:ICK1/KRP1K84/86A under the control of the GL2 promoter generally displayed the previously described ICK1/KRP1 misexpression phenotype with smaller and less branched trichomes (Schnittger et al., 2003; Table II; data not shown). However, the phenotype of the NLS mutated YFP:ICK1/KRP1K84/86A misexpression lines was significantly weaker (χ2 test, P ≤ 0.001) than that of the nonmutated YFP:ICK1/KRP1 lines; the weaker phenotype can best be seen in the distribution of the three-branched trichomes: approximately 42% of trichomes with only three branches in the ProGL2:YFP:ICK1/KRP1K84/86A lines versus approximately 16% in ProGL2:YFP:ICK1/KRP1 (Table II). This shows that a compromised nuclear import reduces ICK1/KRP1 action and suggests that its targets reside in the nucleus.
Table II.
Trichome branch numbers
| Line | Average No. of Trichome Branches in Percentages per Leafa
|
nb | |||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| Landsberg erecta ecotype | 0.0 ± 0.0 | 0.5 ± 1.4 | 95.2 ± 10.3 | 4.3 ± 10.3 | 20/402 |
| Columbia ecotype | 0.0 ± 0.0 | 0.0 ± 0.0 | 93.2 ± 4.4 | 6.8 ± 4.4 | 20/507 |
| ProGL2:YFP:ICK1/KRP1 (#1 and #2)cef | 18.4 ± 7.4 | 65.6 ± 9.0 | 16.0 ± 6.3 | 0.0 ± 0.0 | 18/484 |
| 18.0 ± 9.8 | 64.8 ± 9.5 | 17.2 ± 7.2 | 0.0 ± 0.0 | 20/521 | |
| ProGL2:YFP:ICK1/KRP1K84/86A (#1 and #2)cef | 5.7 ± 4.3 | 52.4 ± 11.0 | 41.9 ± 9.7 | 0.0 ± 0.0 | 19/453 |
| 4.7 ± 5.9 | 50.1 ± 10.9 | 45.2 ± 14.0 | 0.0 ± 0.0 | 17/327 | |
| ProGL2:YFP:ICK1/KRP1109–191 (#1 and #2)d | 15.5 ± 6.6 | 65.3 ± 8.8 | 19.2 ± 7.7 | 0.0 ± 0.0 | 20/392 |
| 10.0 ± 7.5 | 58.1 ± 12.4 | 31.9 ± 11.7 | 0.0 ± 0.0 | 19/407 | |
| ProGL2:GUS:YFP:ICK1/KRP1109–191 (#1 and #2)d | 23.8 ± 7.4 | 61.8 ± 10.8 | 14.4 ± 7.2 | 0.0 ± 0.0 | 20/330 |
| 12.2 ± 7.0 | 60.7 ± 9.6 | 27.1 ± 10.5 | 0.0 ± 0.0 | 20/428 | |
| ProGL2:YFP:ICK1/KRP11–108d | 0.0 ± 0.0 | 1.4 ± 2.7 | 97.0 ± 4.8 | 1.6 ± 3.8 | 19/438 |
| ProGL2:NLS:GFP:GUSd | 0.0 ± 0.0 | 3.3 ± 4.4 | 96.5 ± 4.7 | 0.2 ± 0.9 | 20/335 |
| ProGL2:ICK1/KRP1d | 14.1 ± 9.4 | 59.7 ± 11.0 | 26.2 ± 14.1 | 0.0 ± 0.0 | 18/168 |
| ProGL2:ICK1/KRP1109–191d | 12.6 ± 11.8 | 64.5 ± 15.2 | 22.9 ± 10.9 | 0.0 ± 0.0 | 20/161 |
| ProGL2:YFP:ICK1/KRP11–108 ×ProGL2:ICK1/KRP1d | 0.0 ± 0.0 | 8.1 ± 5.8 | 91.9 ± 5.8 | 0.0 ± 0.0 | 20/406 |
| ProGL2:NLS:GFP:GUS ×ProGL2:ICK1/KRP1d | 0.3 ± 1.5 | 16.8 ± 7.4 | 82.9 ± 7.9 | 0.0 ± 0.0 | 18/293 |
| ProGL2:YFP:ICK1/KRP11–108 × ProGL2:ICK1/KRP1109–191d | 1.7 ± 2.9 | 53.3 ± 12.1 | 45.0 ± 12.1 | 0.0 ± 0.0 | 20/385 |
| ProGL2:NLS:GFP:GUS ×ProGL2:ICK1/KRP1109–191d | 3.1 ± 4.4 | 56.6 ± 9.2 | 40.3 ± 10.8 | 0.0 ± 0.0 | 20/315 |
All trichomes on rosette leaf numbers 3 and 4 were counted from at least 10 plants per line, the average ± sd is given, the branch number with the highest percentage is shown in bold.
Total numbers of counted leaves/trichomes.
In Columbia-0 accession.
In Landsberg erecta accession.
χ2 tests for ProGL2:YFP:ICK1/KRP1 line 1 and ProGL2:YFP:ICK1/KRP1K84/86A line 1 or line 2 retrieved χ2 numbers of 89.512 and 96.272, respectively; with this both genotypes are significantly different, P ≤ 0.001.
χ2 tests for ProGL2:YFP:ICK1/KRP1 line 2 and ProGL2:YFP:ICK1/KRP1K84/86A line 1 or line 2 retrieved χ2 numbers of 90.549 and 96.781, respectively; with this both genotypes are significantly different, P ≤ 0.001.
The N Terminus of ICK1/KRP1 Contains Not Only an NLS But Also Mediates a Specific Intranuclear Localization Pattern and May Interact with CYCLIN D3;1
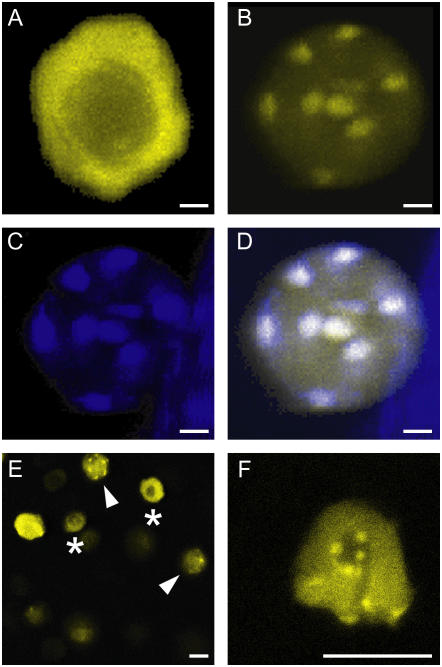
To explore the function of the N-terminal part of ICK1/KRP1 further, plants were generated that expressed C- and N-terminal fusions of YFP to the first 108 amino acids of ICK1/KRP1 under control of the GL2 promoter (Fig. 1). Since this N-terminal part of the ICK1/KRP1 protein harbors the above identified NLS, we expected to find YFP fluorescence in the nucleus. Indeed, only a nuclear YFP signal was detected for both constructs (Fig. 2H; data not shown). Interestingly, YFP fluorescence showed a specific subnuclear localization pattern. The full-length ICK1/KRP1 protein fused to YFP is absent from the nucleolus but otherwise accumulates uniformly in the nucleus (Figs. 2A and 4A). In contrast, both ICK1/KRP11–108:YFP and YFP:ICK1/KRP11–108 displayed a punctuate localization in the nucleus (Fig. 4B; data not shown). Simultaneous staining with the DNA-specific dye 4′,6-diamino-phenylindole (DAPI) showed that the YFP signal completely overlaps with areas of high DNA compaction, the so-called chromocenters (Fig. 4, C and D). This localization pattern could be a unique property of the N terminus of ICK1/KRP1. Alternatively, the full-length protein might have different subnuclear localization domains and may accumulate in the nucleoplasm so strongly that other more subtle localization patterns are obscured. To discriminate between these two scenarios, we analyzed in detail the subnuclear localization pattern of the full-length ICK1/KRP1 in all cells targeted by our expression constructs, i.e. stomata cells, hypocotyl cells, and root cells. Indeed, we found a few cells in which the full-length fusion protein showed the same punctuate localization pattern within the nucleus (Fig. 4, E and F). Intriguingly, this association of the full-length fusion protein with chromocenters was only found in a few dividing cells and never in endoreplicating trichome cells, suggesting that the subnuclear localization depends on the cell cycle program and/or cell cycle phase.
