Abstract
The genetic diversity of 123 Streptococcus suis strains of capsular types 2, 1/2, 3, 7, and 9, isolated from pigs in France and from humans in different countries, was evaluated by pulsed-field gel electrophoresis (PFGE) of DNA restricted with SmaI. The method was highly discriminative (D = 0.98), results were reproducible, and the PFGE analysis was easy to interpret. Among all S. suis strains, 74 PFGE patterns were shown. At 60% homology, three groups (A, B, and C) were identified, and at 69% homology, eight subgroups (a to h) were observed. Strains isolated from diseased pigs or from humans were statistically clustered in group B, especially in subgroup d. By contrast, S. suis strains isolated from clinically healthy pigs were preferentially included in subgroup b of group A. Relationships could be established between capsular types 1/2, 3, and 9 and groups A, e, and B, respectively. S. suis strains isolated from humans were homogeneous, and a very high level of association between these strains and four DNA patterns was observed. The PFGE used in this study is a very useful tool for evaluating the genetic diversity of S. suis strains, and it would be used for epidemiological investigations.
Streptococcus suis is recognized as an important swine pathogen worldwide and is associated with cases of meningitis, arthritis, septicemia, and sudden death (12, 21). Pigs can also be clinically healthy carriers, and S. suis has been isolated from the upper respiratory tract, nasal cavities, and palatine tonsils of these animals (2). Thirty-five capsular types have been described for this microorganism, with serotype 2 being the most prevalent capsular type in France, followed by types 1/2, 3, 9, and 7 (4). S. suis is also a zoonotic agent, responsible for meningitis and septicemia in humans (12).
Virulence markers, such as muramidase released protein, extracellular factor, and suilysin (7, 23), were described for S. suis, but their presence does not always correlate with virulence of S. suis strains (4, 8). Other virulence factors have been suggested, but their role in the pathogenesis of the infection has not been demonstrated (4, 10).
Molecular typing of S. suis was previously carried out to define the diversity of strains and to distinguish virulent from nonvirulent strains. Multilocus enzyme electrophoresis was used with Australian capsular type 2 strains to determine association between genetic patterns and specific isolates responsible for clinical problems in piggeries (15). Canadian S. suis type 2 isolates were also investigated by restriction endonuclease analysis, followed by hybridization with a ribosomal DNA probe to detect correlation between the source of the isolates and DNA patterns (3, 11). The 16S rRNA of S. suis was also studied to elicit relationships between virulence of S. suis strains (types 2 and 1) and ribotypes (18, 20). A random amplified polymorphic DNA (RAPD) analysis was described for S. suis to define whether strains from diseased pigs exhibited particular RAPD patterns (6). These studies have described the diversity among S. suis isolates, and some relationships between genetic patterns and isolates specific to clinical problems were shown, but such results have been obtained by studying limited parts of the bacterial genome. More recently, a macrorestriction of the whole S. suis genome associated with pulsed-field gel electrophoresis (PFGE) was used to study German S. suis isolated from swine (1). The present study employs PFGE analysis with French strains isolated from swine and from human cases of meningitis in different countries.
MATERIALS AND METHODS
S. suis strains.
Ninety-seven epidemiologically unrelated S. suis strains isolated from swine and 26 strains isolated from human meningitis cases worldwide were studied. S. suis strains of capsular types 2, 1/2, 9, 7, and 3 and autoagglutinable strains were isolated from pigs suffering from meningitis, septicemia, or arthritis (n = 67) and from nasal cavities or palatine tonsils (n = 30) of clinically healthy pigs. The S735 reference strain capsular type 2 obtained from M. Gottschalk, Faculté de Médecine Vétérinaire, Université de Montréal, Saint-Hyacinthe, Québec, Canada, was included in the study. Biochemical and capsular typings of these strains were carried out as previously described (4).
To verify the pattern stability of S. suis after in vivo passages, two porcine field strains of capsular type 2 (strains 166" and 65) were inoculated into specific-pathogen-free pigs from a closed experimental environment as previously described (5). Both strains isolated from different organs of pigs during the first week postinfection were analyzed by PFGE.
PFGE.
