Abstract
The shoot apical meristem contains cells that undergo continual growth and division to generate the building blocks for the aerial portion of the plant. As cells leave the meristem, they undergo differentiation to form specific cell types. Most notably, heterotrophic cells of the meristem rapidly gain autotrophic capability by synthesis and assembly of components of the chloroplast. At the same time, cells undergo enlargement via vacuolation. Despite significant advances in the characterization of transcriptional networks involved in meristem maintenance and leaf determination, our understanding of the actual mechanism of meristem cell differentiation remains very limited. Using a microinduction technique, we show that local, transient overexpression of a retinoblastoma-related (RBR) protein in the shoot apical meristem is sufficient to trigger cells in the meristem to undergo the initial stages of differentiation. Taken together with recent data showing that RBR protein plays a key role in restricting stem cell differentiation in the root apical meristem, our data contribute to an emerging picture of RBR proteins as a central part of the mechanism controlling meristem cell differentiation.
The shoot apical meristem (SAM) consists of a group of proliferating cells whose progeny provide the building blocks for all aerial organs of the plant. The continual generation of new cells does not, however, normally lead to a continual increase in the size of the SAM. Instead, cells toward the proximal base of the SAM differentiate and become incorporated into the plant stem, whereas groups of cells on the meristem flank become incorporated at regular intervals in time and space into new organs, the leaves (for review, see Tsiantis and Hay, 2003; Byrne, 2005; Fleming, 2005). This process of meristem cell differentiation is associated with and, indeed, in many ways, defined by specific changes in cytology. Thus, whereas cells in the SAM are distinguished by being relatively small and cytoplasmically dense, as cells become incorporated into the stem and leaf structures they become larger as a result of vacuole enlargement, with the cytoplasm being pushed into a layer around the periphery. Simultaneously, the cell proliferation rate tends to decrease and patterns of cell division emerge that eventually define the histology of the leaves and stem (Donnelly et al., 1999). In addition to this change in size and division pattern, cells leaving the SAM domain gain a specific biochemistry. Cells in the SAM are heterotrophic and depend on imported carbon for the catabolic processes that occur in this part of the plant. Most cells adjacent to the SAM rapidly gain autotrophic capability by differentiation of small proplastids into chloroplasts (Fleming et al., 1996). This involves the synthesis and assembly of various components of the photosynthetic apparatus. Despite our in-depth knowledge of photosynthesis, the process of proplastid differentiation remains essentially a black box. Most work in this area has focused on the transdifferentiation of etioplasts into chloroplasts and has revealed key insights into the role of light in regulating the assembly of functional chloroplasts (for review, see Strand, 2004). It is, however, very unclear to what extent environmentally triggered etioplast differentiation relates to developmentally controlled proplastid differentiation. Exposure of the SAM to light does not trigger chloroplast differentiation in these cells (Fleming et al., 1996), indicating a fundamental difference in the situation with etioplasts.
With respect to differentiation in general, one key aspect that has emerged over the last few years is the potential role of the retinoblastoma protein (pRb) as part of the switching process by which cells cease proliferation and enter a phase of differentiation (Inzé, 2005). Originally based on data from the analysis of mammalian systems, the paradigm is that specific cyclin/cyclin-dependent kinase (CDK) complexes regulate the phosphorylation of a pRb and that the phosphorylation status of this protein regulates the G1 to S phase transition (Weinberg, 1995; Harbour and Dean, 2000). Hypophosphorylated pRb is thought to repress the action of a family of E2F-related transcription factors that are necessary for entry into the S phase, whereas after the action of cyclin/CDK complexes hyperphosphorylated pRb can no longer suppress E2F activity, thus allowing progression from the G1 to the S phase to occur. pRb has thus been characterized as a canonical growth repressor and its misexpression implicated in various aspects of neoplasia (Weinberg, 1995; Harbour and Dean, 2000).
With respect to plants, sequencing data and cloning experiments indicate that plants possess genes that encode retinoblastoma-related (RBR) proteins (Xie et al., 1996; Ach et al., 1997; Durfee et al., 2000; Sabelli et al., 2005). Biochemical data show that RBR proteins can interact with potential partners, such as cyclin/CDK complexes (Huntley et al., 1998; Nakagami et al., 2002), and transgenic and mutational approaches have revealed a role for RBR protein in influencing the cell cycle in a manner generally consistent with the paradigm described above. For example, total abrogation of RBR protein function is gametophytic-lethal due to the proliferation of nuclei in the embryo sac (Ebel et al., 2004), but experiments in which RBR protein activity has been inducibly down-regulated during postembryonic growth led to increased cell proliferation (Park et al., 2005; Wildwater et al., 2005) and increased expression of RBR protein suppressed cell proliferation in cell cultures (Gordon-Kamm et al., 2002). Interestingly, in the root apical meristem (RAM) of Arabidopsis (Arabidopsis thaliana), loss of RBR activity led to an increased number of stem cells and overexpression of RBR protein led to stem cell differentiation (Wildwater et al., 2005), implicating RBR protein as a part of the central mechanism controlling stemness in the RAM. Whether RBR protein performs a similar function in the SAM is unknown.
In this article, we show that local and transient overexpression of RBR protein in the SAM is sufficient to trigger cells toward a more differentiated state, including elements of proplastid differentiation. These data are consistent with the emerging paradigm of RBR protein as an important nexus in plant meristems for the decision to either maintain a meristem state or to enter a pathway of differentiation.
RESULTS
Generation of Transgenic Plants in Which RBR Gene Expression Can Be Transiently Induced
A series of transgenic tobacco (Nicotiana tabacum cv Samsun) plants was generated in which the expression of a full-length cDNA encoding RBR protein from Arabidopsis (Ebel et al., 2004) was under inducible transcriptional regulation by the tetracycline (Tet) operator sequence. In the absence of the inducer (anhydrotetracycline [AhTet]), transcription is repressed (Gatz et al., 1992). This repression is competitively lifted by the addition of the inducer either in the medium or via microinduction in which lanolin beads loaded with the inducer are positioned onto small regions of dissected apices (Pien et al., 2001). The small amount of inducer in these beads diffuses into the surrounding tissue, leading to a local and transient induction of the target gene.
