Abstract
Developmental progression and differentiation of distinct cell types depend on the regulation of gene expression in space and time. Tools that allow spatial and temporal control of gene expression are crucial for the accurate elucidation of gene function. Most systems to manipulate gene expression allow control of only one factor, space or time, and currently available systems that control both temporal and spatial expression of genes have their limitations. We have developed a versatile two-component system that overcomes these limitations, providing reliable, conditional gene activation in restricted tissues or cell types. This system allows conditional tissue-specific ectopic gene expression and provides a tool for conditional cell type- or tissue-specific complementation of mutants. The chimeric transcription factor XVE, in conjunction with Gateway recombination cloning technology, was used to generate a tractable system that can efficiently and faithfully activate target genes in a variety of cell types. Six promoters/enhancers, each with different tissue specificities (including vascular tissue, trichomes, root, and reproductive cell types), were used in activation constructs to generate different expression patterns of XVE. Conditional transactivation of reporter genes was achieved in a predictable, tissue-specific pattern of expression, following the insertion of the activator or the responder T-DNA in a wide variety of positions in the genome. Expression patterns were faithfully replicated in independent transgenic plant lines. Results demonstrate that we can also induce mutant phenotypes using conditional ectopic gene expression. One of these mutant phenotypes could not have been identified using noninducible ectopic gene expression approaches.
Advances in inducible gene expression technologies will facilitate more precise functional analyses of endogenous and exogenous genes, revealing new roles for genes that act at multiple stages in the plant life cycle. Such analyses will assist the development of new, improved crop varieties. Conditional and cell type-specific gene expression systems allow precise functional complementation of mutants, disclosing the spatial and temporal significance of a gene's expression profile at different stages in development. Eukaryotic genomes have evolved through numerous rearrangements, producing duplicated genes with functional redundancy, a characteristic that is particularly evident in plants. In Arabidopsis (Arabidopsis thaliana; for review, see Curtis and Grossniklaus, 2005), duplications comprise more than 60% of the genome (Blanc et al., 2000). The functional significance of redundantly acting genes can be determined only by combining several loss-of-function mutations in genes of the same family, or by expressing one such gene ectopically (Eshed et al., 2001). Typically, genes are ectopically expressed using a constitutive and broadly active promoter, such as the cauliflower mosaic virus (CaMV) 35S promoter (Odell et al., 1985). This approach has been the basis of many gene function studies, including investigations of single-copy genes (Mizukami and Ma, 1992; Jack et al., 1994). Ubiquitous and constitutive gene expression can, however, result in lethality if a gene of critical importance to an early stage in development is misexpressed. Problems can also occur in complementation studies if a gene is expressed not only in the tissue type required, but also in other tissues (Laufs et al., 2003). The combination of cell type-specific complementation and loss-of-function mutation provides powerful tools to elucidate the role of genes, particularly those acting at multiple stages of development (Gross-Hardt et al., 2002), and can expose the noncell-autonomous nature of a gene's activity (An et al., 2004). Genetic complementation at early stages of development can be achieved using the promoter of another gene with a similar early expression profile (Gross-Hardt et al., 2002); however, alternative promoters are not always available. In such cases, a system that allows cell type-specific conditional complementation is invaluable. Such a system that could also allow conditional cell type-specific activation of randomly tagged genes would provide the versatility required to identify classes of mutants with tissue-specific effects or mutants with early lethal effects. Such mutants could be rescued in noninduced plants or plant sectors. None of the currently available systems provides such versatility (for review, see Curtis and Grossniklaus, 2006). Although inducible activation-tagging systems that allow temporal control of gene expression have been developed (Matsuhara et al., 2000; Zuo et al., 2000), none allows tissue-specific activity, which can be critically important, as demonstrated by the induced ectopic expression of expansin in restricted tissue (meristem fractions) leading to leaf formation (Pien et al., 2001). Here, gene activity was controlled by the careful, manual application of a chemical to a subset of meristematic cells. This would be impractical for experiments in less accessible cell types, such as reproductive tissues within the gynoecium.
