Abstract
A restriction fragment length polymorphism (RFLP) detection assay was developed to examine the genetic relationship(s) among VP7-encoding genes from 100 Irish rotavirus isolates and 30 randomly selected global rotavirus isolates (from the current databases). RFLP analysis of the VP7 gene segments was performed independently with three enzymes (RsaI, AluI, and EcoRV) in separate reactions by direct digestion of the DNA product amplified by reverse transcriptase (RT)-mediated PCR (RT-PCR) or by using computational methods. Thirty-six RFLP patterns were identified for all 130 strains, and of these, only nine patterns were associated with the Irish isolates. A correlation between the G type of the Irish isolates and certain single or combined enzyme profiles was apparent. These data suggested that the Irish wild-type rotavirus population was homogeneous and could be distinguished by RFLP analysis from global isolates of the same serotype(s). The deduced amino acid sequences of the VP7 RT-PCR products from six Irish isolates known to be of the G serotype revealed significant amino acid substitutions within major antigenic regions. In addition, these data identified the existence of at least two genetic lineages within serotype G1 strains which were distinguishable by RFLP analysis.
Human rotavirus is the most common cause of severe diarrhea in young children worldwide (17). Two rotavirus vaccines, a live polyvalent bovine vaccine and a human monovalent strain, are being tested in clinical trials (2, 5). Both of these vaccines and the recently withdrawn rhesus rotavirus vaccine were developed to provide protection against the four predominant G serotypes of rotavirus, serotypes G1 to G4 (2, 5, 18). However, other less common serotypes, such as serotypes G8 and G9, are circulating in Ireland (22) and other countries (6, 27, 32, 37). In addition, there is evidence for genetic diversity within rotavirus serotypes that may potentially alter the immune response to vaccination in some settings, but the significance of these findings has yet to be determined. Surveillance of strains is essential to determine whether vaccines will work efficiently against circulating strains and if vaccine coverage for additional serotypes will be required.
The rotavirus genome consists of 11 double-stranded RNA gene segments that encode five structural and six nonstructural proteins (28). Viral proteins VP4 and VP7 are responsible for the production of a neutralizing antibody response to rotavirus infection. These proteins have distinct antigenic specificities referred to as P and G types, respectively (10). Currently, 9 P serotypes and 10 G serotypes have been identified in humans (9, 23, 33, 36). Natural immunity to rotavirus infection is thought to be serotype specific (15). Antibodies derived from a primary infection are described as homotypic and not sufficiently cross-reactive to prevent infection by heterotypic strains. For any vaccine and subsequent vaccination program to be effective, vaccines must be designed so that they include the neutralization phenotypes of the epidemiologically important serotypes. Several investigators have described the predominant types circulating globally as G1P[8], G2P[4], G3P[8], and G4P[8] (3, 14, 25, 30, 38). However, unconventional G types and unusual combinations of G and P types are now being reported with increasing frequency (6, 19, 22, 26, 30, 31). In addition, the genetic complexity within individual serotypes is now regarded as being more extensive than was previously anticipated. Jin et al. (16) identified four distinct genetic lineages of viruses of the G1 serotype (designated G1-1, G1-2, G1-3, and G1-4), and Piec and Palombo (29) described the existence of genetic and antigenic subtypes of serotype G2. Similarly, subtypes of rotavirus serotype G4 have also been reported (7, 20). These genetic variants have the potential to give rise to antigenically distinct strains with similar serotypes. This feature has important implications for vaccine design.
It is now recognized that a greater understanding of the genetic and antigenic compositions of the wild-type virus populations in different geographical locations is required (25). Molecular biology-based techniques have contributed significantly to the characterization of rotaviral genomes. Methods including PCR typing and hybridization studies have facilitated the identification of several serotypes, and in addition, DNA sequence analysis has provided a complete characterization of many rotavirus gene segments. Previously, Gouvea et al. (12) proposed the use of restriction fragment length polymorphism (RFLP) protocols to uniquely identify and group rotavirus strains. Their studies suggested that this strategy may prove useful in monitoring of the extent of genetic variation among rotavirus strains within a population and may prove valuable in the examination of interspecies transmission (4, 12).
In the present study an RFLP assay was developed to identify and differentiate the G types of 100 Irish rotavirus isolates. The Irish isolates analyzed in the present study represented a collection of 330 strains previously isolated over a 3-year period from 1997 through 1999 (21). In addition, the assay was used to examine the extent of genetic relationships between Irish strains and global strains of the same serotype. As a consequence, novel strains circulating in the Irish population were identified.
MATERIALS AND METHODS
Virus samples.
One hundred randomly selected human fecal specimens positive for rotavirus were collected from children age 2 years or younger. These specimens were collected over a 3-year period from 1997 through 1999. The rotavirus strains collected represented the major G types previously identified in Ireland (22), including 25 type G1 isolates, 34 type G2 isolates, 17 type G4 isolates, and 24 isolates identified as being from mixed G-type infections. A complete listing of this collection is given by O'Halloran (21).
RNA purification and RT-PCR.
Double-stranded rotaviral RNA genomes were extracted by a phenol-chloroform method as described by Gouvea et al. (11). Full-length (1,062-bp) VP7 gene segments encoding the major neutralization protein, VP7, were amplified by reverse transcriptase (RT)-mediated PCR (RT-PCR) with primer pair Beg9 and End9 (11). The amplified products were resolved by conventional agarose gel (1.5%) electrophoresis and were visualized after ethidium bromide staining (0.1 mg/ml).
RFLP analysis.
Each amplified DNA product (amplified as described above) was analyzed separately by direct digestion with 1 U of RsaI (GT"AC), AluI (AG"CT), or EcoRV (GAT"ATC). Following 1.5 h of incubation at 37°C, the digested products were analyzed by conventional agarose gel (2%) electrophoresis (as outlined above).
Database rotavirus sequences.
Thirty strains whose G types were known (serotypes G1 through G4, G8, and G9) were selected from the current GenBank database to represent strains from various geographical locations (see Table 3). These sequences were analyzed for the corresponding RsaI, AluI, and EcoRV restriction sites with DNA Sequencher (version 4.01) sequencing software (Gene Codes Corp., Ann Arbor, Mich.).