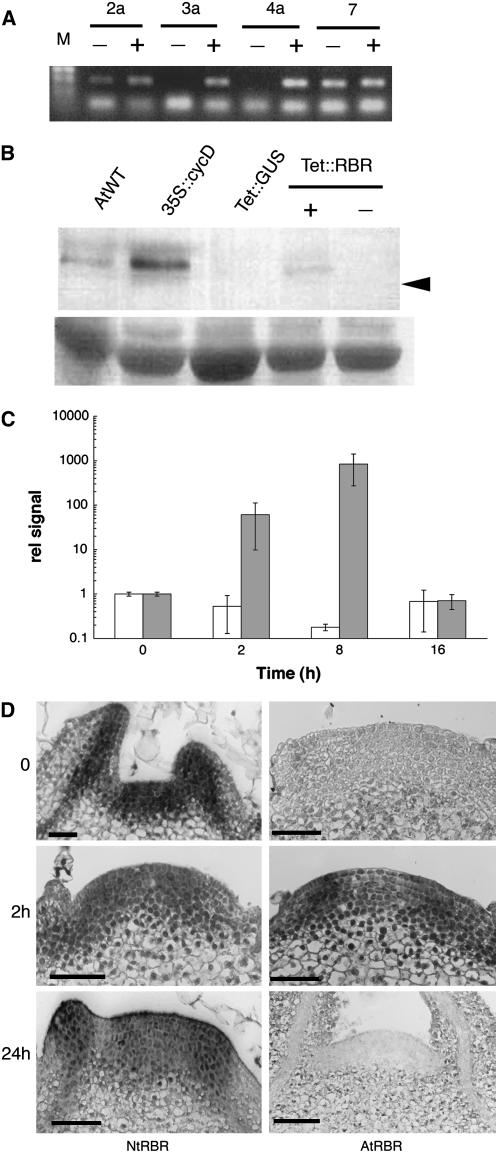
Reverse transcription (RT)-PCR analysis of leaf discs taken from a number of independent transgenic tobacco lines with or without AhTet induction led to the identification of two lines in which AtRBR transcripts were not detectable in uninduced tissue but reached high levels following induction (Fig. 1A). These two lines (Tet∷RBR3a and Tet∷RBR4a) were taken for further characterization by western-blot analysis. Following induction, a low, but detectable, level of RBR protein was detectable within 4 h of treatment with AhTet (Fig. 1B). No signal was detectable in noninduced tissue samples or in induced samples of control plants engineered to contain the β-glucuronidase (GUS) protein under Tet-inducible transcriptional regulation. An intense signal was observed in extracts from Arabidopsis plants engineered to overexpress a cyclinD gene that previous research had shown leads to the accumulation of high levels of the endogenous RBR protein (Dewitte et al., 2003). This sample was used as a positive control to verify that the protein detected in plants in which overexpression of AtRBR had been transiently induced was of the appropriate size. A band of lower signal intensity, but equivalent size, was also observed in proteins extracted from wild-type Arabidopsis plants. In the induced and noninduced tobacco tissue, a number of smaller cross-reacting bands were observed that were of similar intensity in both induced and noninduced tissue. By 16 h after induction, elevated levels of AtRBR protein were not observed in induced tissue, indicating the transient nature of the induction, as observed in previous studies with other gene products using the microinduction system (Pien et al., 2001; Wyrzykowska et al., 2002; Wyrzykowska and Fleming, 2003).
Figure 1.
Characterization of transgenic plants showing inducible expression of the RBR gene. A, RT-PCR analysis of AtRBR expression in leaf discs from independent lines of tobacco transformed with the Tet∷RBR construct. Lines were either induced (+) or not induced (−) with AhTet. M, Molecular size marker. B, Western-blot analysis of protein extracted from tobacco lines transformed with the Tet∷RBR construct and either induced (+) or not induced (−) with AhTet for 4 h. AtWT, Extract from wild-type Arabidopsis; 35S∷CycD, extract from transgenic Arabidopsis engineered to overexpress a cyclinD. These plants have been previously shown to contain elevated levels of the AtRBR protein (Dewitte et al., 2003). Tet∷GUS, Extract from transgenic tobacco plants engineered to inducibly express the GUS reporter gene after treatment with AhTet. Arrowhead indicates migration of a 100-kD marker protein. The bottom image shows amidoblack staining of the blot to ensure approximate equal loading of total protein in all lanes. C, Real-time PCR analysis of NtRBR (white columns) and AtRBR (shaded columns) at different time points in Tet∷RBR apices after induction of AtRBR expression. Expression of each transcript is relative to that of an endogenous actin mRNA (as described in “Materials and Methods”), and this ratio was defined as equal to 1 for the zero time point for each RBR gene. Log data derived from the analysis of three independent biological replicates, with each sample analyzed in triplicate, are shown. D, In situ hybridization analysis of meristems from Tet∷RBR plants hybridized with antisense probes for either NtRBR or AtRBR (as indicated) and at time points 0, 2, and 24 h after AhTet induction (as indicated). Longitudinal sections through the meristems are shown and signal is visible as dark staining. Bar = 50 μm.
To further investigate the temporal change in RBR gene expression in transgenic tobacco plants, a series of microinduction experiments were performed in which the entire SAM of Tet∷RBR plants was induced with AhTet, and the level of both the endogenous NtRBR mRNA and the induced AtRBR mRNA was quantified using real-time PCR. The results (Fig. 1C) indicate that, following this manipulation, there was massive induction of AtRBR gene expression relative to an actin transcript used as an internal control. The timing of the peak of induction varied between 2 and 8 h after apex manipulation, depending on the individual apex analyzed. In all cases, the relative level of AtRBR mRNA returned to an approximately preinduction level within 16 h of the manipulation. Analysis of the mRNA level for an endogenous NtRBR gene indicated that, in the hours following induction, there was a general trend for the expression of this gene relative to the actin control transcript to decrease (Fig. 1C), but, as observed with AtRBR, within 16 h the relative level of the NtRBR transcript had returned to that observed prior to induction. This relative decrease in the NtRBR mRNA level may indicate a mechanism whereby a transient high level of the AtRBR gene product feeds back to the endogenous RBR gene to decrease its expression. Alternatively, due to the similarity of the Arabidopsis and tobacco RBR gene sequences, some limited RNA interference effect cannot be excluded.
These observations on the transient nature of the induction of AtRBR gene expression were supported by in situ hybridization analysis (Fig. 1D). In control apices, there was uniform expression of the endogenous NtRBR transcript throughout the apical meristem and in the young leaf primordia. Hybridization of sections from such uninduced apices revealed no signal with a probe specific for the AtRBR transcript. Within 2 h of induction of Tet∷RBR apices, a distinct signal was apparent in sections hybridized with the AtRBR probe (Fig. 1D). However, by 24 h, only a background signal was apparent in such sections hybridized with the AtRBR probe. In contrast, hybridization with the NtRBR probe of equivalent sections taken at similar time points from induced Tet∷RBR apices revealed a relatively constant and uniform pattern of transcription (Fig. 1D). No overt decrease in the NtRBR mRNA level was observed during this time course (in contrast to the RT-PCR data shown in Fig. 1C), but this might simply reflect the poor quantitative nature of the in situ hybridization technique. Taken together, the western-blot data showing an increased accumulation of RBR protein and the in situ hybridization and quantitative RT-PCR data showing a massive and transient increase in AtRBR mRNA level indicate that the microinduction process led to a transient increase in RBR protein activity in the apex target tissue.
Microinduction of RBR Gene Expression in the Meristem Leads to Repression of Growth
To investigate the outcome of local, transient induction of AtRBR gene expression, aliquots of lanolin impregnated with AhTet were positioned onto either the I1 or I2 position of apical meristems of Tet∷RBR plants and the apices regenerated. The I1 position marks the group of cells that will normally form the next leaf primordium, whereas cells within the I2 position will not normally become incorporated into a leaf until those at I1 have done so. Growth of the apices was observed at various time points up to 5 weeks by counting leaves and imaging plants. Control experiments included both the use of mock-induced Tet∷RBR apices and the AhTet induction of apices dissected from Tet∷GUS plants containing the GUS reporter gene under AhTet-inducible transcriptional regulation (Pien et al., 2001).