There are, of course, systems that transactivate genes using tissue-specific promoters, but these have limitations. The ethanol-inducible system (Deveaux et al., 2003; Maizel and Weigel, 2004), for example, is limited by the volatile nature of the inducer, which can cause unwanted gene activation in neighboring plants (Roslan et al., 2001), toxic effects on the induced plant (Roslan et al., 2001), and can be activated by endogenous inducers under low-oxygen conditions (Salter et al., 1998; Roslan et al., 2001). These features make it difficult to produce distinct sectors of induced and uninduced gene expression essential to determine the cell autonomy of a phenotype and to address hitherto intractable problems encountered during reproductive developmental studies (where gene activation can prevent production of viable seeds). Some systems, based on nonvolatile inducers, have shown leaky activity (Martinez et al., 1999; De Veylder et al., 2000) or an inability to reliably activate responder T-DNAs randomly inserted in the genome (Baroux et al., 2005). This prohibits their use in precise activation-tagging screens. We have solved these problems by developing a system that allows localized, conditional gene induction within sectors of the plant exposed to the inducer. The system conditionally activates randomly integrated responder T-DNAs (regardless of their insertion position) at a high frequency for use in activation-tagging screens and reliably restricts expression to predicted tissue types. This allows the isolation of mutant phenotypes affecting seed development, which could not easily be identified using conventional misexpression approaches. We enhanced our system by incorporating Gateway cloning sites (Hartley et al., 2000) so that researchers and biotechnologists can use the growing number of tissue-specific Gateway-compatible cis-elements (An et al., 2004) and full-length cDNAs (Gong et al., 2004) that are now available for a broad range of applications.
RESULTS
Our aim was to generate a reliable and versatile two-component tissue-specific inducible gene expression system to provide a method by which randomly tagged genes, or candidate genes, could be conditionally activated in restricted sectors of a plant in restricted tissue types. These demands have resulted in the production of a stringent system with broad applications (Fig. 1).
Figure 1.
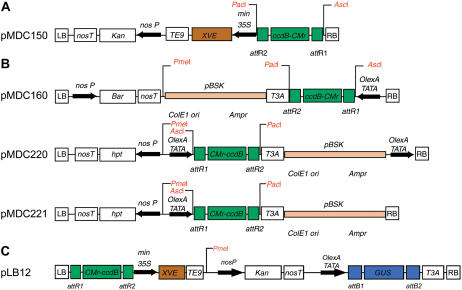
A schematic illustration of the Gateway-compatible constructs. A, Activator vector. B, Responder vectors. C, Activator/responder vector. The activator vector pMDC150 (A) contains a Gateway cloning cassette, flanked by unique AscI and PacI restriction recognition sites upstream of a CaMV 35S minimal promoter (min 35S) and chimeric transcription factor XVE. The inclusion of a minimal promoter allows the insertion of both enhancer sequences and promoters in the constructs (the inclusion of a minimal promoter does not interfere with the specificity of the promoters used in this study). The vector pMDC150 also contains a Nos promoter to drive the expression of a kanamycin-resistance gene for plant selection. All responder vectors (B) contain an XVE-responsive promoter (OlexA-TATA) upstream of a Gateway cloning cassette, also flanked by unique AscI and PacI restriction recognition sites for the easy diagnosis of DNA insertions. Vectors pMDC160, pMDC220, and pMDC221 contain a Nos promoter to drive the expression of the BAR- or hygromycin-resistance genes, respectively, for plant selection. The vectors pMDC160, pMDC220, and pMDC221 also contain the pBluescript vector sequence (CLONTECH), which can be used for plasmid rescue procedures because it encodes an ampicillin-resistance gene for bacterial selection and the ColE1 origin of replication. The pMDC220 vector also contains a second XVE-responsive promoter adjacent to the right border (RB) sequence, so that this vector can be used for conditional activation tagging experiments. The activator/responder vector pLB12 (C) contains both an activator unit and a responder unit, separated by a kanamycin-resistance gene driven by a Nos promoter for plant selection. The sequences and detailed maps of these vectors can be downloaded from http://www.unizh.ch/botinst/Devo_Website/curtisvector. LB, Left border; TE9, TE9 terminator; T3A, terminator; attR1 and attR2, att recombination sites; CMr, bacterial chloramphenicol resistance; ccdB, bacterial toxin gene for negative selection.
Components of the Inducible Transactivation System
The system comprises an activator unit and a responder unit. The activator T-DNA (pMDC150) contains the transcriptional activator, XVE (Zuo et al., 2000), with a minimal CaMV 35S promoter. The responder T-DNA contains an XVE-responsive promoter that can be used to misexpress candidate genes or reporter genes (i.e. pMDC160 and pMDC221) or to activate randomly tagged genes (i.e. pMDC220). Both activator and responder T-DNAs contain Gateway recombination sites (Fig. 1, A and B). The vector pLB12 contains both an activator unit and a β-glucuronidase (GUS) reporter (Jefferson et al., 1987) within a responder unit (Fig. 1C).