TABLE 3.
GenBank sources of VP7-encoding gene segments from group A human rotavirus strains
| Number Straina | G type | GenBank accession no. | Country | RFLP pattern |
|---|---|---|---|---|
| GBR-1 | 1 | M64666 | Australia | R1A2E1 |
| GBR-2 | 1 | D17723 | Japan | R1A6E1 |
| GBR-3 | 1 | D16328 | Japan | R1A6E1 |
| GBR-4 | 1 | K02033 | —b | R1A11E1 |
| GBR-5 | 1 | AF043684 | Australia | R1A6E1 |
| GBR-6 | 2 | AF106295 | Taiwan | R7A7E4 |
| GBR-7 | 2 | D50126 | Japan, China | R8A8E6 |
| GBR-8 | 2 | D50123 | Japan, China | R9A9E7 |
| GBR-9 | 2 | D50116 | Japan, China | R4A7E8 |
| GBR-10 | 2 | U73957 | Australia | R4A8E9 |
| GBR-11 | 3 | U04350 | United States | R10A10E11 |
| GBR-12 | 3 | AB011971 | Japan | R7A13E5 |
| GBR-13 | 3 | AB011970 | Japan | R11A13E10 |
| GBR-14 | 3 | D86284 | Japan, China | R12A12E5 |
| GBR-15 | 3 | D86274 | Japan, China | R13A14E11 |
| GBR-16 | 4 | A01321 | Australia | R14A15E1 |
| GBR-17 | 4 | M86832 | — | R15A16E1 |
| GBR-18 | 4 | AF170837 | South Africa | R14A17E1 |
| GBR-19 | 4 | AB 012078 | Japan | R14A18E1 |
| GBR-20 | 4 | M86490 | — | R14A18E1 |
| GBR-21 | 8 | AF207061 | Australia | R17A20E10 |
| GBR-22 | 8 | L20882 | Finland | R18A21E10 |
| GBR-23 | 8 | AF104104 | Egypt | R17A22E10 |
| GBR-24 | 8 | AF141918 | India | R17A23E13 |
| GBR-25 | 8 | AF143688 | South Africa, United Kingdom | R19A24E14 |
| GBR-26 | 9 | AJ250545 | Malawi | R5A4E12 |
| GBR-27 | 9 | AJ250542 | Bangladesh | R5A19E1 |
| GBR-28 | 9 | AF060487 | United States | R16A5E12 |
| GBR-29 | 9 | AJ250277 | India | R5A5E15 |
| GBR-30 | 9 | AJ250268 | United States | R20A5E12 |
GBR, GenBank reference.
—, not available.
Nucleic acid sequencing.
The VP7 genes of six Irish rotavirus strains were chosen on the basis of their RFLP patterns for complete characterization by DNA sequencing. Initially the RT-PCR-amplified VP7-encoding gene products were directly ligated to pCR2.1 (Invitrogen, Bv, Amsterdam, The Netherlands) and were cloned according to the manufacturer's instructions. The corresponding constructs were screened for the correct insert prior to purification with the Wizard Plus SV Minipreps DNA Purification system (Promega). The products were sequenced as outlined previously (21) with M13 forward and reverse sequencing primers by dye terminator chemistry protocols and cycle sequencing (Beckman Coulter, Fullerton, Calif.). The sequenced products were initially analyzed with DNA Sequencher (version 4.01) software. All sequences were searched for corresponding matches in the databases by using the BLAST suite of programs (1). The deduced amino acid sequences were aligned by use of the CLUSTALW program (35).
Nucleotide sequence accession numbers.
The DNA sequences were directly submitted to GenBank and were assigned the following GenBank accession numbers: AF254137, AF254138, AF254139, AF254140, AF254141, and AF281044.
RESULTS
Analysis of VP7-encoded cDNA fragments with RsaI, AluI, and EcoRV enzymes.
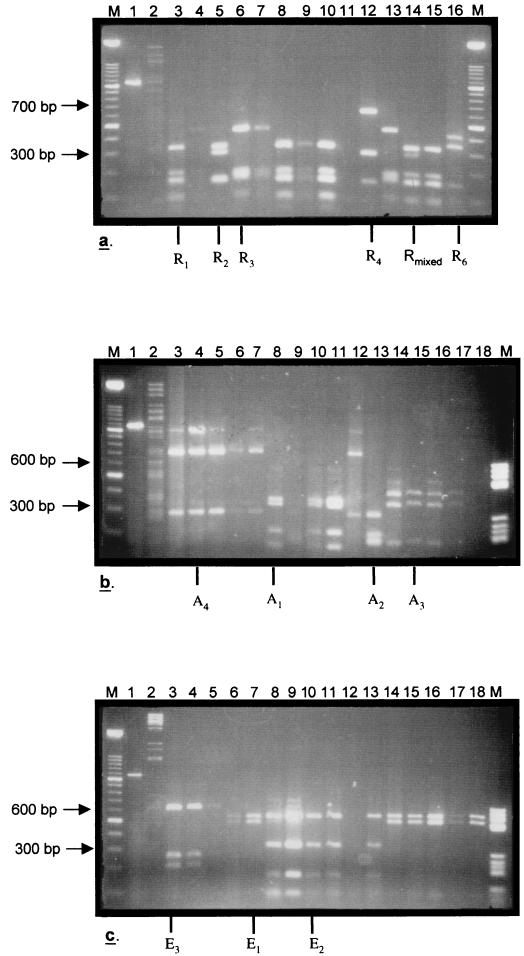
Restriction enzyme-digested VP7 cDNA products generated from randomly selected Irish strains of known serotype after RT-PCR were analyzed by agarose gel (2%) electrophoresis. Each amplified DNA fragment was subjected to separate but sequential digestion with the RsaI, AluI, and EcoRV enzymes. Restriction enzymes were chosen on the basis of their ability to reproducibly digest the VP7 cDNA fragment, in each case producing consistent restriction profiles (Table 1). These fragment arrays correlated with specific G types. The corresponding profiles were defined for each restriction enzyme (profiles R, A, and E for restriction enzymes RsaI, AluI, and EcoRV, respectively) on the basis of the DNA fragment pattern produced (Fig. 1 and 2 and Table 1). Furthermore, all enzymes cleaved within the VP7 gene segments, increasing the sensitivity of the assay, permitting the detection of nucleotide polymorphisms. Later, all enzyme profiles were combined to produce a characteristic RFLP pattern for each strain whose G serotype was determined (Table 2).