As shown in Figure 2, local induction of the meristems of Tet∷RBR apices led to dramatic retardation of plant growth. A range of phenotype was observed that allowed plants to be scored as showing essentially total repression of growth (no new leaves formed; Fig. 2, A and B), moderate inhibition of growth (four to five leaves formed; Fig. 2, C and D), or essentially no inhibition of growth relative to that observed in AhTet-induced Tet∷GUS apices or mock-induced Tet∷RBR plants (seven to nine leaves formed; Fig. 2, E and F). Independent of whether induction was performed at the I1 or I2 position of the meristem, 12/46 (26%) of induced Tet∷RBR apices showed severe growth inhibition (Table I). Such severe growth inhibition was never observed in either mock-induced Tet∷RBR apices or AhTet-induced Tet∷GUS apices. Moderate growth inhibition was observed in 4/28 (14%) AhTet-induced Tet∷GUS apices, whereas 33/46 (72%) of Tet∷RBR apices showed this phenotype after treatment with AhTet. The vast majority of mock-induced Tet∷RBR apices (100%) and AhTet-induced Tet∷GUS apices (86%) showed normal growth after manipulation, whereas only 2% (1/48) of AhTet-induced Tet∷RBR apices displayed a normal rate of growth. These data indicate that local transient overexpression of the AtRBR protein in the tobacco SAM via microinduction was sufficient to induce an extended (5 week) repression of growth. These plants did not display any obvious symptoms of senescence. Instead, they remained green and apparently viable, but with an extremely limited rate of growth.
Figure 2.
Microinduction of RBR gene expression in the SAM leads to repression of growth. A and B, Apices from Tet∷RBR plants showing total repression of growth after microinduction. C and D, Apices from Tet∷RBR plants showing intermediate growth retardation after microinduction. E, Normal growth of apices from Tet∷GUS plants induced with AhTet. F, Normal growth of an apex from a Tet∷RBR plant after mock induction. Plant growth was recorded 5 weeks after microinduction. Bar in A and B = 2 mm.
Table I.
Growth inhibition following microinduction of RBR gene expression
Meristems from Tet∷RBR or Tet∷GUS transgenic plants were either treated with lanolin impregnated with AhTet (+AhTet) or mock induced (−AhTet). Growth response after 5 weeks was classified as either total repression (no new leaf primordia formed); growth retardation (four to five new leaves formed); or normal growth (seven to nine new leaves formed).
| Growth Response | Tet∷RBR + AhTet | Tet∷RBR − AhTet | Tet∷GUS + AhTet |
|---|---|---|---|
| Total repression | 12 | 0 | 0 |
| Growth retardation | 33 | 0 | 4 |
| Normal growth | 1 | 6 | 24 |
Microinduction of RBR Gene Expression Leads to Dramatic Changes in Meristem Cytology
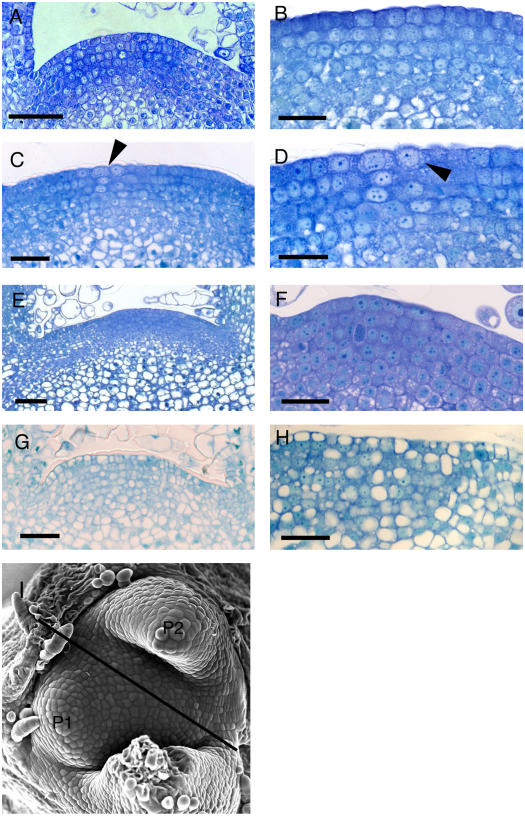
To investigate the nature of growth repression induced by the induction of AtRBR gene expression, we performed a histological and cytological analysis of induced and control apices. As shown in Figure 3A, the SAM of a control plant consists of outer cell layers (the tunica) surrounding an inner corpus in which cell division orientation is not uniform. The cells in the tunica are characterized by being relatively uniform in size, densely cytoplasmic, with a large central nucleus (Fig. 3B). Cells deeper in the corpus tend to be larger and show the first signs of vacuolation. After microinduction of RBR gene expression in the meristem, the first visible change in cytology occurred within 24 h (Fig. 3, C and D). In the area of induction, cells in the outer tunica layers became enlarged and a chain of vesicles became apparent around the central nucleus. By 72 h, the meristem became enlarged and flattened (compare Fig. 3, E and A). There was a clear gradient of cells from the flank of the meristem to the center, with cells on the flanks being vacuolated and cells in the center of the meristem being compact and intensely stained. Closer observation of these cells (Fig. 3F) revealed that they were enlarged and showed a staining pattern distinct from that observed in the tunica of control apices (compare Fig. 3, F and B). Three weeks after induction of RBR gene expression, the meristems contained a mixture of highly vacuolated cells intermingled with smaller, more compact cells (Fig. 3, G and H). The tightly ordered pattern of cell division observed in control meristems tended to disappear and the entire appearance of the SAM was very different from that of a control SAM (compare Fig. 3, G and H, and A and B). The altered meristem histology and cytology correlated with the growth phenotype of the plants (i.e. apices showing the most severe growth retardation showed the most severe cytological and histological defects).
Figure 3.
Induction of RBR gene expression leads to altered meristem histology. A, Longitudinal section of a control apex showing the meristem dome. B, Detail of the outer cell layers from a control meristem. C, As in A, but from a Tet∷RBR meristem in which RBR gene expression has been induced by microinduction 24 h previously (arrowhead). D, Detail from the section in C showing the enlarged cells (arrowhead) observed after microinduction of RBR genes. E, As in A, but from a Tet∷RBR meristem in which RBR gene expression has been induced by microinduction 72 h previously. F, Section to show the histology of the central region of a meristem treated as in E. G, As in A, but from a Tet∷RBR meristem in which RBR expression has been induced by microinduction 3 weeks previously. H, Section to show the histology of the central region of a meristem treated as in G. I, Scanning electron micrograph of a tobacco apex to show approximate orientation (black line) of the sections shown in A to H. Primordia numbers are given as P1 and P2. Bars in A, C, E, F, G, and H = 40 μm; bars in B and D = 20 μm.
To assay the changes in meristem cell size, the cross-sectional areas of cells in micrographs of mock-treated and induced apices were measured (Table II). Cells in control meristems had a mean area of 145 μm2, whereas 24 and 72 h after microinduction the cells in the tunica layers had mean areas of 209 and 205 μm2, respectively. Statistical analysis of these data indicated a significant increase in mean cell size after induction of RBR gene expression (t test; P < 0.01).