Figure 4.
Subnuclear localization of ICK1/KRP1 (A–F). Confocal laser-scanning micrographs of plants expressing ProGL2:YFP:ICK1/KRP1 (A, E, and F) and ProGL2:YFP:ICK1/KRP11–108 (B–D). A, Apart from the nucleolus the fluorescence of YFP:ICK1/KRP1 is evenly distributed in trichome nuclei. B, In contrast, YFP:ICK1/KRP11–108 localizes to some patches in trichome nuclei. C, Staining of the same nucleus shown in B with DAPI; the bright spots are the chromocenters. D, Overlay of the YFP and DAPI signals from B and C, demonstrating that YFP:ICK1/KRP11–108 localizes to chromocenters in trichome nuclei. E, Epidermis of a root expressing ProGL2:YFP:ICK1/KRP1 containing nuclei with different fluorescence patterns; asterisks indicate nuclei with homogenous fluorescence as seen in A, arrowheads indicate nuclei with a punctuate appearance as seen in B. F, Close up of a nucleus of a root expressing ProGL2:YFP:ICK1/KRP1 with a punctuate fluorescence pattern. Scale bars in A to F represent 10 μm.
Next, ProGL2:YFP:ICK1/KRP11–108 and ProGL2:ICK1/KRP11–108:YFP plants were analyzed for their phenotypes. Both transgenic lines displayed trichomes of similar size and branch number in comparison to wild-type plants (Table II; data not shown). To explore a possible function of the N-terminal part of ICK1/KRP1 we crossed plants expressing ProGL2:YFP:ICK1/KRP11–108 and ProGL2:ICK1/KRP11–108:YFP to various other trichome mutants and misexpression lines. Remarkably, when we combined plants expressing ProGL2:YFP:ICK1/KRP11–108 with plants expressing ProGL2:ICK1/KRP1, the KRP-misexpression phenotype was slightly reduced (Table II). In control crosses of the ProGL2:ICK1/KRP1 misexpressing plants with plants expressing ProGL2:NLS:GFP:GUS no phenotypic reduction could be observed (Table II). However, the reduction of the KRP-misexpression phenotype could only be seen in crosses with plants expressing the full-length ICK1/KRP1 protein and not the truncated ICK1/KRP1109–191(Table II). One possible explanation for this finding is that the N-terminal part of ICK1/KRP1 might function as an intramolecular inhibitor and binds to itself. Intramolecular inhibitory domains have been identified in a number of proteins, for instance in caspases that only become active after a prodomain has been cleaved off (Cohen, 1997). However, yeast (Saccharomyces cerevisiae) two-hybrid interaction assays did neither reveal interactions of the full-length protein ICK1/KRP1 with itself nor with the N-terminal part ICK1/KRP11–108 (data not shown).
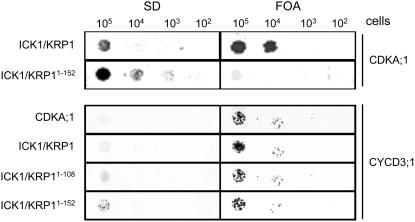
Another possible explanation is that the N terminus of ICK1/KRP1 is involved in substrate binding. If so, the expression of only the N terminus may compete with binding of the ICK1/KRP1 full-length protein to the targets and thus reduce the phenotype of ICK1/KRP1 misexpression. To test this, we analyzed the interaction of ICK1/KRP1, ICK1/KRP11–152, and ICK1/KRP11–108 with CDKA;1 and CYCLIN D3;1 (CYCD3;1) in a split ubiquitin yeast two-hybrid system. Consistent with previous experiments reported by Wang et al. (1998) we also observed in this yeast assay that the full-length ICK1/KRP1 protein interacted with CDKA;1, whereas ICK1/KRP11–152 did not bind to CDKA;1 (Wang et al., 1998); in addition, we found that ICK1/KRP11–108 did not interact with CDKA;1. In contrast to this previous study, we found that not only the full-length ICK1/KRP1 interacted with CYCD3;1 but also ICK1/KRP11–152 and remarkably ICK1/KRP11–108 (Fig. 5). This suggests that in addition to the previously identified cyclin- and CDK-binding domain between the amino acids 152 and 191, the N terminus also harbors a cyclin-binding site.
Figure 5.
Interaction of ICK1/KRP1 with CYCD3;1. The left column shows yeast cells growing on SD medium without uracil, the right column shows yeast cells on SD media plates containing 5-FOA (FOA). Protein-protein interaction results in growth on FOA but not on SD plates; no interaction results in growth on SD but not on FOA. All yeast lines grew on SD with uracil (data not shown). Prey vectors (pNui) are listed on the left, bait vectors (pMet) on the right. CDKA;1 interacts with ICK1/KRP1 but not with ICK1/KRP11–152. In contrast, CYCD3;1 interacts with all three ICK1/KRP1 variants, full-length ICK1/KRP1, ICK1/KRP11–152, and ICK1/KRP11–108.
ICK1/KRP1 Is Subject to Nuclear Export
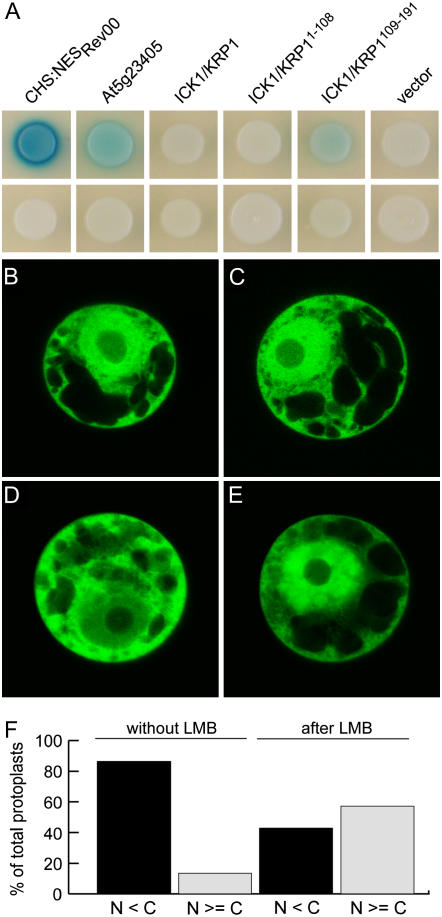
Since presence in the nucleus seems to be crucial for ICK1/KRP1 function, we wondered whether ICK1/KRP1 is also controlled by active nuclear export similar to animal CDK inhibitors (Connor et al., 2003). First, full-length ICK1/KRP1 and ICK1/KRP1109–191 were tested for interaction with the Arabidopsis nuclear export receptor EXPORTIN 1 (XPO1) in yeast two-hybrid interaction assays. As positive controls we used a fragment of the HIV Rev protein containing a nuclear export signal and the Arabidopsis protein At5g23405 that has recently been shown to interact with XPO1 (Haasen et al., 1999; Grasser et al., 2006).