TABLE 1.
Profiles of VP7 gene segments from Irish strains obtained with restriction enzymes RsaI, AluI, and EcoRV
| Enzyme profile | Restriction site (bp) | Fragment size (bp) | Na | Associated G type(s) |
|---|---|---|---|---|
| R1 | 318, 409, 467, 621, 789, 968 | 318, 91, 58, 154, 159, 188, 94 | 7 | G1, mixed types (G1 + G2, G1 + G4) |
| R2 | 318, 467, 780 | 318, 149, 313, 282 | 4 | G1 |
| R3 | 448, 621, 780, 968 | 448, 173, 159, 188, 94 | 5 | G4, mixed types (G1 + G4, G4 + G9) |
| R4 | 621, 774, 780 | 621, 153, 6, 282 | 4 | G2, mixed types (G2 + G4, G1 + G2, G2 + G8) |
| R5 | 467, 621, 780 | 467, 154, 159, 282 | 4 | Mixed types (G1 + G3) |
| R6 | 318, 409, 467, 621, 780, 968 | 318, 91, 58, 154, 159, 188, 94 | 7 | G1 |
| A1 | 39, 196, 514, 721 | 39, 157, 318, 207, 341 | 5 | G1, mixed types (G1 + G8) |
| A2 | 39, 196, 514, 721, 823, 1009 | 39, 157, 318, 207, 102, 186, 53 | 6 | G1, G4, mixed types (G1 + G4, G1 + G2) |
| A3 | 39, 360, 444, 514, 685 | 39, 321, 84, 70, 171, 377 | 6 | G4, mixed types (G1 + G4, G4 + G9) |
| A4 | 39, 313, 1006 | 39, 274, 693 | 4 | G2; mixed types (G2 + G4) |
| A5 | 39, 495, 514, 1009 | 39, 456, 19, 495, 53 | 5 | Mixed types (G1 + G3) |
| E1 | 89, 602 | 89, 513, 460 | 3 | G1, G4, mixed types (G1 + G4, G1 + G2, G1 + G3, G1 + G8) |
| E2 | 89, 602, 762 | 89, 513, 160, 300 | 4 | G4, mixed types (G1 + G4, G4 + G9) |
| E3 | 602, 858 | 602, 256, 204 | 3 | G2, mixed types (G2 + G4, G2 + G8, G1 + G2) |
N, number of restriction fragments generated.
FIG. 1.
Enzyme profiles of VP7 segments digested with three restriction enzymes, RsaI, AluI, and EcoRV. All products were analyzed by agarose gel (2%) electrophoresis and visualized by staining with ethidium bromide (0.1 mg/ml). (a) RsaI digestion. Lanes M, molecular size marker, 100-bp ladder; lane 1, undigested VP7 fragment; lane 2, DNA control digested with RsaI; lane 3, CIT-54RV (G1); lane 4, CIT-124RV (G4); lane 5, CIT-179RV (G1); lane 6, CIT-202RV (G4); lane 7, CIT-222RV (G4); lane 8, CIT-70RV (G1); lane 9, CIT-76RV (G1); lane 10, CIT-78RV (G1); lane 11, CIT-79RV (G1); lane 12, CIT-95RV (G2); lane 13, CIT-155RV (G1 + G4); lane 14, CIT-109RV (G1); lane 15, CIT-112RV (G1); lane 16, CIT-313RV (G1). (b) AluI digestion. Lanes M, molecular size marker, 100-bp ladder, and molecular size marker, grade V; lane 1, undigested VP7 fragment; lane 2, DNA control digested with AluI; lane 3, CIT-68RV (G2); lane 4, CIT-245RV (G2); lane 5, CIT-128RV (G2); lane 6, CIT-152RV (G2); lane 7, CIT-123RV (G2); lane 8, CIT-132RV (G1); lane 9, CIT-176RV (G4); lane 10, CIT-178RV (G4); lane 11, CIT-224RV (G4); lane 12, CIT-273RV (G2); lane 13, CIT-278RV (G4); lane 14, CIT-289RV (G4); lane 15, CIT-293RV (G4); lane 16, CIT-298RV (G4); lane
FIG. 2.
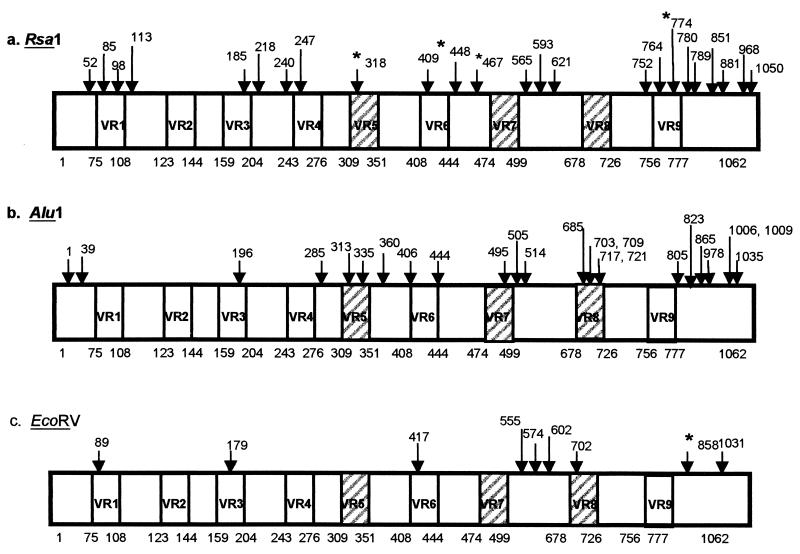
Schematic representations of the restriction maps identified for VP7-encoding gene segment 9 of group A human rotaviruses. The approximate positions of the nine variable regions (VR1 through VR9) are represented by boxes (with the locations of each indicated beneath each panel); and major antigenic regions VR5, VR7, and VR8 are highlighted by hatched boxes. The corresponding restriction sites and nucleotide positions identified for RsaI (a), AluI (b), and EcoRV (c) are shown and are indicated by arrows.