Vector-Dependent Regulation of Gene Expression
While developing this technology, we identified that responder T-DNAs containing the 35S promoter (i.e. a pCAMBIA-derived vector [http://www.cambia.org]), which regulates the antibiotic resistance marker, can lead to uninduced transgene expression in responder constructs (data not shown). This uninduced expression was observed even when a 3-kb fragment containing the entire pUC vector sequence (Invitrogen) was introduced between the 35S promoter and the responder cassette (data not shown). Similar interference by the 35S promoter has recently been reported in the pCAMBIA vector series (Yang et al., 2005) and the pPZP series (Yoo et al., 2005). No uninduced expression of reporter genes was observed with vectors pMDC160, pMDC221, pMDC220, or pLB12, which were derived from the pMoa vector series containing a Nos (nopaline synthase) promoter to regulate the antibiotic resistance marker. As a result, all plant vectors used to develop the two-component inducible system were derived from the pMoa vector series with the Nos promoter (rather than the 35S promoter) to regulate the expression of the selectable marker.
Conditional Gene Expression
To test the system, an enhancer fragment from the CaMV 35S promoter was inserted upstream of XVE, producing the activator T-DNA, pMDC150-35S. This was used to transactivate the GUS reporter in Arabidopsis plants previously transformed by a pMDC160-GUS responder construct. Sectors of induced gene activity were observed when leaf material was treated with 2 μm 17-β-estradiol (0.01% Silwet 77) using an artist's paint brush and GUS stained 24 h later (Fig. 2).
Figure 2.
Arabidopsis leaves showing sectors of induced GUS expression 24 h after induction with 2 μm 17-β-estradiol (0.01% Silwet 77) using an artist's paint brush.
Varying the 17-β-estradiol exposure time resulted in altered reporter gene activity, peaking between 24 to 48 h. A similar peak in activity was reported using the PER8 vector (Zuo et al., 2000). Although GUS activity remains largely in the tissue directly beneath the site of 17-β-estradiol application, there is slow spreading, which results in a halo of weak GUS activity that gradually spreads throughout the entire leaf over a 72- to 96-h period (data not shown). Seedlings grown on 10 μm 17-β-estradiol plates showed strong induced GUS activity throughout the plant tissue when compared to seedlings grown on mock-inoculated plates (which show no expression), demonstrating that 17-β-estradiol can permeate the aerial parts of the plant when only the roots are exposed to the inducer (Supplemental Fig. 1); however, when inflorescences are allowed to take up 17-β-estradiol by transpiration (2 μm 17-β-estradiol in water), the inducer tends to promote most GUS activity in the vasculature and adjacent tissue, but will eventually permeate the stem, cauline leaves, and even the ovules within the gynoecium of the flower after 96 h of exposure (Supplemental Fig. 2). The chosen method of application will depend on the developmental stage of interest to be studied in the plant (i.e. 17-β-estradiol application in media would be most appropriate for early seedling development studies, whereas inducer application to inflorescence tissue by transpiration or topical application using a paint brush and a spreading agent [Silwet 77 or Break thru S240] might be more appropriate for floral or reproductive developmental studies).
Efficiency of Transactivation
The value of a two-component inducible transactivation system depends on its ability to deliver reliable and conditional tissue type-specific gene expression at a high frequency in independent transgenic plant lines, particularly when screening for inducible phenotypes in restricted cell types that result from activation-tagging approaches. A pMDC150-35S activator plant line was used to establish the frequency with which random pMDC160-GUS responder insertions can be activated (Table I). Leaves were excised from transformants and analyzed histochemically for GUS activity, with and without induction by 2 μm 17-β-estradiol (Fig. 3A). Plant lines (87%), with responders inserted at 23 independent loci, gave inducible ubiquitous expression. The reciprocal experiment was also performed (Table I). Fourth-generation pMDC160-GUS responder plant lines were supertransformed with the pMDC150-35S activator. Here, 95.8% of plants showed predictable induced expression with activators inserted at 48 independent loci.
Table I.