While the full-length protein did not interact, we could detect a weak but significant interaction for ICK1/KRP1109–191 with XPO1 (Fig. 6A). Conversely, no interaction of XPO1 with the N-terminal 108 amino acids of ICK1/KRP1 could be observed (Fig. 6A). To examine a possible regulation of ICK1/KRP1 by nuclear export we transfected tobacco (Nicotiana tabacum) Bright-Yellow 2 protoplasts with Pro35S:YFP, Pro35S:YFP:ICK1/KRP1R80/81A, Pro35S:YFP:ICK1/KRP1K84/86A, Pro35S:YFP:ICK1/KRP1109–191, and Pro35S:GUS:YFP:ICK1/KRP1109–191 and treated them with the nuclear export inhibitor leptomycin B (LMB). In all experiments the Pro35S:YFP control did not display an altered subcellular localization pattern after LMB treatment (Fig. 6, B and C). In contrast, ICK1/KRP1 exhibited sensitivity to LMB. The effect was visible for all constructs tested but most pronounced in protoplasts transfected with Pro35S:GUS:YFP:ICK1/KRP1109–191 (Fig. 6, D and E). Here, the fraction of cells with a nuclear fluorescence equal to or stronger than their cytoplasmic fluorescence increased from 14% in untreated protoplasts to 57% after treatment with LMB (Fig. 6F). Thus, we conclude that the plant CDK inhibitor ICK1/KRP1 can also be subject to nuclear export.
Figure 6.
Interaction of ICK1/KRP1 with the nuclear export machinery (A). Yeast strain EGY48[p8op-lacZ] was cotransformed with pGilda bait and pB42AD prey plasmids as indicated and transformed yeast cells were replated on indicator plates containing Gal and X-gal to assay protein interactions. Top row, yeast colonies containing XPO1 in the bait vector and the indicated prey constructs; bottom row, yeast colonies transformed with an empty bait vector and the indicated prey constructs. The two positive controls, a fragment of the HIV Rev protein containing the NES (NESRev) and At5g23405, displayed a strong and medium-strong interaction with XPO1, respectively. Whereas ICK1/KRP1 and ICK1/KRP11–108 did not interact with XPO1, a weak but significant and reproducible interaction of XPO1 with ICK1/KRP1109–191 was detected. B to F, Nuclear export by XPO1 was assayed in tobacco Bright-Yellow 2 protoplasts using the inhibitor LMB. B and C, Protoplasts transfected with either YFP or GUS:YFP:ICK1/KRP1109–191 (D and E) were incubated with ethanol (B and D; 0.7% final concentration) or with LMB (C and E; 0.5 μm in ethanol) for 1 h and analyzed for YFP fluorescence using confocal laser-scanning microscopy. While YFP localization did not change during the treatment, the localization of GUS:YFP:ICK1/KRP1109–191 changed to a more nuclear localization after incubation with LMB. F, Protoplasts producing GUS:YFP:ICK1/KRP1109–191 were analyzed for the percentage of those that showed equal or more YFP fluorescence in the nucleus than in the cytoplasm without (8 out of 59) or with LMB treatment (36 out of 63).
ICK1/KRP1 Is Degraded in the Nucleus Facilitated by an N-Terminal Domain
After seeing that ICK1/KRP1 can be regulated by nuclear import and export, we next asked whether this subcellular localization is linked to protein turnover. Previous studies suggested that ICK1/KRP1 might be an unstable protein that could be regulated by protein degradation (Zhou et al., 2003; Weinl et al., 2005). In particular, the accumulation of ICK1/KRP1109–191 protein in the cytoplasm as well as in the nucleus gave rise to the hypothesis that ICK1/KRP1 might be degraded in the cytoplasm mediated by a motif in the N-terminal region.
To test this hypothesis, we blocked the 26S proteasome with the drug MG132 in plants expressing ICK1/KRP1. Since the Arabidopsis root is readily accessible to the application of drugs and the GL2 promoter is also active in roots we chose to use the root system to address the control of ICK1/KRP1 by protein degradation.
First we attempted to detect a possible accumulation of YFP-tagged ICK1/KRP1 after treatment with MG132 by using an antibody against GFP that also recognizes YFP. However, ICK/KRPs appear to be low abundance proteins and consistent with previous experiments we were unable to detect a band for the full-length ICK1/KRP1 in protein extracts from Arabidopsis root in western blots (data not shown). We therefore explored the possibility to quantify fluorescence signals in planta as a more sensitive detection assay.
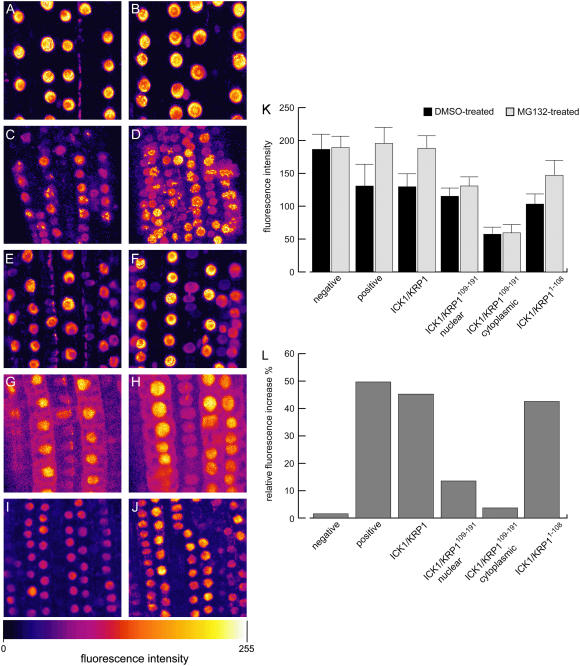
As controls we used previously generated plants expressing ProCPC:SV40NLS:2xGFP or ProGL2:NLS:GFP:GUS (Schnittger et al., 2002; Wada et al., 2002). Both reporter lines showed no altered expression of the fusion proteins after application of MG132 (Fig. 7, A and B). To obtain a positive control, we fused YFP to the destruction box of the mitotic cyclin CYCLIN B1;1 that mediates anaphase-promoting complex/cyclosome-dependent degradation via the 26S proteasome (King et al., 1996; Genschik et al., 1998). This construct was expressed under the control of the CDKA;1 promoter that has been shown to rescue a cdka;1 mutant when driving the CDKA;1 cDNA (Nowack et al., 2006). The CDKA;1 promoter is active in all cells competent to undergo cell cycle progression (M. Nowack, N. Dissmeyer, and A. Schnittger, unpublished data). After treating roots of ProCDKA;1:YFP:DB expressing plants with MG132 we observed a strong increase in nuclear fluorescence of many root cells (Fig. 7, C and D).
Figure 7.