TABLE 2.
RFLP patterns identified for Irish isolates by enzyme profiles
| RFLP pattern | No. of strains associated | G type (no. of isolates) |
|---|---|---|
| R1A1E1 | 17 | G1 (17) |
| R1A2E1 | 7 | G1 (4), G1 + G4 (1), G1 + G2 (2) |
| R2A2E1 | 3 | G1 (3) |
| R3A2E1 | 2 | G4 (2) |
| R3A3E1 | 9 | G4 (7), G1 + G4 (2) |
| R3A3E2 | 13 | G4 (8), G1 + G4 (1), G4 + G9 (4) |
| R4A4E3 | 45 | G2 (34), G2 + G4 (1), G2 + G8 (3), G1 + G2 (6), G2 + G4 + G8 (1) |
| R5A5E1 | 1 | G1 + G3 (1) |
| R6A1E1 | 1 | G1 (1) |
| Mixed patterns | 2 | G1 (1), G1 + G2 (1) |
Six profiles were noted after digestion with RsaI and were designated R profiles R1 through R6 (Fig. 1a and Table 1). Briefly, all G2 viruses were associated with the R4 profile (Fig. 1a, lane 12), and G4 viruses were associated only with the R3 profile (Fig. 1a, lane 6). G1 serotypes were more diverse and generated a number of R profiles including profiles R1 and R2 (Fig. 1a, lanes 3 and 5, respectively). In addition, a single G1 isolate had a unique R6 profile (Fig. 1a, lane 16) not previously recognized among any of our other isolates. The majority of isolates that were part of mixed infections were associated with an R profile indicative of one of the coinfecting viruses (Table 1). There were some exceptions to this observation, including two strains that had mixed R profiles. For example, the isolate in Fig. 1a, lane 14, was associated with both the R1 and the R2 profiles, suggesting dual infection with two unique G1 serotypes. A single isolate, isolate CIT-254RV (21), was identified by PCR serotyping as a mixture of serotypes G1 and G3, and this isolate had a unique R profile, profile R5 (Table 1).
Five profiles were obtained by digestion with AluI (profiles A1 through A5; Table 1). A brief analysis of Irish G2 viruses demonstrated that these strains were associated with a single enzyme profile, profile A4 (Fig. 1b, lane 4). Serotype G1 and G4 strains usually generated profiles A1 and A3, respectively, after resolution by gel electrophoresis (Fig. 1b, lanes 8 and 15). Furthermore, both serotypes were also associated with the A2 profile (Fig. 1b, lane 13, and Table 1). The majority of isolates that were part of mixed infections demonstrated single profiles, which suggested that one of the coinfecting G-type strains was dominant (Table 1). Some exceptions were noted, particularly in the case of two isolates that had mixed A profiles (data not shown), and again, strain CIT-254RV (as noted above) had a unique profile, profile A5 (Table 1). A particular feature observed after AluI digestion was the production of a predominant profile on a background of DNA fragments consistent with partial digestion (Fig. 1b, lanes 4, 11, and 14). Gouvea et al. (12) previously noted this feature when VP7 gene segments were similarly digested with BstYI. This feature did not hinder assignment of a profile to these isolates.
Similarly, the E profiles generated with EcoRV were also informative. Serotype G2 isolates were associated with the E3 profile alone (Fig. 1c, lane 3), and serotype G1 strains were associated only with the E1 profile (Fig. 1c, lane 7). A majority of serotype G4 viruses (n = 9; 53%) were also associated with the E1 profile; however, some strains (n = 8; 47%) also generated a profile designated E2 (Fig. 1c, lane 10). Isolates that were part of mixed infections again had either mixed E profiles or single E profiles indicative of one of the strains responsible for the infection. Isolate CIT-254RV, which previously demonstrated unique enzyme profiles with RsaI and AluI, was in this case associated with the E1 profile, similar to serotype G1 and G4 strains.
Computational analysis of the sequences of 30 randomly selected global rotavirus strains from the GenBank database permitted the construction of the corresponding R, A, and E profiles. An additional 14 R-profile banding patterns were noted after RsaI digestion, and these were designated R7 through R20 (data not shown). These were distinct from the R profiles associated with the Irish strains. Nineteen distinct A profiles (profiles A6 to A24) were associated with the database strains, producing a total of 24 profiles for the complete collection, and similarly, 12 additional E profiles, denoted profiles E4 through E15, were obtained after computational analysis of the preselected global strains (data not shown).
Combined RFLP profiles.
The enzyme profiles obtained previously were then combined for each strain to generate individual RFLP patterns. The 130 human rotavirus VP7-encoding genes were classified into a total of 36 RFLP patterns. Only 9 patterns were observed among the Irish strains, and all of these are listed in Table 2.
For the serotype G1 viruses, 17 of the 25 (68%) strains analyzed had the pattern R1A1E1, Four strains (16%) had the pattern R1A2E1, and three strains (12%) had the pattern R2A2E1. A single G1 isolate had a unique pattern of R6A1E1. Seventeen of the serotype G4 viruses produced three RFLP patterns: R3A3E1 (n = 7; 41%), R3A3E2 (n = 8; 47%), and R3A2E1 (n = 2; 12%). Specifically, the R3A3 combination was evident in the majority of G4 Irish strains (n = 15; 88%). All of the Irish serotype G2 isolates had a single RFLP pattern, pattern R4A4E3 (Table 2). The RFLP profiles of the global strains showed marked genetic variations, with only a single isolate (isolate GBR-1) having an RFLP profile that corresponded to those of the Irish strains (Table 3).
Isolates that were part of mixed G-type infections could also be assigned to specific RFLP profiles. The majority of these corresponded to the RFLP profile for one of the apparently dominant coinfecting viruses. Some exceptions included strains with obvious single enzyme profiles, such as CIT-254RV (21). This isolate was identified by PCR typing as having a mixture of serotypes, serotypes G1 and G3 (21), and had a unique RFLP pattern, pattern R5A5E1 (Table 2). Examination of the electropherotype pattern for this isolate by polyacrylamide gel electrophoresis (PAGE) also suggested that this strain was unique, as the PAGE pattern identified was distinct from those of the other strains in this collection of Irish strains (data not shown).