Comparative expression analysis of independent activator and responder T-DNAs positioned throughout the genome
| Primary Plant Line Contains T-DNA | Supertransformed with T-DNA | Expected | Aberrant | None | Total Lines |
|---|---|---|---|---|---|
| PMDC150-35S activator | pMDC160-GUS responder | 20 | 0 | 3 | 23 |
| PMDC150-SUC2 activator | pMDC160-GUS responder | 28 | 0 | 2 | 30 |
| PMDC150-GL2 activator | pMDC160-GUS responder | 27 (2 weak) | 0 | 3 | 30 |
| Subtotal | 75 | 0 | 8 | 83 | |
| PMDC160-GUS responder | pMDC150-35S activator | 46 | 0 | 2 | 48 |
| PMDC160-GUS responder | pMDC150-SUC2 activator | 12 | 0 | 1 | 13 |
| PMDC160-GUS responder | pMDC150-TobRB7 activator | 21 | 0 | 5 | 26 |
| Subtotal | 79 | 0 | 8 | 87 | |
| PMDC150-GL2 activator | pMDC220-GUS tagging | 1,040 (289a) | 271 | 154 | 1,465 |
| Total lines | 1,194 | 271 | 170 | 1,635 |
Plants with expression in trichomes and other tissues. These patterns of expression were also observed in some control transgenic plant lines containing pMDC163-GL2 promoter-GUS T-DNAs.
Figure 3.
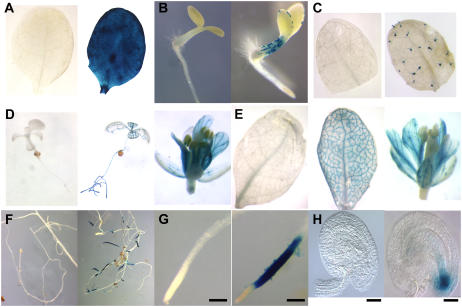
Cell-specific expression patterns of a GUS reporter gene after induction. Uninduced and induced GUS expression (uninduced shown on the left of each image, respectively) in Arabidopsis plant lines 24 h after induction (2 μm 17-β-estradiol in 0.01% [v/v] ethanol) or mock induction (0.01% [v/v] ethanol). A, Leaves of a plant line containing pMDC150-35S and pMDC160-GUS (induced ubiquitous expression can be seen across the leaf). B and C, GUS expression in plant lines containing pMDC150-GL2 and pMDC160-GUS. B, 3-d-old seedlings (induced expression is restricted to the atrichoblast cells). C, Mature leaves of 15-d-old plants (induced expression restricted to the trichomes). D, Whole plants at 7-to-10-d old (left and middle) and a flower (right) from plant lines containing pMDC150-RolC and pMDC160-GUS (induced expression is restricted to the vascular tissue). E, Mature leaves (left and middle) and a flower (right), including petals, from plant lines containing pMDC150-SUC2 and pMDC160-GUS (induced expression is observed in the vascular tissue [companion cells]). F and G, Roots in mature plant lines containing pMDC150-TobRB7 and pMDC160-GUS (induced expression is restricted to the tissue above the root apical meristem [RAM]). H, Plant line containing pLB12-EASE (induced expression is observed in the egg apparatus). Bars = 0.5 mm (G) and 20 μm (H).
Stringency of Gene Expression System
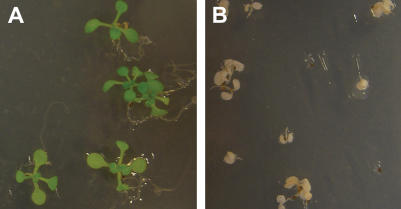
The stringency of the two-component Gateway-compatible system was tested using pMDC150-35S activator lines supertransformed with the responder pMDC221, containing the cytotoxic diphtheria A-chain (DT-A; Maxwell et al., 1986; Harrison et al., 1991; pMDC221-DT-A). The DT-A gene kills cells by ribosylating elongation factor-2, leading to the inhibition of protein synthesis (Collier, 1967). Here, 13 independent transgenic plant lines containing both the pMDC150-35S activator and a pMDC221-DT-A responder showed no phenotypic effects in the absence of the 17-β-estradiol inducer. However, induction with as little as 2 μm 17-β-estradiol leads to signs of cell death in all 13 plant lines (Fig. 4). This demonstrates the tight regulation of genes adjacent to the XVE-responsive promoter. The DT-A toxin has been used as a tool to ablate specific plant tissues (Weijers et al., 2003; Yang et al., 2005). By replacing the 35S promoter in pMDC150 with tissue-specific promoters, expression of the DT-A gene could be restricted to specific tissues allowing inducible cell type-specific ablation.
Figure 4.