Degradation of ICK1/KRP1 in nuclei (A–J). False color confocal laser-scanning micrographs in false colors of Arabidopsis roots; color scale giving at the bottom. Left column shows roots treated with DMSO only, right column displays roots treated with MG132 in DMSO. A and B, In the negative control, NLS:GFP:GUS expressed from the GL2 promoter shows no altered subcellular distribution or increased fluorescence when treated with DMSO, n = 142 (A) in comparison to treatment with MG132, n = 142 (B). C and D, The positive control, YFP:DB expressed from the CDKA;1 promoter, strongly accumulates after treatment with MG132, nDMSO = 117 cells and nMG132 = 113 cells. E and F, YFP:ICK1/KRP1 accumulates in nuclei after treatment with MG132, nDMSO = 334 cells and nMG132 = 439 cells; no accumulation in the cytoplasm was found. G and H, YFP:ICK1/KRP1109–191 displays a slight accumulation of nuclear fluorescence, nDMSO = 216 cells and nMG132 = 235 cells; no cytoplasmic accumulation could be observed, nDMSO = 179 cells and nMG132 = 189 cells. I and J, The N terminus of ICK1/KRP1, ICK1/KRP11–108, strongly accumulates after treatment with MG132, nDMSO = 614 cells and nMG132 = 645 cells. K, Quantification of nuclear and, if applicable, cytoplasmic fluorescence of the above shown untreated (DMSO only) or treated (MG132 in DMSO) roots. Student's t tests were performed to analyze the statistical significance of the observed distributions: No statistical significant change was found for the DMSO-treated versus MG132-treated negative control (P = 0.7285). Also, no significant difference could be found for the fluorescence in the cytoplasm of the solvent- versus the MG132-treated YFP:ICK1/KRP1109–191 lines (P = 0.7957). For all other genotypes a statistically significant difference could be found between the DMSO and the MG132 treatments (P ≤ 0.001). L, Normalization of the fluorescent intensities shown in K; the fluorescence intensities observed with the DMSO treatment were set to 100% and the relative increase by the MG132 treatment is given in percentages. For ICK1/KRP1, 439 nuclei of MG132-treated roots and 334 nuclei of DMSO-treated roots were analyzed. For ICK1/KRP1109–191, the nuclear fluorescence of 235 nuclei of MG132-treated and 216 nuclei of DMSO-treated roots and the cytoplasm of 189 MG132- and 179 DMSO-treated cells was quantified. For ICK1/KRP11–108, 645 nuclei of MG132- and 614 nuclei of DMSO-treated roots were analyzed. For the negative control 142 nuclei of MG132- and 142 nuclei of DMSO-treated roots were analyzed. For the positive control 113 nuclei of MG132- and 117 nuclei of DMSO-treated plants were analyzed.
Next, we treated the roots of different lines misexpressing the various ICK1/KRP1 mutant versions with MG132 or solvent only (dimethyl sulfoxide [DMSO]) for 3 h and quantified nuclear and cytoplasmic fluorescence. Plants expressing ProGL2:YFP:ICK1/KRP1 displayed a stronger YFP signal in the nucleus after MG132 treatment (Fig. 7, E and F); notably, no cytoplasmic accumulation of YFP fluorescence was found. Nuclear accumulation of the fusion proteins was confirmed by quantifying fluorescence intensities (Fig. 7, K and L). Whereas the fluorescence in the negative controls, ProCPC:SV40NLS:2xGFP and ProGL2:NLS:GFP:GUS expressing plants, increased only by 1.5%, in YFP:ICK1/KRP1 expressing plants the signal increased by 45% in roots treated with MG132 (Fig. 7, K and L).
To address whether or not the N terminus is involved in the degradation of ICK1/KRP1, we next applied MG132 to roots of plants producing ICK1/KRP1109–191 and ICK1/KRP11–108 fused to YFP. For plants producing YFP:ICK1/KRP1109–191 we also found accumulation of nuclear fluorescence after MG132 treatment (Fig. 7, G and H). Even though this enhancement was much weaker than in lines producing the full-length YFP:ICK1/KRP1, with an increase in fluorescence intensity of only 13.6%, it was statistically significantly different from DMSO-treated roots of the same line (t test, P ≤ 0.001). Remarkably, no significant cytoplasmic accumulation could be found (3.7%; t test, P = 0.7957; Fig. 7, K and L). In contrast, YFP:ICK1/KRP11–108 strongly accumulated in nuclei upon MG132 treatment, leading to a 42.6% increase in fluorescence intensity, similar to the full-length YFP:ICK1/KRP1 protein (Fig. 7, I–L).
Taken together, these results provide evidence for ICK1/KRP1 degradation in the nucleus being facilitated by the first 108 amino acids in ICK1/KRP1. However, the observation that ICK1/KRP1109–191 also accumulated after MG132 treatment suggests that multiple degradation signals are present in ICK1/KRP1.
DISCUSSION
In this study, we have addressed the posttranslational regulation of ICK1/KRP1, i.e. the interrelationship between its subcellular localization, function, and protein turnover. In addition, we identified domains that are critical to the function and posttranscriptional control of ICK1/KRP1. An emerging theme is that substrate binding as well as protein localization and abundance are regulated by multiple pathways, suggesting that CDK inhibitors are tightly controlled in plants. These manifold regulatory pathways might offer plants the possibility to fine tune CDK activity throughout growth and development.
The Nuclear Localization of ICK1/KRP1
Whereas the human p27Kip1 protein contains a classic bipartite NLS, none of the Arabidopsis ICK/KRPs show this canonical motif (De Veylder et al., 2001; Sekimoto et al., 2004). Previously, an NLS could be predicted for only four out of the seven members of the Arabidopsis KRP family (ICK2/KRP2, KRP4, KRP5, and KRP7; De Veylder et al., 2001). However, to our knowledge, nothing is known about the actual subcellular localization pattern of these four inhibitors in vivo.
Here, we have functionally characterized an NLS for ICK1/KRP1 residing between the amino acid residues 80 to 87 based on the web service predict NLS (Cokol et al., 2000). However, for the remaining two KRP members, KRP3 and KRP6, we could not pinpoint a putative NLS with this prediction tool. However, ICK1/KRP1 also has at least one other less clearly defined NLS since a GUS:YFP fusion containing ICK1/KRP1109–191 that lacks the NLS at residues 80 to 87 localized to the nucleus; its Mr of 105 kD is much too high to explain nuclear entry simply by diffusion. The second NLS in ICK1/KRP1 appears to overlap with the CDK-cyclin binding domain, rendering a functional analysis difficult. At the moment, we favor a scenario in which the N-terminally truncated ICK1/KRP1 is cotransported into the nucleus via piggybacking on a CDK-cyclin complex. Such a cotransport mechanism is consistent with our observation that YFP:ICK1/KRP1109–191 can bind to CDKA;1 in the cytoplasm. In contrast to the C-terminal NLS, we observed that the N-terminal signal is very strong. Various attempts to exclude ICK1/KRP1 from the nucleus, for instance by fusing it with the rat glucocorticoid receptor, were unsuccessful (S. Pusch and A. Schnittger, unpublished data). This strong NLS may have functional relevance by driving target proteins, e.g. a CDK-cyclin complex, into the nucleus. A function in promoting nuclear import has been found for the animal CDK inhibitor p57Kip2 that can bind and translocate the LIM Kinase-1 from the cytoplasm into the nucleus (Yokoo et al., 2003).