For each S. suis strain, two independent extractions of DNA were performed to verify the reproducibility of patterns. Bacteria were grown for 18 h in 8 ml of Todd-Hewitt Broth (Difco Laboratories, Detroit, Mich.). Preparation of the genomic DNA for PFGE analysis was further performed as described by Rolland et al. (19) with some modifications. The lysis solution did not contain mutanolysin (0.01 M Tris-HCl [pH 7.6], 1 M NaCl, 0.5 Sarkosyl, 1 mg of lysozyme/ml). The DNA was digested with 25 U of SmaI (Roche Diagnostics, Meylan, France) at 25°C for 24 h and washed in 0.1 M EDTA and was subjected to electrophoresis in 1% agarose gel (Tebu, Le Perray en Yvelines, France) in Tris-borate-EDTA (50 mM Tris, 45 mM Borate, 0.5 mM EDTA [pH 8.4]) (Gibco-BRL, Cergy Pontoise, France) using a contour-clamped homogenous electric field apparatus (CHEF-DRIII; Bio-Rad Laboratories). Pulse times were ramped from 1.2 to 30 s over 18 h at 200 V. PFGE patterns were detected by UV transillumination after ethidium bromide staining (0.1 μg/ml) for 1 h followed by water washing for 1 h. Lambda phage concatemers were used as the DNA size standard (Ozyme, Montigny Le Bretonneux, France).
Statistical analysis of PFGE patterns.
The dendrogram representing the genetic relationships between the 123 S. suis strains was drawn using the Biogene package (Vilber-Lourmat, Marne la Vallée, France) as previously described (14). The unweighted pair group method with arithmetic mean was used with a confidence interval of 7.5%. The numerical index of discrimination (D) was calculated using the equation defined by Hunter and Gaston (13):
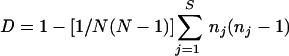 |
D is the probability of two unrelated strains being placed into different typing groups. N is the total number of strains in the sample population, S is the total number of described types, and nj is the number of strains belonging to the jth type.
Statistical analysis was performed to analyze relationships between S. suis PFGE patterns and virulence, capsular types, and species (pig or human origins). The Omnistat program (Hauer-Jensens, Little Rock, Ark.) was used with the Chi-square test (n > 5) or the Fisher exact test (n ≤ 5). Differences between groups were considered significant when probabilities were lower than 0.05.
RESULTS
Reproducibility and stability of PFGE patterns.
Reproducible results were observed because a similar pattern was shown for each S. suis strain after the two independent DNA extractions. The in vivo stability was also verified after experimental infection. The same PFGE patterns with respect to size and number of fragments were obtained for isolates from different organs for each of two strains (strains 166" and 65).