Stringent regulation of gene expression using the system is demonstrated by induction of DT-A in seedlings. Arabidopsis plant lines containing both the pMDC150-35S activator T-DNA and a pMDC221-DT-A responder T-DNA after 13 d of growth under uninduced conditions (A; mock inoculated Murashige and Skoog media) and under induced conditions (B; 2 μm 17-β-estradiol in Murashige and Skoog medium).
Inducible Tissue-Specific Transactivation
To test the system's ability to deliver inducible tissue-specific gene expression, five activator constructs were generated using promoter or enhancer elements with different tissue specificities. These included elements with vascular-specific (RolC from Agrobacterium tumefaciens and AtSUC2 companion cell specific; An et al., 2004), root-specific (NtTobRB7; An et al., 2004), trichome/atrichoblast-specific (AtGL2; Szymanski et al., 1998), and egg apparatus-specific (AtEASE; Yang et al., 2005) activities. These elements were all tested in the pMDC150 activator T-DNA, except for AtEASE, which was tested in the vector pLB12 (Fig. 1). The T-DNA pLB12-AtEASE contains both the activator and responder units, greatly facilitating expression analysis of AtEASE by placing both units within the same haploid cells of the T1 female gametophytes. This provides the most stringent of tests for the system because the egg apparatus is inaccessible, located at the micropylar end of the ovule, deep within the gynoecium, surrounded by the reproductive tissue of the flower. Each activator construct was tested for inducible cell type-specific activity. After induction, the pattern of expression was shown to be identical to those obtained with the same cis-elements in the GUS reporter construct pMDC163 (Curtis and Grossniklaus, 2003; data not shown) and to previously published patterns of expression (Szymanski et al., 1998; An et al., 2004; Yang et al., 2005). Inducible transactivation patterns of GUS expression are shown in Figure 3.
Activation in Trichomes
Inducible GUS activity observed in Arabidopsis transformed with pMDC150-GL2 and a pMDC160-GUS responder T-DNA was consistent with patterns of expression previously observed (Hung et al., 1998; Szymanski et al., 1998). The AtGL2 promoter fragment used in this study contained the 500-bp EcoRV/XbaI DNA fragment previously identified as necessary to direct GUS expression in differentiating hairless epidermal cells in the hypocotyls and roots (atrichoblasts). After induction, GUS expression was observed in the differentiating hairless epidermal cells that form the atrichoblast cells (Fig. 3B). In leaf primordia and developing leaves, pMDC150-GL2 was able to inducibly transactivate the GUS responder T-DNA in developing trichomes and in surrounding epidermal cells. At later stages in leaf development, this pattern of expression became more tightly restricted to trichome cells, with GUS activity also observed at the petiole and base of the leaf. In mature leaves, expression was restricted entirely to trichomes and was maintained throughout the lifetime of these cells (Fig. 3C). Similar patterns of expression (data not shown) were also observed in Arabidopsis plants transformed by the same AtGL2 promoter fragment inserted upstream of a GUS reporter in the vector pMDC163 (Curtis and Grossniklaus, 2003).
Activation in Vascular Tissue
The previously reported phloem-specific expression pattern of the RolC promoter (Booker et al., 2003) was faithfully reproduced using pMDC150-RolC and a pMDC160-GUS responder T-DNA. Upon induction, GUS activity was observed in the phloem of the roots, stem, leaves, and floral organs (such as sepals, petals, anther filaments, and style; Fig. 3D). The promoter fragment used (An et al., 2004) is nearly identical to that described by Booker and colleagues (2003). Similarly, when using the AtSUC2 promoter to drive XVE expression using pMDC150-SUC2, the GUS gene was conditionally expressed in companion cells, as described by Truernit and Sauer (1995; Fig. 3E). GUS expression was also detected in the phloem throughout the plant; but, whereas Truernit and Sauer (1995) observed no GUS activity in the petals, we observed induced activity in the vascular tissue of all floral organs, including petals (Fig. 3E). In contrast to the promoter fragment of Truernit and Sauer (1995), which extends 156 bp into the protein coding sequence, the promoter fragment we used (An et al., 2004) ends at the start codon of the AtSUC2 gene.
Activation in Roots
Arabidopsis plants, transformed with pMDC150-TobRB7 and pMDC160-GUS responder T-DNA, showed induced reporter gene expression in mature plants that mimicked the expression pattern described for tobacco (Nicotiana tabacum) plants transformed with TobRB7-GUS constructs (Yamamoto et al., 1991; Fig. 3F). GUS activity in mature roots was restricted to a region of the root above the root apical meristem (Fig. 3G).