In the nucleus, ICK1/KRP1 has a distinct localization pattern and can be found in at least two subdomains, namely the nucleoplasm and at the chromocenters. Chromocenters are heterochromatic regions, often located around the centromeres (Fransz et al., 2000). Interestingly, other cell cycle regulators are also present at these regions; for example, YFP fusions with the mitotic cyclin CYCLIN B1;1 also localize to chromocenters (Fig. 7C; F. Roodbarkelari, N. Dissmeyer, and A. Schnittger, unpublished data). The centromeres are known from animal and yeast systems to be focal points of cell cycle regulation, especially during spindle checkpoint control (Lew and Burke, 2003; Chan et al., 2005). At the moment not much is known about the spindle checkpoint in plants and it will be interesting to find out what the function of the chromocenter association of the N-terminal KRP1 fragment or CYCLIN B1;1 is.
The localization to chromocenters appears to be mediated by a domain in the N terminus of ICK1/KRP1. Intriguingly, we found that the N terminus of ICK1/KRP1 can bind to CYCD3;1 in yeast two-hybrid assays. Possibly, this domain may participate in binding to the target, e.g. a CDK-cyclin complex. Since ICK1/KRP1 can bind to cyclins without this N-terminal domain (Wang et al., 1998; S. Pusch and A. Schnittger, unpublished data), this region might function to discriminate between different substrates or affect binding kinetics. Alternatively, this domain might be involved in mediating a possible regulation of ICK1/KRP1 by a CDK-cyclin complex. Recently, it has been found that the CDK inhibitor ICK2/KRP2 from Arabidopsis is regulated by B-type CDKs (see below; Verkest et al., 2005a).
Nuclear versus Cytoplasmic Functions of ICK1/KRP1
In animals, the general consensus is that CDK inhibitors exert their function in the nucleus and the control of nuclear access represents an important mechanism of CDK inhibitor regulation. Disturbances of the import and export mechanisms can have dramatic consequences. For instance in many cancer variants, the Akt kinase pathway is up-regulated and one outcome is that p27Kip1 is not imported into the nucleus and thus cannot execute its antiproliferative function; in fact, cytoplasmic localization of p27Kip1 indicates a poor prognosis in cancer patients (Viglietto et al., 2002; Min et al., 2004; Rosen et al., 2005).
We show that ICK1/KRP1 already has the potential to bind to a CDK-cyclin complex in the cytoplasm. However, our data suggest that ICK1/KRP1 mainly functions in the nucleus in common with animal CDK inhibitors. This was seen in plants expressing a ICK1/KRP1 protein version that had a compromised NLS and concomitantly displayed a weaker phenotype than the full-length ICK1/KRP1 version. An exclusive role of ICK1/KRP1 in the nucleus is consistent with its function as CDK inhibitor at the G1-S and the G2-M checkpoints since the regulatory actions during these stages are predominantly nuclear.
We observed that ICK1/KRP1 action might be controlled by export from the nucleus into the cytoplasm. However, we could only find evidence for regulation by nuclear export with the N-terminally truncated ICK1/KRP1 version, and it remains to be seen whether, and under what circumstances, the full-length ICK1/KRP1 is exported from the nucleus. One possible scenario is that the export signal in ICK1/KRP1 is only exposed after a conformational change, as seen in the nuclear export of human p27Kip1, which depends on phosphorylation of Ser-10 (Ishida et al., 2002). Candidate regulators of ICK/KRPs by phosphorylation are B-type CDKs, and it has previously been found that phosphorylation of ICK2/KRP2 by CDKB1;1 interferes with its ability to inhibit CDKA;1 kinase activity (Verkest et al., 2005a). Conversely, Pettko-Szandtner et al. (2006) have shown that phosphorylation of the Medicago inhibitor KRPMt by the Ca2+-dependent MsCPK3 kinase resulted in reduced CDK activity (Pettko-Szandtner et al., 2006). However, whether subcellular localization or other aspects of ICK/KRP function are affected by these phosphorylations remains to be determined; altered protein turnover is likely to be another consequence of modifying the phosphorylation status of ICK/KRPs.
Degradation of ICK1/KRP1
In animals, p27Kip1 is degraded via two different pathways. One pathway is found in the cytoplasm and involves the action of the recently identified KPC ubiquitin ligase (Kamura et al., 2004). However, we could not find an obvious homolog of KPC in the Arabidopsis genome, indicating that this regulatory pathway might not be conserved between animals and plants. The second degradation pathway in animals relies on the action of an Skp1-Cdc53/cullin-F-box-SKP2 complex and takes place in the nucleus (Carrano et al., 1999; Tsvetkov et al., 1999).
The Arabidopsis genome contains several genes with sequence similarities to the animal SKP2 gene. However, the similarity mostly results from Leu-rich repeats present in the human and Arabidopsis proteins. Moreover, none of the amino acids from the human SKP2 known to be crucial for mediating the degradation of p27Kip1 are conserved in the Arabidopsis proteins, i.e. amino acids involved in binding of the cofactor CKS and for making contact with p27Kip1 (Hao et al., 2005). In addition, ICK/KRPs and p27Kip1 share only a stretch of about 20 amino acid residues containing the cyclin- and the CDK-binding domain. Thr-187, which is crucial for Skp2-mediated degradation, is not conserved. Thus, it remains to be seen which E3 ligase is involved in ICK/KRP degradation and which cofactors are required.
Here we have demonstrated that the N-terminal 108 amino acid residues harbor a degradation signal. As discussed above, this part of the protein also mediates localization to the chromocenters when fused to YFP. Thus, it is possible that this localization pattern represents a negative stain, i.e. that ICK1/KRP1 is rapidly degraded in the nucleoplasm but is protected at the chromocenters. In addition to this N-terminal degradation domain, there might be at least one more motif that could be involved in degradation since ICK1/KRP1109–191 was also slightly stabilized in our MG132 experiments. Consistently, we found a much stronger signal for YFP or GFP than for the truncated ICK1/KRP1 protein version ICK1/KRP1109–191, fused to YFP in western blots (data not shown). Thus, the regulation of ICK1/KRP1 stability appears to be quite complex, possibly involving multiple degradation signals, and it promises to be an exciting task to unravel the underlying regulatory pathways controlling ICK1/KRP1 abundance.