DNA sequence analysis and genetic variation within the variable-region domains of VP7.
Sequence analysis of VP7 DNA fragments cloned from six Irish isolates with defined G serotypes confirmed the location and subsequent enzyme profiles for RsaI, AluI, and EcoRV (Table 1 and Fig. 2). The isolates chosen for sequence analysis were selected on the basis of their corresponding RFLP arrays (i.e., R, A, and E profiles) and included the following: CIT-4RV (R1A1E1), CIT-6RV (R2A2E1), CIT-313RV (R6A1E1), CIT-220RV (R3A3E2), CIT-176RV (R4A4E3), and CIT-254RV (R5A5E1) (21). Open reading frames were identified in all cases, and the deduced amino acid sequences were compared to each other and the VP7 sequences of selected global isolates and were subsequently aligned by use of the CLUSTALW program (35).
Careful inspection of the restriction maps of the VP7-encoding genes of the Irish strains revealed several interesting features (Fig. 2). In this study some restriction sites identified were associated with a specific G type, including the RsaI site located at nucleotide position 318 bp (the asterisk labeled 318 in Fig. 2a), which was unique to the Irish serotype G1 viral population (Table 1). The RsaI site at nucleotide position 774 bp (the asterisk labeled 774 in Fig. 2a) was uniquely linked with G2 strains (Table 1). These sites exist within variable regions VR5 and VR9, respectively. Other restriction sites were located outside the variable regions, and these also appeared to be associated with specific G types. Some of these included the EcoRV site at nucleotide position 858 bp (the asterisk labeled 858 in Fig. 2c), which was identified with serotype G2 strains (Table 1), and the RsaI sites at nucleotide position 448 and 467 bp (the asterisks labeled 448 and 467, respectively, in Fig. 2a), which appeared to be conserved among serotype G4 and G1 strains, respectively (Table 1).
Comparison of the amino acid lineages among VP7 proteins from Irish and global strains whose G serotypes were determined.
Rotavirus strains of the G1 serotype are the most prevalent strains circulating in the Irish population (22, 24). Of all Irish isolates studied in our laboratory, G1 strains produced the greatest RFLP-associated diversity (Table 2). It was therefore of interest to further characterize the VP7 proteins from these viruses. The deduced amino acid sequences of 3 Irish serotype G1 isolates selected on the basis of different RFLP profiles (R1A1E1 [isolate CIT-4RV], R2A2E1 [isolate CIT-6RV], and R6A1E1 [isolate CIT-313RV]) were compared to 11 global serotype G1 viruses from various geographical locations (Table 4).
TABLE 4.
Amino acid alignments of 14 G1 VP7 protein sequencesa
| Strain designation or yr (country) of isolation | GenBank accession no. | VR5 (aa 87 to 101)b | VR7 (aa 142 to 151) | VR8 (aa 208 to 221) | Lineage identification code for position:
|
Lineage | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 29 | 37 | 41 | 49 | 55 | 57 | 65 | 66 | 68 | ||||||
| Wa strain | M21843 | TEASTQINDGDWKDS | MKYDQSLKLDM | QTTNVDSFEMIAEN | I | F | T | R | L | L | A | V | T | G1-4 |
| 1999 (Australia) | AF043684 | -------S--E---- | -----N-E--- | --------DTV--- | I | F | S | K | L | I | T | V | S | G1-3 |
| 1999 (Australia) | AF043678 | ----------E---- | -----N-E--- | ---------MV--- | I | S | Y | R | L | L | T | V | A | G1-3 |
| 1999 (Australia) | AF043680 | -------S--E---- | -----N-E--- | --------GRQ--- | I | F | S | T | L | I | T | V | S | G1-3 |
| 1999 (Taiwan) | AF183858 | ----------E---- | -----N-E--- | ---------TV--- | I | F | S | K | L | I | T | V | S | G1-3 |
| 1993 (Japan, China) | D17723 | -------S--E---- | -----NFE--- | ---------TV--- | I | F | S | K | L | I | T | V | S | G1-3 |
| 1999 (Japan, China) | D16327 | -------S--E---- | -----N----- | ---------TV--- | I | F | S | K | L | I | T | V | S | G1-3 |
| 2000 (China) | AF260951 | ----A--S--E---- | -----N-E--- | ---------TV--- | I | F | S | K | L | I | T | V | S | G1-3 |
| 1999 (Thailand) | AF181863 | -------S--E---- | -----N-E--- | ---------TV--- | I | F | F | K | L | I | T | V | S | G1-3 |
| 1998 (Finland) | Z80271 | ----------E---- | -----N-E--- | -------------- | M | S | S | R | L | L | A | A | A | G1-1a |
| 1998 (Finland) | Z80272 | ----------E---- | -----N-E--- | -------------- | I | S | T | R | L | L | A | I | A | G1-2 |
| 2000 (Ireland) | AF254138 | -------S--E---- | -----N-E--- | ---------TV--- | I | F | S | K | L | I | T | V | A | G1-3 |
| 2000 (Ireland) | AF254137 | ---------E---- | -----N-EI-- | ----------V--- | I | S | Y | R | L | I | T | V | A | G1-1c |
| 2000 (Ireland) | AF254141 | -------S--E---- | -----N-E--- | ---------TV--- | I | F | S | K | L | I | T | V | S | G1-3 |
The amino acids in major antigenic regions VR5, VR7, and VR8 are compared to those for reference Wa strain. The nine amino acids involved in defining the lineages of G1 viruses, according to Maunula and von Bonsdorff (20), are also indicated.
The boldface letters indicate significant amino acid residues.