Table II.
Microinduction of RBR gene expression leads to altered cell size
Apices of Tet∷RBR plants were microinduced with AhTet (+AhTet) or mock induced (−AhTet). At times after induction (24 or 72 h), samples were analyzed for cross-sectional cell area in either the apical meristem or P3 stage leaf primordium. Images were taken from at least three independent apices and the mean area and sd calculated from a total number of cells (n).
| Tissue Analyzed | Treatment | Mean (n) | sd |
|---|---|---|---|
| Apical meristem | −AhTet | 145.35 (214) | 43.17 |
| +AhTet, 24 h | 209.26 (137) | 69.51 | |
| +AhTet, 72 h | 204.94 (256) | 61.29 | |
| Primordium | −AhTet | 566.83 (50) | 202.57 |
| +AhTet, 72 h | 251.13 (65) | 65.59 |
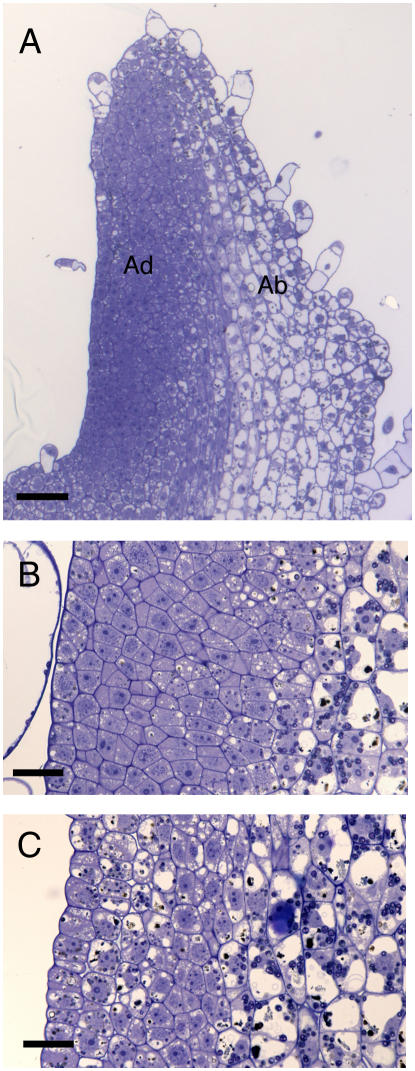
Following microinduction of RBR gene expression, changes in cytology were observed not only in the SAM, but also in the leaf primordia immediately adjacent to the induction site on the meristem. Thus, as shown in Figure 4A, 72 h following induction of RBR gene expression, the entire adaxial face of the primordium closest to the site of microinduction consisted of cells that were relatively small and cytoplasmically dense compared with cells on the abaxial face, which underwent the normal process of cell expansion and vacuolation characteristic of cells in this position at this stage of leaf development. The histology of the adaxial face of primordia in induced apices is more clearly shown in Figure 4B, which can be compared with that of tissue in this region from noninduced apices (Fig. 4C). The adaxial cells in the primordia of induced apices are smaller, lack large vacuoles, and possess a cytology more reminiscent of meristematic cells, whereas the adaxial cells of control primordia undergo cell expansion and vacuolation, with cell division orientation being maintained to generate the layered structure characteristic of the maturing leaf. Measurement of cell cross-sectional areas in the adaxial tissue of primordia from induced and noninduced apices confirmed the visual impression of altered cell size, with induced adaxial cells being significantly smaller (t test; P < 0.01) than noninduced cells (Table II).
Figure 4.
Histology of primordia from apices in which RBR gene expression has been locally induced. A, Longitudinal section through a primordium showing abnormal histology (retarded vacuolation and enlargement) on the induced adaxial (Ad) face of the organ compared to the abaxial (Ab) face in which a normal progression of leaf cell differentiation is occurring. B, Longitudinal section through the adaxial face of a primordium from an induced Tet∷RBR apex. C, As in B, but from a control mock-induced apex. Bar in A = 200 μm; bar in B and C = 40 μm.
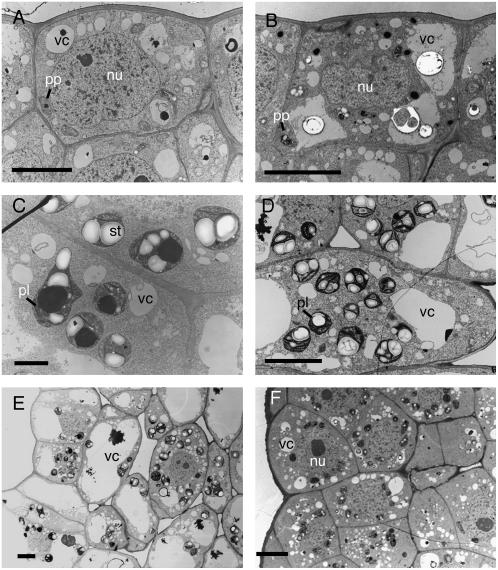
To further analyze the cytological response to microinduction of RBR gene expression, we performed a transmission electron microscopy (TEM) analysis of the tissue. A typical cell in the LI layer of the tunica is relatively small (diameter approximately 10 μm) and contains a large nucleus surrounded by dense cytoplasm within which a few small vacuoles and proplastids are visible (Fig. 5A). After induction of RBR gene expression, significant changes in cytology were apparent within 24 h (Fig. 5B). The most noticeable change was an agglomeration of vacuoles to form larger bodies and an increase in cell size. In addition, electron-opaque, globular bodies were frequently observed within the vacuoles (which may represent lipid bodies) and electron-dense bodies were apparent within the cytoplasm. Already at these early stages of response to RBR gene induction, changes in plastid differentiation were detectable and these became obvious within 72 h (Fig. 5C). Lamellae were observed to form within the proplastids, which also tended to accumulate large electron-opaque starch granules, as well as electron-dense bodies. Such plastids were never observed in cells in control apices at this position within the meristem. The structure of the differentiating plastids within the induced meristems can be compared with plastids in a normally developing leaf primordium (Fig. 5D). At this stage of leaf differentiation, the plastids contain stacks of lamellae and prominent starch granules. Electron-dense bodies are not observed in these differentiated plastids. The cells themselves also contain relatively large vacuoles.
Figure 5.
Electron microscopy reveals promotion of differentiation in meristem cells after microinduction of RBR gene expression. A, Cell from the outer layer of a control meristem. B, Cell from the outer layer of a Tet∷RBR meristem in which RBR gene expression has been induced 24 h previously. C, Plastid differentiation in a tunica cell of a Tet∷RBR plant in which RBR gene expression has been induced 24 h previously. D, Plastid differentiation in a primordium from a control apex. E, Adaxial cell differentiation from a control leaf primordium. F, Adaxial cells from a primordium of a Tet∷RBR apex has been microinduced to express RBR 72 h previously. nu, Nucleus; pl, plastid; pp, proplastid; st, starch granule; vc, vacuole. Bars in A, B, D, E, and F = 5 μm; bar in C = 1 μm.