Activation in the Egg Apparatus
Reporter gene expression in the egg apparatus was conditionally activated in Arabidopsis plants transformed with pLB12-AtEASE. The activation unit of this construct contains five tandem repeats of 77-bp AtEASE (a modified version of that previously described by Yang et al., 2005). Upon induction with 17-β-estradiol, GUS activity was observed in the egg apparatus (Fig. 3H). Induced expression was occasionally observed in the entire embryo sac, which is consistent with the previous report that AtEASE is sometimes active prior to cellularization of the female gametophyte (Yang et al., 2005).
Efficiency of Tissue-Specific Transactivation
To determine whether tissue-specific expression patterns are maintained in independent plant lines, regardless of the genomic position of pMDC160-GUS responder insertions, numerous transformants were generated. The plants used had activation T-DNAs in fixed positions in the genome, showing different patterns of XVE expression (pMDC150-SUC2 and pMDC150-GL2; Table I). Leaves were excised from transformants and analyzed histochemically for GUS activity, with and without induction by 2 μm 17-β-estradiol; 91.7% of plant lines, with responders inserted at 60 independent loci, gave inducible expression in the expected tissue type (Table I). This confirms that our system could be used to create an inducible activation-tagging system to activate randomly tagged genes in specific tissues or cell types. The reciprocal experiment was also performed, using the AtSUC2 and NtTobRB7 promoters to investigate the ability of randomly inserted pMDC150-promoter T-DNAs to activate a pMDC160-GUS responder plant line (Table I). Fourth-generation responder plant lines were supertransformed with pMDC150-promoter activator T-DNAs. Again, leaves were excised from transformants and analyzed histochemically for GUS activity with and without induction by 17-β-estradiol. Here, 84.6% of plant lines, with pMDC150-promoter activators inserted at 39 independent loci, showed inducible expression in the expected tissue type (Table I).
In a pilot study, we produced a T-DNA activation-tagging construct, pMDC220-GUS, and analyzed the GUS expression of 1,465 independent insertions for induced activity in trichomes of a plant line containing the pMDC150-GL2 activator T-DNA (Table I). Here, 71% of plant lines with randomly inserted activation-tagging constructs produced tissue-specific expression that faithfully mimicked both the previously described expression patterns (Szymanski et al., 1998) and that observed in control plant lines: 18.5% showed aberrant expression and 10.5% showed no expression. If we also take the results obtained with other specific promoters into account, our data suggest that 73% of transformants produce an inducible pattern of expression that faithfully mimics that of the promoter/enhancer selected to transactivate the responder T-DNA. A low number of insertions (10.4%) show no expression, suggesting that the activator and the responder are equally likely to insert into regions of the genome that affect their activity. Furthermore, a proportion (16.6%) of transgenic plant lines show aberrant expression, a percentage similar to that observed in experiments with promoter-GUS constructs.
Induction of Specific cDNAs Using the 35S XVE Activator
To demonstrate the value of the system for high-throughput gene analysis, cDNAs were inducibly expressed in plant tissues corresponding to the expression pattern of the CaMV 35S promoter during seedling development. These cDNAs were selected for their variety of clearly visible phenotypes early in development. They included KNOTTED-LIKE FROM ARABIDOPSIS 1 (KNAT1; Lincoln et al., 1994; Fig. 5), which produces lobed leaves when misexpressed using the 35S promoter, BABY BOOM (BBM; Boutilier et al., 2002; Fig. 6), and LEAFY COTYLEDON 2 (LEC2; Stone et al., 2001; Fig. 7), both triggering a conversion from vegetative to embryonic growth when misexpressed using the 35S promoter. Using this system, we were able to inducibly reproduce phenotypes previously described in the literature.
Figure 5.
Inducible gain-of-function phenotypes: Overexpression of KNAT1 leads to lobed leaf formation. A, 27 d of growth under uninduced conditions. B, 27 d of growth under induced conditions. A and B, Siblings from a plant line containing both the pMDC150-35S activator T-DNA and a pMDC221-KNAT1 responder T-DNA.
Figure 6.