Jin et al. (16) described four distinct lineages (G1-1, G1-2, G1-3, and G1-4) on the basis of the amino acid sequences of specific and limited regions of the VP7 proteins of serotype G1 viruses. Defined amino acid substitutions in the major antigenic regions outlined previously (the shaded regions in Fig. 2) have also been reported to correlate with a change in G1 serotype lineage. Specifically, amino acid substitutions Asn-94-Ser (within VR5; Fig. 2) and Met-217-Thr (within VR8) signal a change to the G1- lineage (Table 4) (7, 20). The G1-4 lineage is defined by the Asp-97-Glu substitution (within VR5; Fig. 2). On the basis of these and other comparisons, two G1 lineages were found to be cocirculating in the Irish rotavirus population. The VP7 gene segment from isolate CIT-4RV (GenBank accession number AF254138) had the amino acid identification code IFSKLITVA, with Ser-94 within the VR5 region (Fig. 2 and Table 4), identifying this strain as being of the G1-3 lineage. Similarly, isolate CIT-6RV (GenBank accession number AF254137) had the amino acid identification code ISYRLITVA and an Asn residue at position 94, defining this strain as being of the G1-1c lineage. The third Irish isolate, isolate CIT-313RV (GenBank accession number AF254141), had an identification code that indicated that it was of the G1-3 lineage (Table 4). However, examination of the complete amino acid sequence of this protein revealed that it was identical to the amino acid sequences of the proteins of both CIT-4RV and CIT-6RV. The corresponding RFLP pattern associated with strain CIT-313RV was also unique, pattern R6A1E1 (Table 2).
The deduced protein sequences of representative G2 and G4 Irish strains were also examined and compared to those of global strains (data not shown). Comparison of these sequences demonstrated a high level of amino acid sequence similarity between Irish and global strains of similar serotypes. However, amino acid substitutions were identified within the variable regions of the Irish isolates only. These data, together with the results of RFLP analysis, further supported the view that the Irish population of serotype G2 and G4 strains was homogeneous and could be distinguished from global strains.
The amino acid sequence of the VP7 protein of isolate CIT-254RV was also examined and aligned with those of other VP7 proteins of isolates representing serotypes G1 to G4, G8, and G9 (data not shown). Sequence similarities to more than one serotype were identified in variable regions VR1 through VR9, including serotypes G1, G3, and G9. However, unique amino acid substitutions which may affect antigenic determinants were also identified in VR6 and VR7. This information, together with the unique RFLP and PAGE patterns associated with this isolate, suggested that CIT-254RV was a novel strain circulating in the population.
DISCUSSION
In the present study the VP7-encoding gene segments of 130 group A human rotaviruses were examined by RFLP analysis. Careful examination of restriction profiles revealed several interesting features of strain diversity in Ireland. The restriction enzyme profiles obtained upon digestion with RsaI, AluI, and EcoRV suggested that when single G-type infections occurred there was an obvious association between the specific restriction profiles and the G types. Some strains demonstrated a single enzyme profile, while others had a combination of enzyme profiles or a unique RFLP pattern. In addition, comparison of RFLP data for Irish isolates to those for isolates of global origin suggested that the Irish rotavirus population is homogeneous and distinguishable from global strains. For example, 34 Irish G2 strains analyzed in the present study were associated with a single RFLP pattern, pattern R4A4E3. The RFLP patterns associated with the global serotype G2 strains demonstrated genetic variation between the global and Irish strains, and none of their associated RFLP patterns were identified in the Irish serotype G2 strains. Similarly, the 17 Irish serotype G4 strains analyzed were associated with three RFLP patterns. Specifically, a single profile (profile R3) was observed among all serotype G4 viruses. The RFLP patterns associated with the global G4 strains were again very different from those associated with the Irish strains. Significantly, the Irish serotype G2 and G4 strains analyzed represented a collection of strains from a 3-year study (21), and the RFLP data suggested that the Irish population of serotype G2 and G4 strains remained genetically stable over this period.
The most significant genetic variation among Irish strains occurred within the serotype G1 virus population, among which four different RFLP patterns were identified. This finding indicated that, compared to serotype G2 and G4 strains, the serotype G1 viruses were genetically more diverse. To investigate this further the nucleotide and deduced amino acid sequences of three G1 isolates associated with three different RFLP patterns were identified. These data revealed that at least two distinct G1 lineages were cocirculating in the Irish population, including G1-1 (isolate CIT-6RV) and G1-3 (isolate CIT-4RV). Both of these strains were isolated in the first year of the original study (21), indicating that both lineages were cocirculating at that time. Significantly, the RFLP pattern identified for the latter strain (strain CIT-4RV) was R1A1E1, which was associated with 68% of the Irish serotype G1 isolates examined. This may imply that the G1-3 lineage predominated. A single G1 isolate, CIT-313RV, was associated with the unique RFLP pattern R6A1E1. Examination of the deduced VP7 amino acid sequence of this strain revealed similarities to the VP7 amino acid sequences of both the G1-1 and the G1-3 lineages (data not shown). Significantly, strain CIT-313RV was isolated in 1999, near the end of the study, and thus may be the product of an interserotypic recombination event (34) that may have occurred between the VP7 genes of two distinct G1 lineages, G1-1 and G1-3. The RFLP data informed our choice of VP7 genes for sequencing, and subsequently, two lineages, G1-1 and G1-3, were detected. It would not be unreasonable to suggest that had all 25 serotype G1 strains been sequenced, additional lineages may have been identified.
A restriction enzyme's potential to identify the presence of mixed G-type infections appeared to be dependent on the dosage of the gene for VP7 from coinfecting strains and whether recombination events occurred between strains. The majority of isolates with mixed serotypes were associated with single R, A, and E profiles. It may be reasonable to suggest that one of the coexisting VP7-encoding gene segments was present at a higher copy number in one strain than in other strains. However, some isolates with mixed serotypes, including isolate CIT-254RV, were associated with unique restriction profiles. Further examination of the protein sequence confirmed that this strain was antigenically distinct from other Irish isolates. The two major mechanisms believed to be responsible for the production of antigenic variants of rotavirus strains are nucleotide substitution and gene reassortment (14, 34). However, the latter mechanism occurs in an all-or-none fashion, and the deduced protein sequence of CIT-254RV revealed similarities to the sequences of more than one serotype. Intragenic recombination is a third mechanism by which rotavirus strains escape the neutralizing effect of the host immune system. This involves the exchange of antigenic regions between strains of different serotypes and can potentially create antigenic variants with dramatic changes in their antigenicities (34). This mechanism may be more suited as an explanation for the evolution of strain CIT-254RV in the Irish population. In these settings the RFLP assay was capable of detecting novel antigenically distinct strains.