MATERIALS AND METHODS
Plant Material, Growth Conditions, Plant Transformation, and Crosses of Transgenic Lines
Arabidopsis (Arabidopsis thaliana) plants were grown under long-day conditions (16 h of light, 8 h of darkness) between 18°C and 25°C under standard greenhouse conditions. The Arabidopsis accessions Landsberg erecta and Columbia-0 were used as wild-type controls. At least 20 transgenic plants were generated for all expression constructs. A number of representative reference lines displaying a typical phenotype were chosen for further analysis. The same transgenic reference line was used for all data obtained for one expression construct. For ProGL2:YFP:ICK1/KRP1R80/81A, ProGL2:YFP:ICK1/KRP1K84/86A, ProTMM:YFP:ICK1/KRP1R80/81A, and ProTMM:YFP:ICK1/KRP1K84/86A, approximately 50% of the approximately 100 primary transformants resembled the ICK1/KRP1 misexpression phenotype. T2 plants were tested for segregation and used for analysis. ProGL2:ICK1/KRP11–108:YFP, ProGL2:YFP:ICK1/KRP11–108, ProTMM:YFP:ICK1/KRP1, ProTMM:ICK1/KRP11–108:YFP, ProTMM:YFP:ICK1/KRP11–108, ProTMM:GFP5ER, and ProCDKA;1:YFP:DB expressing T2 plants were tested for segregation and homozygous T3 plants were established and analyzed. More than 100 T1 transgenic plants of ProGL2:GUS:YFP:ICK1/KRP1109–152 and ProTMM:GUS:YFP:ICK1/KRP1109–152 were generated, analyzed for their phenotype, and for each construct at least 10 lines were checked for their YFP localization. The ProGL2:ICK1/KRP1, ProGL2:ICK1/KRP1109–191, ProGL2:YFP:ICK1/KRP1, ProGL2:YFP:ICK1/KRP1109–191, ProGL2:GUS:YFP:ICK1/KRP1109–191, ProGL2:NLS:GFP:GUS, ProTMM:YFP:ICK1/KRP1109–191, and ProCPC:SV40NLS:2xGFP expression lines have been described previously (Wada et al., 2002; Schnittger et al., 2003; Weinl et al., 2005). Plants were transformed by a modified version of the floral dipping method according to Clough and Bent (1998). Flower buds were submerged for approximately 10 s in 500 mL Agrobacterium tumefaciens overnight culture to which 25 g Suc and 100 μL Silwet L-77 had been added. Plants were kept under a plastic cover overnight and then allowed to grow under standard greenhouse conditions. For crosses between transgenic lines homozygous T3 plants were used and the F1 generation was analyzed. For analysis of roots, Arabidopsis seeds were sterilized with chlorine gas overnight and grown in liquid culture medium (one-half Murashige and Skoog, 1% Suc) with rotation (50 rpm) for 5 d.
Expression Constructs
All primer sequences used for cloning are listed in Supplemental Table I. To achieve expression in plants, the previously described plant transformation vectors pAM-PAT-GWPro35S (GenBank AY436765), pAM-PAT-GWProGL2, and pAM-PAT-GWProTMM were used (Weinl et al., 2005). To generate ICK1/KRP11–108:YFP and YFP:ICK1/KRP11–108 fusion constructs, gene-specific primers C-ICK_108-S and C-ICK_108-AS were used for the cDNAs of ICK1/KRP1 and primers C-YFP_108-AS and C-YFP_108-S were used for EYFP-N1 (CLONTECH), and a two-step PCR was performed. The upper primers contained a BamHI and the lower primers a SacI recognition site. After the second PCR step the obtained fragments were subcloned into pGEM-T vector (Promega), sequenced, and transferred via BamHI and SacI into a modified pENTR1A vector (Invitrogen) containing a SacI restriction site in its MCS. Thereafter a recombination into the respective plant transformation vector was performed using Gateway LR Clonase enzyme mix (Invitrogen). The putative NLS in ICK1/KRP1 was mutated using the QuikChange site-directed mutagenesis kit (Stratagene). For the exchange of R80 and R81 to A, primers krpR8081Ar and krpR8081Af were used, for the exchange of K84 and K86 to A, krpK8486Ar and krpK8486Af were used with pENTR1a:YFP:ICK1/KRP1 (Weinl et al., 2005) as a template following the manufacturer's recommendations. The fusion construct GUS:YFP:ICK1/KRP1109–152 was amplified as a subfragment from GUS:YFP:ICK1/KRP1109–191 (Weinl et al., 2005) with an attB1 GUS primer and attB2_krp1_152 and cloned into pDONR201. For the construction of the Gateway compatible BiFC plant expression vector pSYN and the minimal promoter and GUS gene of pATGUS, a BASTA-resistant derivative of pPAM (GenBank AY027531), were deleted by digestion with SapI/SacI and replaced with the 35S promoter, the Gateway cassette, a Myc tag, and the N-terminal part of YFP1–155 of pUC-SPYNEG (Walter et al., 2004). For pSYC the 35S minimal promoter and the Gateway cassette of pBENDER, a kanamycin-resistant derivative of pPAM were excised by digestion with SdaI/AvaIII and replaced with the 35S promoter, the Gateway cassette, the HA-tag, and the C-terminal part of YFP156–239 of pUC-SPYCEG (Walter et al., 2004). To introduce ICK1/KRP1, ICK1/KRP1109–191, ICK1/KRP1K84/86A, and CDKA;1 into the Split-Ubiquitin assay vectors (pMet and pNuI) as well as into the BiFC assay vectors (pSYN and pSYC) STOP codons of the respective cDNAs had to be removed. The cDNAs were amplified via PCR using the following primers: AttB1-KRP1 and AttB2-KRP1 on the pENTR1a:YFP:ICK1/KRP1 template, primers AttB1-KRP1 109 to 191 and AttB2-KRP1 on the pENTR1a:YFP:ICK1/KRP1 template, primers AttB1-KRP1and AttB2-KRP1 on the pENTR1a:YFP:ICK1/ KRP1K84/86A template, primers AttB1-CDKA1 and AttB2-CDKA1 on the CDKA;1 cDNA (Nowack et al., 2006), and primers AttB1-CYCD3,1 and AttB2-CYCD3,1 on the CYCD3;1 cDNA template (pART61; Schnittger et al., 2002). To generate the CDKA;1:YFP fusion construct, gene-specific primers ss_attB1:CDKcoreN and as_CDKcoreC:YFPovlpN were used on the CDKA;1 cDNA and primers ss_YFPcoreN:CDKovlpC and as_YFPcoreC:attB2 were used for EYFP-N1 and a two-step PCR was performed. The fusion construct was cloned into pDONR201 via a Gateway BP reaction, sequenced, and transferred into pAM-PATGW-ProCDKA;1 (Nowack et al., 2006).
MG132 Treatment
Five-day-old seedlings grown in liquid medium were treated with 50 μg/mL MG132 (Sigma and Biomol; dissolved to 50 mg/mL in DMSO) or with 0.1% DMSO alone as control. Whole seedlings were submersed in media and incubated for 3 h at 20°C in the light. Fluorescence intensity measurements were performed on CLSM image stacks using Leica LCSlite 2.0 (Leica Microsystems) and Amira 3.1.1 (Mercury Computer Systems) software.
BiFC Assays
For infiltration of Nicotiana benthamiana leaves the A. tumefaciens strain GV3101 pMP90RK was used. The Agrobacterium strains containing the BiFC vectors were infiltrated as described (Walter et al., 2004). Infiltration was always performed on the abaxial leaf side of 2-month-old tobacco (Nicotiana tabacum) plants and analyzed 3 to 5 d later.
Gal4 Yeast Two-Hybrid Tests
Yeast (Saccharomyces cerevisiae) two-hybrid assays were performed as described in Haasen et al. (1999). Yeast strain EGY48[p8op-lacZ] was transformed with pGilda bait and pB42AD prey plasmids (CLONTECH), and interactions were tested on induction plates containing X-gal (5-Bromo-4-chloro-3-indolyl β-d-galactopyranoside) according to the manufacturer's protocol. The plasmids pGilda-XPO1 and pB42AD-NLS-CHS-NESRev containing the nuclear export signal of the HIV Rev protein as a positive control have been described in Haasen et al. (1999). The prey vectors were constructed by recombining pDONR vectors containing the cDNA fragments of ICK1/KRP1, ICK1/KRP11-108, and ICK1/KRP1109-191 into pB42AD-GWY. This vector was constructed by digestion of pB42AD with EcoRI and treatment with Klenow fragment. After ligation of the rfB Gateway cassette into the opened vector, in-frame insertion was verified by sequencing.