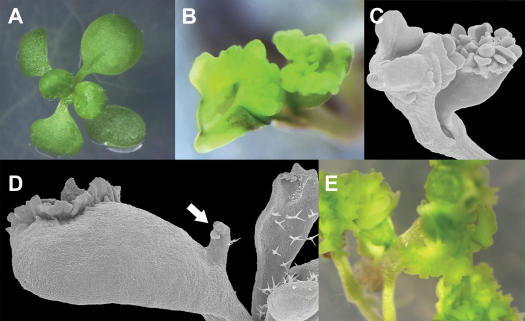
Inducible expression of BBM leads to the formation of somatic embryos on cotyledons and leaves. A, 13 d of growth under uninduced conditions (mock inoculated Murashige and Skoog medium). B, 13 d of growth under induced conditions (5 μm 17-β-estradiol in Murashige and Skoog medium). A and B, Sibling plant lines containing both the pMDC150-35S activator T-DNA and a pMDC221-BBM responder T-DNA. C and D, Scanning electron micrographs of induced somatic embryos (C) in plant shown in B. D, Scanning electron micrographs of cotyledons and leaves with induced somatic embryos and a leaf-like outgrowth with a trichome on a cotyledon (arrow). E, 13-d-old plants that constitutively express BBM under the control of the CaMV 35S promoter (seeds kindly provided by Kim Boutilier).
Figure 7.
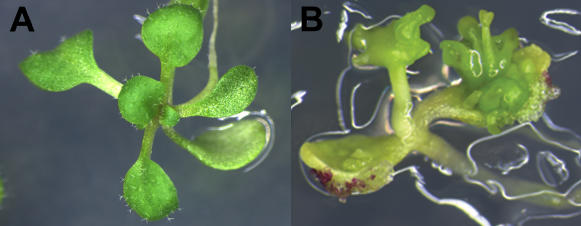
Induced ectopic LEC2 expression results in the formation of somatic embryos on cotelydons. A, 29 d of growth under uninduced conditions (mock inoculated Murashige and Skoog medium). B, 29 d of growth under induced conditions (5 μm 17-β-estradiol in Murashige and Skoog medium). A and B, Sibling plant lines containing both the pMDC150-35S activator T-DNA and a pMDC221-LEC2 responder T-DNA.
To determine whether this system could identify the phenotype of a gene that would be overlooked by conventional ectopic gene expression methods, we inducibly expressed the FUSCA3 (FUS3) gene. Because mutations in this gene can cause viviparous seed development (Raz et al., 2001), our prediction was that ectopic expression of FUS3 would cause seed dormancy. In conventional ectopic gene expression approaches, dormant seeds would be indistinguishable from nontransformants on selection plates after transformation. Plants containing both pMDC150-35S and pMDC221-FUS3 were selected in the absence of induction and showed no mutant phenotype (Fig. 8A). The T1 generation double transformants showed delayed germination when exposed to 2 μm 17-β-estradiol (Fig. 8B), with a stronger seed dormancy phenotype when exposed to 5 μm 17-β-estradiol (Fig. 8C). Similar plant lines misexpressing the GUS reporter gene, instead of FUS3, in the presence of 2 and 5 μm 17-β-estradiol grow normally. Seedlings containing both pMDC150-35S and pMDC221-FUS3 T-DNAs that were able to germinate on 2 μm 17-β-estradiol showed abnormal growth, with extended hypocotyls and a tendency to produce reduced leaves (data not shown). Seeds that showed prolonged seed dormancy when incubated in the presence of the inducer occasionally germinated several weeks later when left on the same plates. Degradation of the light-sensitive 17-β-estradiol may account for the initiation of this low germination rate. When dormant seeds exposed to 5 μm 17-β-estradiol were transferred to noninductive media, within a short period of 5 d, 65% of seeds germinated and looked normal, no longer showing any aberrant growth phenotypes associated with ectopic FUS3 expression. When these transferred seeds were examined 9 d later, 92% had germinated; however, some of the later germinating seeds (17% of the total) showed the abnormal growth phenotype observed earlier, with extended hypocotyls and a tendency to produce reduced leaves. In fact, our findings are consistent with previously published data (Zuo et al., 2006), which suggest that induced phenotypes, in general, return to wild type in the absence of the inducer after a period of 5 to 7 d. The time taken, however, to return to a wild-type phenotype is gene dependent, perhaps reflecting the stability of a transcript or the type of downstream effects that result from its ectopic expression.
Figure 8.