Genetic diversity of strains defined by PFGE.
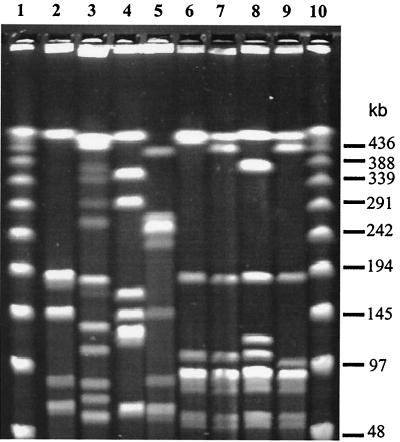
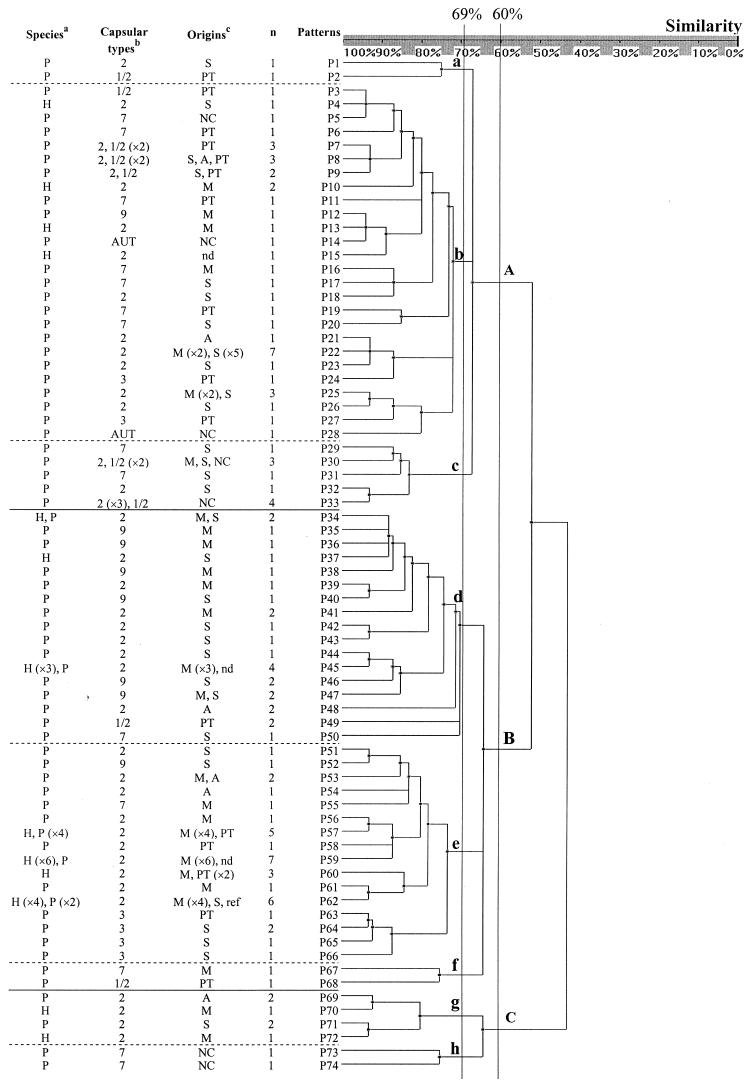
PFGE patterns after restriction with SmaI were characterized by 5 to 12 bands (Fig. 1, lanes 2 and 3) in a 48.5- to 508-kb size range. Among the 123 S. suis isolates, 74 PFGE patterns were identified, each corresponding to one to seven strains (Fig. 2). The index of discrimination was 0.986. The genetic relationships between the 123 isolates of S. suis are presented in the dendrogram (Fig. 2), and they diverged by up to 56% (44% homology). At 60% homology, three PFGE groups, A, B, and C, were identified, and at 69% homology, eight subgroups, a to h, were observed (Fig. 2).
FIG. 1.
PFGE patterns generated after SmaI macrorestriction of S. suis genome. Lanes 1 and 10, DNA molecular size marker; lanes 2 and 3, patterns 55 and 11 presenting 5 and 12 bands, respectively; lanes 4 and 5 correspond to strains isolated from healthy carrier pigs and belonging to patterns P23 and P73, respectively; lanes 6, 7, and 8, patterns P57, P59, and P60, respectively, corresponding to S. suis strains isolated from humans; lane 9 corresponds to S. suis strain belonging to the pattern P62.
FIG.2.
Genetic relationships between 123 S. suis strains, as estimated by clustering analysis of PFGE patterns, was obtained after macrorestriction with SmaI. The classification and divergence of strains were calculated by the unweighted pair group method with arithmetic mean, and a confidence interval of 7.5% was used. The species where the strains have been isolated, capsular types, origins, and numbers of strains for each PFGE patterns are reported in the dendrogram. Footnote-style, superscript letters indicate that the abbreviations in the corresponding columns are defined here, as follows. (a) P, pig; H, human. (b) AUT, autoagglutinable. (c) S, septicemia; PT, palatine tonsils; NC, nasal cavities; A, arthritis; M, meningitis; nd, not done; ref, reference strain.
Genetic diversity of S. suis strains isolated from diseased or clinically healthy pigs and from humans.
The 90 strains isolated from cases of meningitis, arthritis, or septicemia of diseased pigs and humans presented 55 different PFGE patterns (Fig. 2, Table 1). Most of them (77 of 90), were in subgroups b, d, and e. Relationships between these strains and group B (P = 0.002), especially subgroup d (P = 0.018), were significant. Twenty-three PFGE patterns were identified from S. suis strains isolated from the nasal cavities or palatine tonsils of healthy animals, and they were preferentially clustered in group A (P = 0.002), in subgroup b.
TABLE 1.