Split Ubiquitin Yeast Two-Hybrid Assays
The yeast strains used were JD53 and YM4271. The bait vector pMet (a kind gift of Laurent Deslandes and Imre E. Somssich) was transformed into JD53, the prey vector pNuI (a kind gift of Laurent Deslandes and Imre E. Somssich) into YM4271. After transformation yeast cells were streaked out on synthetic drop-out (SD) medium. The strains were mated (Mata:Matα, 2,5: 1) in YPAD (YPD plus adenine) medium for 3 h and plated on SD. A dilution series of the yeast strains starting with an OD600 of 1 (approximately 105 cells) were plated onto SD plates for growth control, on SD without uracil supplemented with 100 μm copper sulfate, and on SD supplemented with 1 mg/mL 5-FOA and 100 μm copper sulfate. Yeast cells were grown at 30°C for 3 d.
Transient Protoplast Transfection Assays with YFP Fusion Constructs
Protoplasts were prepared from dark-grown tobacco Bright-Yellow 2 cells and transiently transfected with the indicated plasmids by polyethylene glycol-mediated transfection as described previously (Haasen et al., 1999). Analysis of the localization of the YFP fusion proteins was performed using both fluorescence and laser-scanning microscopy in three independent experiments, representing approximately 60 to 80 transfected protoplasts. The effect of LMB on the localization of the GUS:YFP fusion containing ICK1/KRP1109–191 was analyzed by confocal laser-scanning microscopy in transfected Bright-Yellow 2 protoplasts that were incubated with 0.5 μm LMB for 1 h versus mock-treated protoplasts. Protoplasts in either sample were grouped into two categories according to the localization of the GUS:YFP fusion protein. Protoplasts grouped in category N ≤ C showed an approximately equal distribution of the GUS:YFP fusion protein between the cytoplasm and the nucleus, whereas protoplasts of category N > C showed a clearly higher YFP fluorescence in the nucleus than in the cytoplasm. In this way, the effect of LMB on the localization of the GUS:YFP:ICK1/KRP109–191 fusion protein could be assessed by comparison of the percentage of protoplasts grouped into the two categories.
Microscopy
Light microscopy was performed with an Axiophot microscope (Zeiss) or a Leica DM RA2 microscope (Leica) equipped with differential interference contrast (Nomarski) and epifluorescence optics using a JVC 3CCD camera. Confocal laser-scanning microscopy was performed with a Leica TCS SP2 AOBS CLSM system equipped with an argon-krypton laser and a 405 nm diode laser (Leica) or a Zeiss LSM 510 META equipped with an argon-krypton laser (Zeiss). Images were processed using Adobe Photoshop CS 8.0, Adobe Illustrator CS 11.0, and ImageJ 1.36 (http://rsb.info.nih.gov/ij/index.html).
Sequence data from this article are as follows: ICK1/KRP1 has the Arabidopsis gene code At2g23430 and GenBank accession number NM_127907; AtGL2 has the Arabidopsis gene code At1g79840 and GenBank accession number NM_106633; AtTMM has the Arabidopsis gene code At1g80080 and GenBank accession number NM_106657. pAM-PAT-GWPro35S is a gateway-compatible derivative of the pAMPAT MCS binary plant transformation vector (GenBank accession number AY436765). AtXPO1 has the gene code At5g17020 and GenBank accession number Y18469. CYCD3;1 has the gene code At4g34160 and GenBank accession number X83371. CDKA;1 has the gene code At3g48750 and GenBank accession number X57839.
Supplementary Material
Acknowledgments
The authors thank Tom Colby (Max-Planck-Institute for Plant Breeding Research, Cologne), Lieven DeVeylder (Flanders Interuniversity Institute for Biotechnology, Gent), Pascal Genschik (Institut de Biologie Moléculaire des Plantes, Strasbourg), Sebastian Marquardt, and Richard O'Connell (Max-Planck-Institute for Plant Breeding Research, Cologne) for critical reading and helpful comments on the manuscript. We thank Oliver Hofmann for his help in generating plant material used in this work. The authors thank Laurent Deslandes and Imre E. Somssich from the Max-Planck-Institute for Plant Breeding Research in Cologne for providing Gateway-based split ubiquitin vectors. The authors are grateful to Klaus Harter from the University of Tübingen for providing the pUC-SPYNEG and pUC-SPYCEG BiFC vectors. We thank Takuji Wada from the RIKEN Plant Science Center for providing ProCPC:SV40NLS:2xGFP seeds used in this analysis.
This work was supported by grants from the Deutsche Forschungsgemeinschaft SFB 572 and the Volkswagen-Stiftung (to A.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Arp Schnittger (schnitt@mpiz-koeln.mpg.de).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.081406.
References
- Bisbis B, Delmas F, Joubes J, Sicard A, Hernould M, Inze D, Mouras A, Chevalier C (2006) Cyclin-dependent kinase inhibitors regulate the CDK/cyclin complex activities in endoreduplicating cells of developing tomato fruit. J Biol Chem 281: 7374–7383 [DOI] [PubMed] [Google Scholar]
- Carrano AC, Eytan E, Hershko A, Pagano M (1999) SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol 1: 193–199 [DOI] [PubMed] [Google Scholar]
- Chan GK, Liu ST, Yen TJ (2005) Kinetochore structure and function. Trends Cell Biol 15: 589–598 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coelho CM, Dante RA, Sabelli PA, Sun Y, Dilkes BP, Gordon-Kamm WJ, Larkins BA (2005) Cyclin-dependent kinase inhibitors in maize endosperm and their potential role in endoreduplication. Plant Physiol 138: 2323–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen GM (1997) Caspases: the executioners of apoptosis. Biochem J 326: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokol M, Nair R, Rost B (2000) Finding nuclear localization signals. EMBO Rep 1: 411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor MK, Kotchetkov R, Cariou S, Resch A, Lupetti R, Beniston RG, Melchior F, Hengst L, Slingerland JM (2003) CRM1/Ran-mediated nuclear export of p27(Kip1) involves a nuclear export signal and links p27 export and proteolysis. Mol Biol Cell 14: 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inze D (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz PF, Armstrong S, de Jong JH, Parnell LD, van Drunen C, Dean C, Zabel P, Bisseling T, Jones GH (2000) Integrated cytogenetic map of chromosome arm 4S of A. thaliana: structural organization of heterochromatic knob and centromere region. Cell 100: 367–376 [DOI] [PubMed] [Google Scholar]
- Fujita N, Sato S, Katayama K, Tsuruo T (2002) Akt-dependent phosphorylation of p27Kip1 promotes binding to 14-3-3 and cytoplasmic localization. J Biol Chem 277: 28706–28713 [DOI] [PubMed] [Google Scholar]
- Fujita N, Sato S, Tsuruo T (2003) Phosphorylation of p27Kip1 at threonine 198 by p90 ribosomal protein S6 kinases promotes its binding to 14-3-3 and cytoplasmic localization. J Biol Chem 278: 49254–49260 [DOI] [PubMed] [Google Scholar]
- Genschik P, Criqui MC, Parmentier Y, Derevier A, Fleck J (1998) Cell cycle -dependent proteolysis in plants: identification of the destruction box pathway and metaphase arrest produced by the proteasome inhibitor mg132. Plant Cell 10: 2063–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Kutay U (1999) Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15: 607–660 [DOI] [PubMed] [Google Scholar]