Comparative sequencing studies previously identified nine variable regions (VR1 to VR9) within the gene segment encoding the VP7 protein (13). These regions are known to be divergent among different serotypes but highly conserved within a given serotype and are thus useful for prediction of the serotype of any isolate by sequence comparison. In particular three of these regions, VR5 (including amino acid residues 87 to 101), VR7 (amino acid residues 142 to 152) and VR8 (amino acid residues 208 to 221) are designated the major antigenic regions within VP7 (Fig. 2, hatched boxes) (7, 8). As stated above, some restriction sites located within the variable regions were associated with a specific G type, supporting the hypothesis that the sequences of the variable regions are highly conserved within a given serotype and are divergent between serotypes. However, some restriction sites located outside the variable regions were G-type specific. These regions encoded the corresponding conserved amino acid residues that were not directly involved in antigenic specificity but, rather, that predicted a specific G type. In earlier studies, Gouvea et al. (12) suggested that these sites are probably reminiscent of a G-type ancestor, and as they do not appear to be influenced by direct selective pressure from neutralizing antibodies, they are stable over time and thus may be useful as potential epidemiological markers for Irish isolates.
Finally, recent reports identified the major circulating G serotypes in Ireland as G1 and G2, with a lower incidence of serotype G4 strains (22). It was previously noted that mixed infections occurred in large numbers, and this feature had the potential to lead to the emergence of novel strains. In the present study restriction enzyme analysis in conjunction with sequencing studies identified genetic and antigenic variants circulating in the population. Thus, RFLP analysis could be a useful epidemiological tool for monitoring the emergence of new strains, assessing the efficacy of vaccination, and identifying strains for ancestral evolution.
Acknowledgments
We thank our colleagues at the Departments of Medical Microbiology at Cork University Hospital, Limerick Regional Hospital, Waterford Regional Hospital, and Temple Street Children's Hospital for supplying rotavirus strains. We also thank Jon Gentsch for valuable comments on the manuscript.
Financial support from Irish Government Scientific Funding Agencies is gratefully acknowledged (grants GTP 96/CR/028 and ARG 98/226). Wyeth Lederle (Ireland) is also acknowledged for financial support.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein, D. I., D. A. Sack, E. Rothstein, K. Reisinger, V. E. O'Sullivan, D. R. Spriggs, and R. L. Ward. 1999. Efficacy of live, attenuated, human rotavirus vaccine 89-12 in infants: a randomised placebo-controlled trial. Lancet 354:287-290. [DOI] [PubMed] [Google Scholar]
- 3.Cardoso, D. D., M. L. Racz, M. S. Azevedo, R. M. Martins, and C. M. Soares. 2001. Genotyping of group A rotavirus samples from Brazilian children by probe hybridization. Braz. J. Med. Biol. Res. 34:471-473. [DOI] [PubMed] [Google Scholar]
- 4.Chang, K. O., A. V. Parwani, and L. J. Saif. 1996. The characterization of VP7 (G type) and VP4 (P type) genes of bovine group A rotaviruses from field samples using RT-PCR and RFLP analysis. Arch. Virol. 141:1727-1739. [DOI] [PubMed] [Google Scholar]
- 5.Clarke, H. F., P. A. Offit, R. W. Ellis, J. J. Eiden, D. Krah, A. R. Shaw, M. Pichichero, J. J. Treanor, F. E. Borian, L. M. Bell, and S. A. Plotkin. 1996. The development of multivalent bovine rotavirus (strain WC30) reassortant vaccine for infants. J. Infect. Dis. 174(Suppl. 1):S73-S80. [DOI] [PubMed] [Google Scholar]
- 6.Cunliffe, N. A., J. S. Gondwe, S. M. Graham, B. D. Thindwa, W. Dove, R. L. Broadhead, M. E. Molyneux, and C. A. Hart. 2001. Rotavirus strain diversity in Blantyre, Malawi, from 1997 to 1999. J. Clin. Microbiol. 39:836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diwakarla, C. S., and E. A. Palombo. 1999. Genetic and antigenic variation of capsid protein VP7 of serotype G1 human rotavirus isolates. J. Gen. Virol. 80:341-344. [DOI] [PubMed] [Google Scholar]
- 8.Dyall-Smith, M. L., I. Lazdins, G. W. Tregear, and I. H. Holmes. 1986. Location of the major antigenic sites involved in rotavirus serotype-specific neutralization. Proc. Natl. Acad. Sci. USA 83:3465-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estes, M. 1996. Rotaviruses and their replication, p. 1625-1655. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa. [Google Scholar]
- 10.Gentsch, J. R., P. A. Woods, M. Ramachandran, B. K. Das, J. P. Leite, A. Alfrieri, R. Kumar, M. K. Bhan, and R. I. Glass. 1996. Review of G and P typing results from a global collection of rotavirus strains: implications for vaccine development. J. Infect. Dis. 174(Suppl. 1):S30-S36. [DOI] [PubMed] [Google Scholar]
- 11.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z.-Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gouvea, V., C. Ramirez, B. Li, N. Santos, L. Saif, H. F. Clark, and Y. Hoshino. 1993. Restriction endonuclease analysis of the VP7 genes of human and animal rotaviruses. J. Clin. Microbiol. 31:917-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green, K. Y., Y. Hoshino, and N. Ikegami. 1989. Sequence analysis of the gene encoding the serotype-specific glycoprotein (VP7) of two new human rotavirus serotypes. Virology 168:429-433. [DOI] [PubMed] [Google Scholar]
- 14.Iturriza-Gomara, M., B. Isherwood, U. Desselberger, and J. Gray. 2001. Reassortment in vivo: driving force for diversity of human rotavirus strains isolated in the United Kingdom between 1995 and 1999. J. Virol. 75:3696-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobson, R. M. 1999. The current status of the rotavirus vaccine. Vaccine 17:13-14. [DOI] [PubMed] [Google Scholar]
- 16.Jin, Q., R. L. Ward, D. R. Knowlton, Y. B. Gabbay, A. C. Linhares, R. Rappaport, P. A. Woods, R. I. Glass, and J. R. Gentsch. 1996. Divergence of VP7 genes of G1 rotaviruses isolated from infants vaccinated with reassortant rhesus rotaviruses. Arch. Virol. 141:2057-2076. [DOI] [PubMed] [Google Scholar]
- 17.Kapikian, A. Z., and R. M. Chanock. 1996. Rotaviruses, p. 1657-1708. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa. [Google Scholar]
- 18.Kapikian, A. Z., Y. Hoshino, R. M. Chanock, and I. Perez-Schael. 1996. Efficacy of a quadrivalent rhesus rotavirus-based human rotavirus vaccine aimed at preventing severe rotavirus diarrhea in infants and young children. J. Infect. Dis. 174(Suppl. 1):S65-S72. [DOI] [PubMed] [Google Scholar]
- 19.Leite, J. P. G., A. A. Alfieri, P. A. Woods, R. I. Glass, and J. R. Gentsch. 1996. Rotavirus G and P types circulating in Brazil: characterisation by RT-PCR, probe hybridisation and sequence analysis. Arch Virol. 141:2365-2374. [DOI] [PubMed] [Google Scholar]
- 20.Maunula, L., and C.-H. von Bonsdorff. 1998. Short sequences define genetic lineages: phylogenetic analysis of group A rotaviurses based on partial sequence of genome segments 4 and 9. J. Gen. Virol. 79:321-332. [DOI] [PubMed] [Google Scholar]
- 21.O'Halloran, F. 2000. The molecular epidemiology of rotavirus in Ireland. Ph.D. thesis. National Council for Educational Awards, Dublin, Ireland.