Distribution of PFGE patterns and groups in relation to capsular types and origins of S. suis strains
| Parameter | na | Value for group
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
S. suis isolates
|
Capsular type
|
Origin
|
||||||||||||
| Virulentb | Nonvirulentc | Otherd | 2 | 1/2 | 3 | 7 | 9 | AUTe | Human | Pig | ||||
| No. of isolates | 123 | 90 | 30 | 3 | 77 | 13 | 7 | 14 | 10 | 2 | 26 | 97 | ||
| No. of PFGE patterns | 74 | 55 | 23 | 3 | 39 | 9 | 6 | 14 | 8 | 2 | 13 | 66 | ||
| No. in PFGE groupf | ||||||||||||||
| A | ||||||||||||||
| a | 2 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | ||
| b | 40 | 25 | 14 | 1 | 22 | 6 | 2 | 7 | 1 | 2 | 5 | 35 | ||
| c | 10 | 5 | 5 | 0 | 5 | 3 | 0 | 2 | 0 | 0 | 0 | 10 | ||
| B | ||||||||||||||
| d | 26 | 23 | 2 | 1 | 15 | 2 | 0 | 1 | 8 | 0 | 5 | 21 | ||
| e | 35 | 29 | 5 | 1 | 28 | 0 | 5 | 1 | 1 | 0 | 14 | 21 | ||
| f | 2 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | ||
| C | ||||||||||||||
| g | 6 | 6 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 2 | 4 | ||
| h | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | ||
n, total no.
S. suis strains isolated from meningitis, arthritis, and septicemia in pigs and humans were classified as virulent strains.
Strains isolated from palatine tonsils and nasal cavities of clinically healthy pigs were classified as avirulent strains.
Isolation of three strains was not done.
AUT, autoagglutinable strains of S. suis.
The 123 isolates of S. suis diverged, and at 60% homology, three PFGE groups, A, B, and C, were identified, and at 69% homology, eight subgroups, a to h, were observed.
Genetic diversity of S. suis strains in relation to capsular types.
Eighty-four percent (65 of 77) of S. suis type 2 isolates were included in subgroups b, d, and e, but no significant association between capsular type 2 and these PFGE groups was observed (P > 0.05). The same results were observed with S. suis capsular type 7. In contrast, S. suis strains capsular types 1/2, 3, and 9 were clustered in particular groups (Fig. 1). Ten of the thirteen isolates of capsular type 1/2 (77%) were associated with group A (P = 0.009), and capsular types 3 (5/7) and 9 (9/10) were often clustered (P = 0.01) in the subgroup e and group B, respectively.
Genetic diversity of S. suis strains in relation to pig or human origin.
S. suis strains isolated from humans were more homogeneous than swine strains. Indeed, the 26 human strains presented only 13 different PFGE patterns, in subgroups b, d, e, and g (Fig. 2; Table 1). In contrast, the 44 capsular type 2 strains isolated from meningitis, septicemia, or arthritis in diseased pigs presented 28 patterns which were included in all different PFGE subgroups. Interestingly, 14 of the 26 (54%) human strains were included only in four patterns. A significant association (P = 2 × 10−8) was observed between the human strains and the patterns P57, P59, P60, and P62 (group B, subgroup e).
DISCUSSION
The PFGE is an established technique for analyzing the whole genome of bacteria and studying genetic differences among isolates (14, 19, 24). Other molecular typing methods have been used for S. suis, such as multilocus enzyme electrophoresis, ribotyping, and RAPD (6, 15-17, 20). Recently the use of PFGE for the study of the whole S. suis DNA has been reported (1). In our hands, this method seems to greatly discriminate among S. suis strains (D = 0.98). In addition, results are reproducible, showing identical patterns with PFGE products from different extractions for each strain. Because of the stability of S. suis DNA and the low number of bands obtained in the present study, PFGE analysis is also easy to interpret.
Differences in virulence among S. suis strains belonging to capsular type 2 (and other capsular types) were often reported (21). Many studies were carried out to determine virulence factors in S. suis, but at the present time it is very difficult to distinguish virulent from avirulent strains of S. suis (9, 21). In the present study, a significant statistical diversity was recorded between strains isolated from diseased pigs and those recovered from clinically healthy animals, since the two sets were clustered in different PFGE subgroups, subgroups d and b, respectively. A few strains of both categories were clustered in the same PFGE patterns. The possibility that healthy carrier pigs harbor strains capable of causing disease under specific circumstances cannot be ruled out (21). Interestingly, the virulence of three S. suis strains, isolates 93, 166" and 65, was previously evaluated in vivo in specific-pathogen-free pigs (5). They were isolated from diseased (93 and 166") or clinically healthy (65) pigs. The virulent strains (93 and 166") inducing severe disease in piglets experimentally infected were closely related based on the PFGE patterns, both clustering in subgroup e (patterns P57 and P62, Fig. 2). In contrast, the avirulent strain (65) presented a high level of divergence, clustering in the avirulent strain subgroup, pattern P7 in the subgroup b (Fig. 2).