- Grasser M, Lentz A, Lichota J, Merkle T, Grasser KD (2006) The Arabidopsis genome encodes structurally and functionally diverse HMGB-type proteins. J Mol Biol 358: 654–664 [DOI] [PubMed] [Google Scholar]
- Haasen D, Kohler C, Neuhaus G, Merkle T (1999) Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J 20: 695–705 [DOI] [PubMed] [Google Scholar]
- Hao B, Zheng N, Schulman BA, Wu G, Miller JJ, Pagano M, Pavletich NP (2005) Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Mol Cell 20: 9–19 [DOI] [PubMed] [Google Scholar]
- Hengst L (2004) A second RING to destroy p27(Kip1). Nat Cell Biol 6: 1153–1155 [DOI] [PubMed] [Google Scholar]
- Ishida N, Hara T, Kamura T, Yoshida M, Nakayama K, Nakayama KI (2002) Phosphorylation of p27Kip1 on serine 10 is required for its binding to CRM1 and nuclear export. J Biol Chem 277: 14355–14358 [DOI] [PubMed] [Google Scholar]
- Kamura T, Hara T, Matsumoto M, Ishida N, Okumura F, Hatakeyama S, Yoshida M, Nakayama K, Nakayama KI (2004) Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat Cell Biol 6: 1229–1235 [DOI] [PubMed] [Google Scholar]
- King RW, Glotzer M, Kirschner MW (1996) Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol Biol Cell 7: 1343–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Burke DJ (2003) The spindle assembly and spindle position checkpoints. Annu Rev Genet 37: 251–282 [DOI] [PubMed] [Google Scholar]
- Liang J, Slingerland JM (2003) Multiple roles of the PI3K/PKB (Akt) pathway in cell cycle progression. Cell Cycle 2: 339–345 [PubMed] [Google Scholar]
- Meier I (2005) Nucleocytoplasmic trafficking in plant cells. Int Rev Cytol 244: 95–135 [DOI] [PubMed] [Google Scholar]
- Merkle T (2003) Nucleo-cytoplasmic partitioning of proteins in plants: implications for the regulation of environmental and developmental signalling. Curr Genet 44: 231–260 [DOI] [PubMed] [Google Scholar]
- Min YH, Cheong JW, Kim JY, Eom JI, Lee ST, Hahn JS, Ko YW, Lee MH (2004) Cytoplasmic mislocalization of p27Kip1 protein is associated with constitutive phosphorylation of Akt or protein kinase B and poor prognosis in acute myelogenous leukemia. Cancer Res 64: 5225–5231 [DOI] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD (2002) Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Nakai T, Kato K, Shinmyo A, Sekine M (2006) Arabidopsis KRPs have distinct inhibitory activity toward cyclin D2-associated kinases, including plant-specific B-type cyclin-dependent kinase. FEBS Lett 580: 336–340 [DOI] [PubMed] [Google Scholar]
- Nakayama KI, Hatakeyama S, Nakayama K (2001) Regulation of the cell cycle at the G1-S transition by proteolysis of cyclin E and p27Kip1. Biochem Biophys Res Commun 282: 853–860 [DOI] [PubMed] [Google Scholar]
- Nowack MK, Grini PE, Jakoby MJ, Lafos M, Koncz C, Schnittger A (2006) A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat Genet 38: 63–67 [DOI] [PubMed] [Google Scholar]
- Osaki M, Oshimura M, Ito H (2004) PI3K-Akt pathway: its functions and alterations in human cancer. Apoptosis 9: 667–676 [DOI] [PubMed] [Google Scholar]
- Pemberton LF, Paschal BM (2005) Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6: 187–198 [DOI] [PubMed] [Google Scholar]
- Pettko-Szandtner A, Meszaros T, Horvath GV, Bako L, Csordas-Toth E, Blastyak A, Zhiponova M, Miskolczi P, Dudits D (2006) Activation of an alfalfa cyclin-dependent kinase inhibitor by calmodulin-like domain protein kinase. Plant J 46: 111–123 [DOI] [PubMed] [Google Scholar]
- Pines J (1999) Four-dimensional control of the cell cycle. Nat Cell Biol 1: E73–E79 [DOI] [PubMed] [Google Scholar]
- Rosen DG, Yang G, Cai KQ, Bast RC Jr, Gershenson DM, Silva EG, Liu J (2005) Subcellular localization of p27kip1 expression predicts poor prognosis in human ovarian cancer. Clin Cancer Res 11: 632–637 [PubMed] [Google Scholar]
- Schnittger A, Schobinger U, Bouyer D, Weinl C, Stierhof YD, Hulskamp M (2002) Ectopic D-type cyclin expression induces not only DNA replication but also cell division in Arabidopsis trichomes. Proc Natl Acad Sci USA 99: 6410–6415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger A, Weinl C, Bouyer D, Schobinger U, Hulskamp M (2003) Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell 15: 303–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimoto T, Fukumoto M, Yoneda Y (2004) 14-3-3 suppresses the nuclear localization of threonine 157-phosphorylated p27(Kip1). EMBO J 23: 1934–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov LM, Yeh KH, Lee SJ, Sun H, Zhang H (1999) p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr Biol 9: 661–664 [DOI] [PubMed] [Google Scholar]
- Verkest A, Manes CL, Vercruysse S, Maes S, Van Der Schueren E, Beeckman T, Genschik P, Kuiper M, Inze D, De Veylder L (2005. a) The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17: 1723–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest A, Weinl C, Inze D, De Veylder L, Schnittger A (2005. b) Switching the cell cycle: Kip-related proteins in plant cell cycle control. Plant Physiol 139: 1099–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viglietto G, Motti ML, Bruni P, Melillo RM, D'Alessio A, Califano D, Vinci F, Chiappetta G, Tsichlis P, Bellacosa A, et al (2002) Cytoplasmic relocalization and inhibition of the cyclin-dependent kinase inhibitor p27(Kip1) by PKB/Akt-mediated phosphorylation in breast cancer. Nat Med 8: 1136–1144 [DOI] [PubMed] [Google Scholar]
- Wada T, Kurata T, Tominaga R, Koshino-Kimura Y, Tachibana T, Goto K, Marks MD, Shimura Y, Okada K (2002) Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129: 5409–5419 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang H, Qi Q, Schorr P, Cutler AJ, Crosby WL, Fowke LC (1998) ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J 15: 501–510 [DOI] [PubMed] [Google Scholar]
- Weingartner M, Binarova P, Drykova D, Schweighofer A, David JP, Heberle-Bors E, Doonan J, Bogre L (2001) Dynamic recruitment of Cdc2 to specific microtubule structures during mitosis. Plant Cell 13: 1929–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinl C, Marquardt S, Kuijt SJ, Nowack MK, Jakoby MJ, Hulskamp M, Schnittger A (2005) Novel functions of plant cyclin-dependent kinase inhibitors, ICK1/KRP1, can act non-cell-autonomously and inhibit entry into mitosis. Plant Cell 17: 1704–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo T, Toyoshima H, Miura M, Wang Y, Iida KT, Suzuki H, Sone H, Shimano H, Gotoda T, Nishimori S, et al (2003) p57Kip2 regulates actin dynamics by binding and translocating LIM-kinase 1 to the nucleus. J Biol Chem 278: 52919–52923 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Li G, Brandizzi F, Fowke LC, Wang H (2003) The plant cyclin-dependent kinase inhibitor ICK1 has distinct functional domains for in vivo kinase inhibition, protein instability and nuclear localization. Plant J 35: 476–489 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.