As shown in Figure 4, one result of induction of RBR gene expression was the accumulation of small cytoplasmically dense cells on the adaxial face of developing leaf primordia adjacent to the site of microinduction. TEM analysis confirmed the data obtained by light microscopy, as shown in Figure 5F. The adaxial cells of induced Tet∷RBR apices were smaller, less vacuolated, and more cytoplasmically dense than comparable cells in this position from noninduced tissue (Fig. 5E).
RBR Gene-Induced Growth Retardation Is Associated with Altered Expression Patterns of Marker Genes for the Cell Cycle and Meristem Function
A paradigm of RBR function is that it controls entry into the cell cycle via regulation of passage of cells through a point in the G1 to S phase transition. To investigate the outcome of local induction of RBR gene expression on cell division, we performed a series of in situ hybridizations using marker genes for various phases of the cell cycle (Fig. 6). In nontreated control apices, histone H4 mRNA accumulates in spots throughout the SAM and young leaf primordia, marking cells in the S phase (Fig. 6A). Within 24 h of RBR gene induction, transcripts for H4 were virtually undetectable and did not accumulate in their characteristic speckled pattern (Fig. 6B). After 72 h, a slight recovery in H4 mRNA signal was visible in some apices, with a few cells showing a low level of transcript accumulation (Fig. 6C). Nicta;CYCA3;2 (encoding an A-type cyclin that can also be used as a marker for the G1 to S phase transition; Wyrzykowska et al., 2002; Yu et al., 2003) showed an essentially similar pattern of expression (data not shown).
Figure 6.
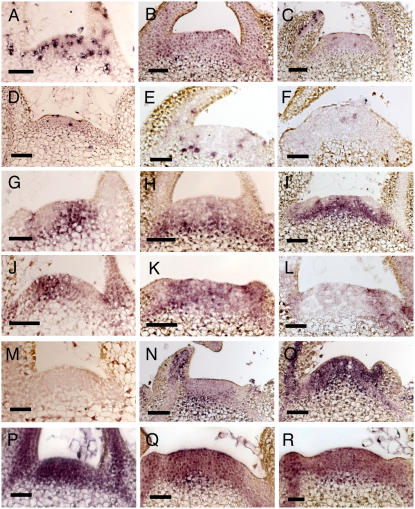
Analysis of marker gene expression in meristems microinduced to overexpress RBR gene expression. A, In situ hybridization with an antisense probe for histone H4 against a longitudinal section of an apex from a Tet∷RBR plant without AhTet induction. B, As in A, but 24 h after microinduction. C, As in A, but 72 h after microinduction. D, As in A, but hybridized with an antisense probe for NtCYCB1. E, As in D, but 24 h after microinduction. F, As in D, but 72 h after microinduction. G, As in A, but hybridized with an antisense probe against NTH15. H, As in G, but 24 h after microinduction. I, As in G, but 72 h after microinduction. J, As in A, but hybridized with an antisense probe against NTPHAN. K, As in J, but 24 h after microinduction. L, As in J, but 72 h after microinduction. M, As in A, but hybridized with an antisense probe against RBCS. N, As in M, but 24 h after microinduction. O, As in M, but 72 h after microinduction. P, As in A, but hybridized with an antisense probe against NteIF4A. Q, As in P, but 24 h after microinduction. R, As in P, but 72 h after microinduction. Bars = 50 μm.
Nt;CYCB1 transcripts (encoding a B-type cyclin) accumulate in plant cells during the G2 to M phase transition. In untreated apices, Nt;CYCB1 transcripts accumulated in a speckled pattern throughout the SAM (Fig. 6D). Within 24 h of RBR gene induction, cyclinB transcripts were virtually undetectable within the meristem (Fig. 6E). After 72 h, Nt;CYCB1 transcripts (as observed for H4 and Nicta;CYCA3;2) showed limited recovery of the normal expression pattern, but with a spectrum of signal intensity depending on the apex analyzed (Fig. 6F).
In addition to the specific expression patterns of genes encoding cell cycle-associated proteins, the SAM is characterized by a number of distinct patterns of gene expression that can be used as diagnostics of meristem function. For example, transcripts encoding homeodomain transcription factors of the KNOTTED class (KNOX genes) accumulate throughout the SAM but are excluded from the region involved in specification of a new leaf primordium (for review, see Tsiantis and Hay, 2003; Byrne, 2005). At the same time, leaf primordium initiation is characterized by accumulation of transcripts encoding a class of MYB transcription factors (encoded by ARP genes), with the pairwise regulation of KNOX and ARP gene families being intimately involved with the process of leaf initiation. In tobacco, NTH15 transcripts (encoding an STM-like KNOX factor) mark all cells in the meristem except the presumptive primordium (Fig. 6G; Sakamoto et al., 2001), and NTPHAN mRNA (encoding an ARP factor) marks the earliest stage of primordium formation (Fig. 6J; Wyrzykowska and Fleming, 2003). Within 24 h of microinduction of the RBR gene in the SAM, disruption of the normal pattern of NTH15 and NTPHAN marker gene expression was observed. NTH15 transcript accumulation in the SAM decreased, although mRNA was still detectable in the submeristem region (Fig. 6H). By 72 h, some expression of NTH15 was reestablished within the SAM (Fig. 6I), although the pattern remained patchy compared with that observed in control apices. At 24 h after induction, NTPHAN transcripts accumulated in a patchy pattern throughout the meristem (Fig. 6K). By 72 h after induction, NTPHAN transcripts were barely detectable within the SAM (in which vacuolated cells were clearly apparent), except toward the flank of the meristem (Fig. 6L).
Meristem cells are nonphotosynthetic and do not normally express genes encoding key components of this metabolic pathway, such as the small subunit of Rubisco (RBCS; Fleming et al., 1996). Thus, transcripts encoding RBCS cannot be detected in the SAM (Fig. 6M). RBCS transcripts were detectable within 24 h of microinduction of RBR gene expression (Fig. 6N), and by 72 h a strong RBCS transcript signal was apparent throughout the SAM (Fig. 6O).
As a final control to investigate the general pattern of transcriptional activity in the responding meristems, we performed hybridizations with a probe against mRNA encoding a translation initiation factor, NteIF4A. The encoded protein is involved in protein translation and can be used as a probe for general transcriptional/translational activity (Mandel et al., 1995). In control apices, a strong signal was observed throughout the SAM (Fig. 6P). Within 24 h of induction of RBR gene expression, the intensity of NteIF4A transcript accumulation was lower (Fig. 6Q), and this decreased level of signal was maintained at 72 h after induction (Fig. 6R), consistent with the observed decrease in tissue growth.