Genetic heterogeneity of S. suis was previously reported, especially among strains of capsular type 2, the capsular type most often associated with disease (21, 22). In the present work, the study was extended to isolates belonging to the five most important capsular types in France (4). No significant statistical association could be shown between capsular type 2 and PFGE patterns, suggesting a considerable variation occurring among isolates of capsular type 2, as previously reported (20, 22). In contrast, relationships between PFGE groups and capsular types 1/2, 3, and 9 were shown, making the use of PFGE of S. suis reliable as an additional mean of strain identification.
In this study, a PFGE analysis with isolates from humans was carried out for the first time. Genetic patterns were more homogeneous than those for strains isolated from pigs, with most of them clustering in patterns P57, P59, P60, and P62. Interestingly, some S. suis strains isolated from humans originated with different countries (The Netherlands and France) and presented the same PFGE pattern, as observed previously with RAPD analysis and ribotyping (6, 18). S. suis strains isolated from humans and from pigs were clustered in the same patterns (group B, patterns P34, P45, P57, P59, and P62) (Fig. 1 and 2). As previously shown in RAPD analysis (6), this agrees with S. suis being a zoonotic agent, one which could be transmitted from pigs to humans.
In conclusion, because of all the advantages presented for the PFGE technique used in this study, it is believed that this technique can be efficiently used during further epidemiological investigations of S. suis infections.
Acknowledgments
We thank Sandrine Gueguen and Thierry Ogel for their technical assistance and Gwennola Ermel for her advice about PFGE.
This research was supported by Fonds Européens d'Orientation et de Garantie Agricole.
REFERENCES
- 1.Allgaier, A., R. Goethe, H. J. Wisselink, H. E. Smith, and P. Valentin-Weigand. 2001. Relatedness of Streptococcus suis isolates of various serotypes and clinical backgrounds as evaluated by macrorestriction analysis and expression of potential virulence traits. J. Clin. Microbiol. 39:445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arends, J. P., N. Harwig, M. Rudolphy, and H. C. Zanen. 1984. Carrier rate of Streptococcus suis capsular type 2 in palatine tonsils of slaughtered pigs. J. Clin. Microbiol. 20:945-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaudouin, M., J. Harel, R. Higgins, M. Gottschalk, M. Frenette, and J. I. MacInnes. 1992. Molecular analysis of isolates of Streptococcus suis type 2 by restriction-endonuclease-digested DNA separated on SDS-PAGE and by hybridization with an rDNA probe. J. Gen. Microbiol. 138:2639-2645. [DOI] [PubMed] [Google Scholar]
- 4.Berthelot-Hérault, F., H. Morvan, A. M. Kéribin, M. Gottschalk, and M. Kobisch. 2000. Production of muramidase released protein (MRP), extracellular factor (EF) and haemolysin by field isolates of Streptococcus suis capsular type 2, 1/2, 9,7 and 3 isolated from swine in France. Vet. Res. 31:473-479. [DOI] [PubMed] [Google Scholar]
- 5.Berthelot-Hérault, F., R. Cariolet, A. Labbé, M. Gottschalk, J. Y. Cardinal, and M. Kobisch. 2001. Experimental infection in specific pathogen free piglets with French strains of Streptococcus suis capsular type 2. Can. J. Vet. Res. 65:196-200. [PMC free article] [PubMed] [Google Scholar]
- 6.Chatellier, S., M. Gottschalk, R. Higgins, R. Brousseau, and J. Harel. 1999. Relatedness of Streptococcus suis type 2 isolates from different geographic origins as evaluated by molecular fingerprinting and phenotyping. J. Clin. Microbiol. 37:362-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottschalk, M., S. Lacouture, and J. D. Dubreuil. 1995. Characterization of Streptococcus suis capsular type 2 haemolysin. Microbiology 141:189-195. [DOI] [PubMed] [Google Scholar]