- 22.O'Halloran, F., M. Lynch, B. Cryan, H. O'Shea, and S. Fanning. 2000. Molecular characterization of rotavirus in Ireland: detection of novel strains circulating in the population. J. Clin. Microbiol. 38:3370-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okada, J., T. Urasawa, N. Kobayashi, K. Taniguchi, A. Hasegawa, K. Mise, and S. Urasawa. 2000. New P serotype of group A human rotavirus closely related to that of a porcine rotavirus. J. Med. Virol. 60:63-69. [PubMed] [Google Scholar]
- 24.O'Mahony, J., B. Foley, S. Morgan, J. G. Morgan, and C. Hill. 1999. VP4 and VP7 genotyping of rotavirus samples recovered from infected children in Ireland over a three-year period. J. Clin. Microbiol. 37:1699-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palombo, E. A. 1999. Genetic and antigenic diversity of human rotaviruses: potential impact on the success of candidate vaccines. FEMS Microbiol. 181:1-8. [DOI] [PubMed] [Google Scholar]
- 26.Palombo, E. A., H. C. Bugg, P. J. Masendycz, and R. F. Bishop. 1997. Sequence of the VP7 gene of an atypical human rotavirus: evidence for genetic and antigenic drift. DNA Sequence 7:307-311. [DOI] [PubMed] [Google Scholar]
- 27.Palombo, E. A., R. Clark, and R. F. Bishop. 2000. Characterisation of a European-like serotype G8 human rotavirus isolated in Australia. J. Med. Virol. 60:56-62. [PubMed] [Google Scholar]
- 28.Patton, J. T. 1995. Structure and function of the rotavirus RNA-binding proteins. J. Gen. Virol. 76:2633-2644. [DOI] [PubMed] [Google Scholar]
- 29.Piec, T. L., and E. A. Palombo. 1998. Sequence comparison of the VP7 of serotype G2 rotaviruses from diverse geographical locations. DNA Sequence 9:369-373. [DOI] [PubMed] [Google Scholar]
- 30.Ramachandran, M., B. K. Das, A. Vij, R. Kumar, S. S. Bhambal, N. Kesari, H. Rawat, L. Bahl, S. Thakur, P. A. Woods, R. I. Glass, M. K. Bhan, and J. R. Gentsch. 1996. Unusual diversity of human rotavirus G and P genotypes in India. J. Clin. Microbiol. 34:436-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramachandran, M., J. R. Gentsch, U. D. Parashar, S. Jin, P. A. Woods, J. L. Holmes, C. D. Kirkwood, R. F. Bishop, H. B. Greenberg, S. Urasawa, G. Gerna, B. S. Coulson, K. Taniguchi, J. S. Bresee, R. I. Glass, and the National Rotavirus Strain Surveillance System Collaborating Laboratories. 1998. Detection and characterization of novel rotavirus strains in the United States. J. Clin. Microbiol. 36:3223-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santos, N., R. C. Lima, C. F. Pereira, and V. Gouvea. 1998. Detection of rotavirus types G8 and G10 among Brazilian children with diarrhea. J. Clin. Microbiol. 36:2727-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sereno, M. M., and M. I. Gorziglia. 1994. The outer capsid protein VP4 of murine rotavirus strain Eb represents a tentative new P type. Virology 199:500-504. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki, Y., T. Gojobori, and O. Nakagomi. 1998. Intragenic recombinations in rotaviruses. FEBS Lett. 427:183-187. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Timenetsky, M. D., V. Gouvea, N. Santos, R. C. Carmona, and Y. Hoshino. 1997. A novel human rotavirus serotype with dual G5-G11 specificity. J. Gen. Virol. 78(Pt. 6):1373-1378. [DOI] [PubMed] [Google Scholar]
- 37.Unicomb, L. E., G. Podder, J. R. Gentsch, P. A. Woods, K. Z. Hasan, A. S. G. Faruque, M. J. Albert, and R. I. Glass. 1999. Evidence of high frequency genomic reassortment of group A rotavirus strains in Bangladesh: emergence of type 9 in 1995. J. Clin. Microbiol. 37:1885-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xin, K.-Q., S. Morikawa, Z.-Y. Fang, A. Mukoyama, K. Okuda, and H. Ushijima. 1993. Genetic variation in VP7 gene of human rotavirus serotype 1 (G1 type) isolated in Japan and China. Virology 197:813-816. [DOI] [PubMed] [Google Scholar]