DISCUSSION
RBR Protein and Control of Meristem Cell Differentiation
Recent work on the RBR gene in Arabidopsis has implicated it both in maintenance of stemness of classically defined initial cells in the RAM and in control of differentiation of these cells (Wildwater et al., 2005). Induced loss of RBR protein using a cre-lox system led to sectors of tissue permanently lacking RBR protein activity. When these sectors were targeted to the stem cell initials around the quiescent center of the RAM, increased stem cell proliferation was observed. In contrast, induced overexpression of RBR protein in the RAM led to stem cells undergoing at least some aspects of differentiation (e.g. acquisition of statoliths). The results reported here confirm and extend this work by showing that overexpression of RBR protein in the SAM is sufficient to switch these cells from proliferation to a state in which the initial events of differentiation occur. Thus, the potential role of RBR protein in controlling stemness in the RAM may be a general feature of meristems in plants. Moreover, because of the nature of the technique used in this investigation, we show that a brief (<24 h) elevation of RBR protein level is sufficient to trigger this change. This suggests that the response to elevated RBR protein in the SAM is unidirectional (i.e. once the cells in the meristem have been triggered to enter the RBR protein-directed pathway, it is not a trivial matter for the process to be reversed). Such unidirectional flow of differentiation is reminiscent of the concept of epigenetic landscapes first promulgated by Waddington almost 50 years ago (Waddington, 1957) in which cell differentiation can be envisaged as a sphere moving by gravity down an undulating hillside in which the different valleys represent alternative pathways of differentiation. Once a particular valley has been entered, it requires specific inputs to push the cell back up the slope to a less differentiated state. Following this analogy, one interpretation is that RBR protein acts as either a modulator of the landscape or as a brake preventing movement of a cell downhill toward differentiation (see also Harris, 2004). Interestingly, pRb-related complexes have been shown to interact with chromatin remodeling factors capable of setting particular patterns of gene expression in an epigenetic fashion (i.e. capable of setting relatively stable patterns of gene expression; Hennig et al., 2003; Narita et al., 2003), and chromatin remodeling factors have recently been implicated as key players in the maintenance of stem cell identity in mammals (Lee et al., 2006). Such a mechanism of action would fit with the observed requirement for only a transient induction of RBR protein in the SAM to set in motion a pathway of differentiation. At the same time, at least some of the induced SAMs in our experiments did eventually reinitiate an approximately normal rate of growth. This implies that reversal of the RBR protein-induced state was possible in at least some cells, or that at least some cells in the SAM retained meristematic potential to reestablish a functional SAM. Further investigation is required to distinguish these possibilities.
Previous investigators have indicated that RBR protein in tobacco can functionally interact with cyclinD/CDK complexes (Huntley et al., 1998; Nakagami et al., 2002; Uemukai et al., 2005) and, taken together with the accepted model of RBR function, it seems likely that an initial outcome of elevated RBR protein level in the SAM in our experiments would be to temporarily swamp the endogenous cell cycle-promoting activity of cyclinD/CDK complexes, thus leading to inhibition of cell proliferation, as observed. It should be noted, however, that induced RBR protein expression led to an almost complete and rapid cessation of cell division (as visualized by cell cycle marker gene expression and subsequent lack of growth), whereas during the normal process of meristem cell differentiation some level of proliferation is maintained for a period of time after exit from the SAM. If RBR protein does act as part of the endogenous system for meristem cell differentiation (discussed below), it is likely to act in a graded fashion mediated by posttranslational modification of a relatively constant local level of protein rather than by an all-or-nothing change in expression level instigated by the genetic methods used here and in other investigations. Investigating such local changes in RBR activity in vivo will be technically challenging.
Although the experiments reported here depended on altered transcription of an introduced RBR gene, the endogenous function of RBR protein is likely to be highly dependent on posttranslational modification of the protein (Kaye et al., 1990; Harbour and Dean, 2000). The importance of such posttranslational events in RBR action makes interpretation of RBR transcription patterns difficult. Thus, although our in situ hybridization data suggest a higher level of RBR expression in the SAM (where cells remain relatively undifferentiated and maintain a relatively high rate of proliferation), they do not necessarily imply that high RBR activity is associated with high rates of cell division. Indeed, due to the limited vacuolization of cells in the SAM, even a gene that is uniformly expressed (on a cell basis) will tend to show a higher signal in the SAM (where cytoplasmic density is higher); that is, our data do not imply SAM-specific expression of RBR. Rather, they suggest that RBR is present to some extent in all cells of the shoot apex and are consistent with the idea that RBR protein acts as a fulcrum around which factors promoting proliferation or differentiation can act. Because, generally, the cellular decision process goes from proliferation to nonproliferation (and differentiation), a relatively high level of transcript encoding a protein involved in this decision in proliferating cells is not unexpected. Our expression data define a region where RBR can act and this area encompasses the region where the decision to maintain or exit proliferation takes place.
The results of our experiments also suggest that modification of cell activity in one part of the SAM (via RBR protein induction) leads to modification of cell activity in other parts of the SAM and, indeed, on the flanks of the leaf primordia adjacent to the induction site on the SAM. Thus, whereas cells induced to express RBR protein underwent initial steps in differentiation in the SAM, adjacent cells in the leaf primordium maintained meristematic cytology. This is consistent with the idea of a signaling system by which appropriate differentiation of cells in the leaf primordium requires a signal from a functioning SAM. In the absence of such a signal, the cells in the leaf do not undergo appropriate growth and differentiation. The mechanisms underlying these observations are at present unknown; however, a number of publications indicate that dynamic signaling events occur both within the SAM and between the SAM and the surrounding leaf primordia to coordinate growth activity (e.g. Waites and Hudson, 1995; Kim et al., 2002; Juarez et al., 2004; Kidner and Martienssen, 2004). The identification and characterization of these signaling systems is a pressing requirement. Our data do not resolve this issue, but they identify appropriate cell proliferation in the SAM as a component of the control system. Our results also highlight the importance of specifying the spatial and temporal boundaries of altered gene expression in transgenic experiments and show how interpreting the function of genes such as RBR simply as favoring differentiation or proliferation may be rather simplistic in the context of a multicellular organism in which altered cellular activity in one region may (via endogenous signaling mechanisms) elicit an apparently different response in neighboring tissue.
RBR Gene and the KNOX/ARP Transcription Factor Module
Down-regulation of the KNOX transcript level in cells undergoing determination to form leaves, and concomitant accumulation of transcripts encoding ARP transcription factors, is a general facet of SAM function (Tsiantis and Hay, 2003; Byrne, 2005). This has led to the proposal that KNOX gene expression is intimately linked to the maintenance of meristem identity (Jackson et al., 1994; Long et al., 1996; Sablowski, 2004). However, at the proximal base of the SAM, the cutoff between KNOX expressing and nonexpressing cells is not sharp, with published images showing both meristem cells and cells undergoing differentiation expressing KNOX genes. After induction of RBR gene overexpression, a loss of transcripts for both NTH15, concomitant ectopic accumulation of NTPHAN transcripts, and cellular differentiation were observed in the SAM. These observations are consistent with the idea that loss of KNOX-like gene expression is linked with the loss of meristem identity/potential and with the initiation of specific aspects of biochemical differentiation (Mele et al., 2003). At later stages after RBR induction, a partial restoration of NTH15 expression occurred, but this was insufficient to restore SAM activity, indicating that KNOX gene expression alone is insufficient to establish meristem activity. These data support the idea that the cellular context of KNOX gene expression is important (Hay et al., 2003) and that epigenetic changes induced by RBR overexpression might render the cells nonresponsive to the normal action of NTH15 in the SAM. Research in the RAM has revealed an interaction between RBR protein-mediated and transcription factor-mediated control of meristem function (Wildwater et al., 2005). Whether a similar relationship exists in the SAM between RBR and KNOX factors (or other transcription factor families, such as WUSCHEL-related homeodomain factors) will require analysis in a genetically more tractable system, such as Arabidopsis, but our data demonstrate the potential for such a linkage.