- 8.Gottschalk, M., A. Lebrun, H. Wisselink, J. D. Dubreuil, H. Smith, and U. Vecht. 1998. Production of virulence-related proteins by Canadian strains of Streptococcus suis capsular type 2. Can. J. Vet. Res. 62:75-79. [PMC free article] [PubMed] [Google Scholar]
- 9.Gottschalk, M., R. Higgins, and S. Quessy. 1999. Dilemma of the virulence of Streptococcus suis strains. J. Clin. Microbiol. 37:4202-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gottschalk, M., and M. Segura. 2000. The pathogenesis of the meningitis caused by Streptococcus suis: the unresolved questions. Vet. Microbiol. 76:259-272. [DOI] [PubMed] [Google Scholar]
- 11.Harel, J., R. Higgins, M. Gottschalk, and M. Bigras-Poulin. 1994. Genomic relatedness among reference strains of different Streptococcus suis serotypes. Can. J. Vet. Res. 58:259-262. [PMC free article] [PubMed] [Google Scholar]
- 12.Higgins, R., and M. Gottschalk. 1999. Streptococcal diseases, p. 563-578. In B. E. Straw, S. Allaire, W. L. Mengeling, and D. J. Taylor (ed.), Diseases of swine, 8th ed. Iowa University Press, Ames, Iowa.
- 13.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marois, C., F. Dufour-Gesbert, and I. Kempf. 2001. Comparison of pulsed-field gel electrophoresis with random amplified polymorphic DNA for typing of Mycoplasma synoviae. Vet. Microbiol. 79:1-9. [DOI] [PubMed] [Google Scholar]
- 15.Mwaniki, C. G., I. D. Robertson, D. J. Trott, R. F. Atyeo, B. J. Lee, and D. J. Hampson. 1994. Clonal analysis and virulence of Australian isolates of Streptococcus suis type 2. Epidemiol. Infect. 113:321-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okwumabua, O. G. I., J. Staats, and M. M. Chengappa. 1995. Detection of genomic heterogeneity in Streptococcus suis isolates by DNA restriction fragment length polymorphisms of RNA genes (ribotyping). J. Clin. Microbiol. 33:968-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Power, E. G. M. 1996. RAPD typing in microbiology--a technical review. J. Hosp. Infect. 34:247-265. [DOI] [PubMed] [Google Scholar]
- 18.Rasmussen, S. R., F. M. Aarestrup, N. E. Jensen, and S. E. Jorsal. 1999. Associations of Streptococcus suis serotype 2 ribotypes profiles with clinical disease and antimicrobial resistance. J. Clin. Microbiol. 37:404-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rolland, K., C. Marois, V. Siquier, B. Cattier, and R. Quentin. 1999. Genetic features of Streptococcus agalactiae strains causing severe neonatal infections, as revealed by pulsed-field gel electrophoresis and hylB gene analysis. J. Clin. Microbiol. 37:1892-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith, H. E., M. Rijnsburger, N. Stockhofe-Zurwieden, H. J. Wisselink, U. Vecht, and M. A. Smits. 1997. Virulent strains of Streptococcus suis serotype 2 and highly virulent strains of Streptococcus suis serotype 1 can be recognized by a unique ribotype profile. J. Clin. Microbiol. 35:1049-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staats, J. J., I. Feder, O. Okwumabua, and M. M. Chengappa. 1997. Streptococcus suis: past and present. Vet. Res. Commun. 21:381-387. [DOI] [PubMed] [Google Scholar]
- 22.Staats, J. J., B. L. Plattner, J. Nietfeld, S. Dritz, and M. M. Chengappa. 1998. Use of ribotyping and hemolysin activity to identify highly virulent Streptococcus suis type 2 isolates. J. Clin. Microbiol. 36:15-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vecht, U., H. J. Wisselink, M. L. Jellima, and H. E. Smith. 1991. Virulence of Streptococcus suis type 2. Infect. Immun. 59:3156-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vicki, A. L., D. B. Jernigan, A. Tice, J. D. Kellner, and M. C. Roberts. 2000. A novel multiresistant Streptococcus pneumoniae serogroup 19 clone from Washington state identified by pulsed-field gel electrophoresis and restriction fragment length patterns. J. Clin. Microbiol. 38:1575-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]