In conclusion, our data indicate that regulation of RBR protein in the SAM might play a similar role to that recently established in the RAM with respect to the control of stem cell differentiation. The function of retinoblastoma-like proteins in controlling entry into the cell cycle has long been established, but the potential role of these proteins in directing differentiation has been less well explored (Dimova et al., 2003; Wikenheiser-Brokamp, 2004). The further characterization of the role of RBR protein in plant cell differentiation will be of great interest. In particular, in the context of the SAM, there is a dearth of information on the earliest steps of proplastid differentiation and on the switch to vacuolar-driven expansion growth, which occurs as cells leave the SAM (Fleming, 2006). The ability to switch meristem cells toward a pathway of differentiation, coupled with the development of techniques that allow the characterization of global patterns of gene expression in minute tissue samples, opens a pathway for the investigation of basic aspects of plant cell differentiation.
MATERIALS AND METHODS
Plant Material and Transformation
R7 tobacco (Nicotiana tabacum cv Samsun) seedlings (a gift from A. Jones, University of North Carolina, Chapel Hill) were transformed (Pien et al., 2001), regenerants grown in a greenhouse, and F1 seeds collected for analysis. For microinduction experiments, plants were grown on soil in a growth chamber (16 h light at 24°C/8 h dark at 20°C cycle) or on one-half-strength Murashige and Skoog medium, pH 5.6, 1% (w/v) agar (16 h light/8 h dark cycle at 24°C, 100 μmol m−2 s−1). For RNA-blot and protein analyses, seedlings were grown on Murashige and Skoog medium.
DNA Manipulation
The Arabidopsis (Arabidopsis thaliana) RBR cDNA was cloned into the pBINHyg-TX vector (Gatz et al., 1992) to generate the clones pBinHyg-Tx-RBR. This was used to transform tobacco seedlings, as described above. All DNA manipulations were performed by standard techniques (Sambrook et al., 1992).
Microinduction
Microinductions were performed as described (Pien et al., 2001) using lanolin/paraffin paste with 10 mg mL−1 AhTet. Controls were performed by using dimethyl sulfoxide/lanolin/paraffin paste without AhTet. After manipulation, apices were grown on one-half-strength Murashige and Skoog medium, pH 5.6, in a growth chamber (16 h light/8 h dark cycle at 24°C, 100 μmol m−2 s−1) as described by Pien et al. (2001).
RNA and Protein Analysis
For RT-PCR, total RNA was extracted from 4-week-old seedlings using RNeasy columns (Qiagen). In situ hybridization was done as described (Pien et al., 2001) using digoxigenin-labeled sense and antisense riboprobes for AtRBR, NtRBR, NTH15, NTPHAN, Nt;CYCB1, Nt;CYCA3;2, Nt;EIF4A, RBCS, and H4. For real-time PCR, total RNA was extracted from 4-week-old seedlings using TRIzol (Invitrogen). Traces of DNA were removed with DNase I (RNase-free), then 2 μg of total RNA was used to prepare cDNA using avian myeloblastosis virus reverse transcriptase (Invitrogen). Quantitative RT-PCR was performed using an ABI PRISM 7700 sequence detection system. PCR reactions for each time point (for target genes and the reference gene) were prepared using SYBR Green PCR master mix (Applied Biosystems) and the following primers: Nt.RBR forward, 5′-gctgggttccggaagcttgtct and reverse, 5′-cacctgtaagcacagcgatgacaa; At.RBR forward, 5′-agatgctgccacctccgtt and reverse, 5′-tcctccacctcctgggttg; and Nt.actin forward, 5′-gccagtggccgtacaacaggtattg and reverse, 5′-tagtggtgaacgagtagcctcgct.
Statistical analyses were performed following the formula of Pfaffl et al. (2002). Western-blot analysis was by standard protocols with a horseradish peroxidase-linked secondary antibody and enhanced chemiluminescence visualization, as described by the manufacturer (Roche Diagnostics). Fifty micrograms of protein from seedling extract were loaded per lane. The primary antibody used was raised (Eurogenetec) in rabbit against a mixture of synthetic peptides from three regions of the AtRBR protein (N terminus, AB domain, and C terminus). In extracts from wild-type Arabidopsis seedlings, the antibody detected a protein with an observed migration close to that predicted for the AtRBR protein (112 kD). Moreover, the intensity of this band was elevated in plants engineered to overexpress the AtRBR gene either directly (via overexpression of the AtRBR gene; L. Mariconti, H. Feiler, J. Futterer, and W. Gruissem, unpublished data) or indirectly via overexpression of a cyclinD gene (shown in Fig. 1A). In addition, a previously characterized antibody raised against maize (Zea mays) RBR protein (Huntley et al., 1998) identified the same band in parallel extracts.
TEM
Tobacco pieces were fixed in 2% (w/v) glutaraldehyde in 0.05 m sodium cacodylate buffer, pH 7.2, for 2 h at room temperature and postfixed with 1% (w/v) OsO4 in sodium cacodylate buffer at 4°C overnight. After dehydration with acetone at room temperature, the material was embedded in Spurr's standard epoxy resin with dibutyl phthalate and the resin was polymerized at 70°C for 19 h. Thin sections were stained with 2% (w/v) uranyl acetate in 50% (v/v) acetone for 30 min and alkaline lead citrate for 30 min (Reynolds, 1963). Micrographs were taken with a Philips CM 100 BIOTWIN electron microscope.
Acknowledgments
We thank Chantal Ebel and Luisa Mariconti (Swiss Federal Institute of Technology [ETH]) for fruitful discussions, Jean-Pierre Metraux for access to the Fribourg TEM facility, and Nikolaus Amrhein (ETH) for providing lab space and encouragement.
This work was supported by the Swiss Federal Institute of Technology-Zürich (grant to A.J.F. and W.G.) and the Swiss National Science Foundation (START Fellow to A.J.F.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Andrew J. Fleming (a.fleming@sheffield.ac.uk).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.083022.
References
- Ach RA, Durfee T, Miller AB, Taranto P, Hanley-Bowdoin L, Zambryski PC, Gruissem W (1997) RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol Cell Biol 17: 5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME (2005) Networks in leaf development. Curr Opin Plant Biol 8: 59–66 [DOI] [PubMed] [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy JMS, Jacqmard A, Kilby NJ, Murray JA (2003) Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15: 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova DK, Stevaux O, Frolov MV, Dyson NJ (2003) Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev 17: 2308–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Durfee T, Feiler HS, Gruissem W (2000) Retinoblastoma-related proteins in plants: homologues or orthologues of their metazoan counterparts? Plant Mol Biol 43: 635–642 [DOI] [PubMed] [Google Scholar]
- Ebel C, Mariconti L, Gruissem W (2004) Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429: 776–780 [DOI] [PubMed] [Google Scholar]
- Fleming AJ (2005) Formation of primordia and phyllotaxy. Curr Opin Plant Biol 8: 53–58 [DOI] [PubMed] [Google Scholar]
- Fleming AJ (2006) The co-ordination of cell division, differentiation and morphogenesis in the shoot apical meristem: a perspective. J Exp Bot 57: 25–32 [DOI] [PubMed] [Google Scholar]
- Fleming AJ, Manzara T, Gruissem W, Kuhlemeier C (1996) Fluorescent imaging of GUS activity and RT-PCR analysis of gene expression in the shoot apical meristem. Plant J 10: 745–754 [DOI] [PubMed] [Google Scholar]
- Gatz C, Frohberg C, Wendenburg R (1992) Stringent repression and homogeneous de-repression by tetracycline of a modified CaMV 35S promoter in intact transgenic tobacco plants. Plant J 2: 397–404 [DOI] [PubMed] [Google Scholar]
- Gordon-Kamm W, Dilkes BP, Lowe K, Hoerster G, Sun X, Ross M, Church L, Bunde C, Farrell J, Hill P, et al (2002) Stimulation of the cell cycle and maize transformation by disruption of the plant retinoblastoma pathway. Proc Natl Acad Sci USA 99: 11975–11980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour JW, Dean DC (2000) Rb function in cell-cycle regulation and apoptosis. Nat Cell Biol 2: E65–67 [DOI] [PubMed] [Google Scholar]
- Harris H (2004) Tumour suppression: putting on the brakes. Nature 427: 201. [DOI] [PubMed] [Google Scholar]
- Hay A, Jackson D, Ori N, Hake S (2003) Analysis of the competence to respond to KNOTTED1 activity in Arabidopsis leaves using a steroid induction system. Plant Physiol 131: 1671–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Taranto P, Walser M, Schönrock N, Gruissem W (2003) Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development 130: 2555–2565 [DOI] [PubMed] [Google Scholar]
- Huntley R, Healy S, Freeman D, Lavender P, de Jager S, Greenwood J, Makker J, Walker E, Jackman M, Xie Q, et al (1998) The maize retinoblastoma protein homologue ZmRb-1 is regulated during leaf development and displays conserved interactions with G1/S regulators and plant cyclin D (CycD) proteins. Plant Mol Biol 37: 155–169 [DOI] [PubMed] [Google Scholar]
- Inzé D (2005) Green light for the cell cycle. EMBO J 24: 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D, Veit B, Hake S (1994) Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413 [Google Scholar]
- Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MCP (2004) microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428: 84–88 [DOI] [PubMed] [Google Scholar]
- Kaye FJ, Kratzke RA, Gerster JL, Horowitz JM (1990) A single amino acid substitution results in a retinoblastoma protein defective in phosphorylation and oncoprotein binding. Proc Natl Acad Sci USA 87: 6922–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA (2004) Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428: 81–84 [DOI] [PubMed] [Google Scholar]
- Kim JY, Yuan Z, Cilia M, Khalfan-Jagani Z, Jackson D (2002) Intercellular trafficking of a KNOTTED1 green fluorescent protein fusion in the leaf and shoot meristem of Arabidopsis. Proc Natl Acad Sci USA 99: 4103–4108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al (2006) Control of developmental regulators by polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Mandel T, Fleming AJ, Krähenbühl R, Kuhlemeier C (1995) Definition of constitutive gene expression in plants: the translation initiation factor 4A gene as a model. Plant Mol Biol 29: 995–1004 [DOI] [PubMed] [Google Scholar]
- Mele G, Ori N, Sato Y, Hake S (2003) The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev 17: 2088–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Kawamura K, Sugisaka K, Sekine M, Shinmyo A (2002) Phosphorylation of retinoblastoma-related protein by the cyclin D/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. Plant Cell 14: 1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Nuñez S, Heard E, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW (2003) Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113: 703–716 [DOI] [PubMed] [Google Scholar]
- Park J-A, Ahn J-W, Kim Y-K, Kim SJ, Kim J-K, Kim WT, Pai H-S (2005) Retinoblastoma protein regulates cell proliferation, differentiation, and endoreduplication in plants. Plant J 42: 153–163 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien S, Wyrzykowska J, McQueen-Mason S, Smart C, Fleming A (2001) Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc Natl Acad Sci USA 98: 11812–11817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds EST (1963) The use of lead citrate at high pH as an electron opaque stain in electron microscopy. J Cell Biol 25: 208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli PA, Dante RA, Leiva-Neto JT, Jung R, Gordon-Kamm WJ, Larkins BA (2005) RBR3, a member of the retinoblastoma-related family from maize, is regulated by the RBR1/E2F pathway. Proc Natl Acad Sci USA 102: 13005–13012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R (2004) Plant and animal stem cells: conceptually similar, molecularly distinct? Trends Cell Biol 14: 605–611 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Kamiya N, Ueguchi-Tanaka M, Iwahori S, Matsuoka M (2001) KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev 15: 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1992) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Strand A (2004) Plastid-to-nucleus signalling. Curr Opin Plant Biol 7: 621–625 [DOI] [PubMed] [Google Scholar]
- Tsiantis M, Hay A (2003) Comparative plant development: the time of the leaf? Nat Rev Genet 4: 169–180 [DOI] [PubMed] [Google Scholar]
- Uemukai K, Iwakawa H, Kosugi S, de Uemukai S, Kato K, Kondorosi E, Murray JA, Ito M, Shinmyo A, Sekine M (2005) Transcriptional activation of tobacco E2F is repressed by co-transfection with the retinoblastoma-related protein: cyclin D expression overcomes this repressor activity. Plant Mol Biol 57: 83–100 [DOI] [PubMed] [Google Scholar]
- Waddington CH (1957) The Strategy of the Genes. A Discussion of Some Aspects of Theoretical Biology. George Allen & Unwin, London
- Waites R, Hudson A (1995) Phantastica: a gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121: 2143–2154 [Google Scholar]
- Weinberg RA (1995) The retinoblastoma protein and cell cycle control. Cell 81: 323–330 [DOI] [PubMed] [Google Scholar]
- Wikenheiser-Brokamp KA (2004) Rb family proteins differentially regulate distinct cell lineages during epithelial development. Development 131: 4299–4310 [DOI] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B (2005) The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123: 1337–1349 [DOI] [PubMed] [Google Scholar]
- Wyrzykowska J, Fleming A (2003) Cell division pattern influences gene expression in the shoot apical meristem. Proc Natl Acad Sci USA 100: 5561–5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrzykowska J, Pien S, Shen WH, Fleming AJ (2002) Manipulation of leaf shape by modulation of cell division. Development 129: 957–964 [DOI] [PubMed] [Google Scholar]
- Xie Q, Sanz-Burgos AP, Hannon GJ, Gutiérrez C (1996) Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J 15: 4900–4908 [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Steinmetz A, Meyer D, Brown S, Shen W-H (2003) The tobacco A-type cyclin, Nicta;CYCA3;2, at the nexus of cell division and differentiation. Plant Cell 15: 2763–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]