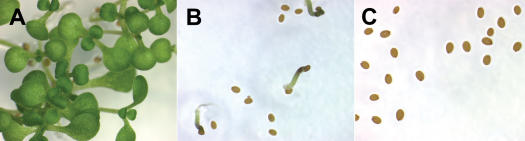
The ectopic expression of FUSCA3 produces a seed dormancy phenotype. Plant lines containing both the pMDC150-35S activator T-DNA and a pMDC221-FUS3 responder T-DNA. A, Uninduced. B, Induced with 2 μm 17-β-estradiol. C, Induced with 5 μm 17-β-estradiol after 14 d.
DISCUSSION
Local Inducible Expression
Here, we describe a highly versatile, inducible gene expression system that provides both spatial and temporal control of gene expression in plants. The system allows rapid production of cell type-specific activation constructs. These activation constructs can faithfully reproduce expression patterns previously described for six promoters/enhancers with different tissue or cell type specificities (Odell et al., 1985; Truernit and Sauer, 1995; Szymanski et al., 1998; Booker et al., 2003; An et al., 2004; Yang et al., 2005). About 90% of plant lines containing the pMDC150-derived activator and pMDC160-derived responder showed inducible expression in the expected tissue type regardless of the position of the T-DNAs in the genome (Table I). These results demonstrate that the system can be used to generate a library of tissue-specific transactivator plant lines for the activation of LexA responder T-DNAs and that the genomic location of the activator or responder is equally likely to affect the reliability of the system. Such efficiency could not be easily achieved using systems in which gene expression is more sensitive to the position of the responder T-DNA (Baroux et al., 2005). In the system described by Baroux et al. (2005), a similar percentage of plant lines (to data presented here) showed transactivated GUS expression (i.e. 86% of plant lines with GUS responders inserted at 37 independent loci), but expression was often weak (e.g. 93.5% of these plant lines showed weak expression). In reciprocal experiments in which one of their strongly responsive pOp-GUS responder plant lines was supertransformed or crossed to a variety of promoter-LhG4 activators, 78.4% of plant lines showed strong GUS expression in predictable patterns with promoter-LhG4 activators at 74 independent loci. The positional effects observed for responders in the LhG4 system may be caused by the relatively weak activity of the Gal4 activation domain used in the LhG4 chimeric transcription factor. The more active VP16 activation domain used in XVE, when tethered to DNA-binding domains, stimulates transcription by targeting histone-acetylating complexes to nucleosomal templates (Tumbar et al., 1999; Vignali et al., 2000), resulting in the decondensation of heterochromatin. Similar decondensation of chromatin structure has been observed around the target sites of the estrogen receptor (Nye et al., 2002), suggesting that elements of XVE's estrogen receptor may also contribute to the higher efficiency of strong gene expression observed in our system. These results suggest that our system could be used to create cell type-specific and inducible activation-tagging technology. Our activation-tagging pilot study showed that 71% of plant lines produced tissue-specific expression that faithfully mimicked both the previously described expression pattern (Szymanski et al., 1998) and that observed in control plant lines; 18.5% showed aberrant expression and 10.5% showed no expression. This frequency is sufficient for use in activation-tagging screens.
Tightly Regulated Gene Expression
A good inducible gene expression system must deliver tightly regulated gene expression. Experiments using the cytotoxin DT-A gene demonstrated that our system is stringently regulated: Despite containing a cytotoxic gene, plants develop normally in the absence of induction. Such a system that provides stringent control of a cytotoxic gene, in combination with the ability to restrict expression to a subset of cells, creates the opportunity to study plants in which certain cell types are ablated.
Production of Induced Sectors of Gene Expression
The nonvolatile nature of the 17-β-estradiol inducer provides a further advantage because it can be applied to restricted sectors of the plant. This means that interesting mutants with lethal effects can be rescued in noninduced sectors. This is of particular importance when attempting to deregulate gene expression in reproductive tissues, where gene induction may produce sterility or lethal effects in the next generation.
Gateway Compatibility and Induced Gene Expression
Our system also benefits from the inclusion of Gateway cloning sites, making the system compatible with the growing collections of full-length cDNA entry clones that are available in the Arabidopsis Stock Center (Gong et al., 2004) and promoter/enhancer cis-elements (An et al., 2004) that allow these genes to be expressed inducibly in any plant cell-type. We used one such cDNA from the Stock Center, pYAT5G17430 (BBM; Gong et al., 2004) to induce somatic embryos on vegetative tissues, faithfully reproducing the previously described phenotype (Boutilier et al., 2002). We also generated Gateway-compatible cDNAs and inducibly misexpressed both KNAT1 (replicating the lobed-leaf phenotype described by Lincoln et al., 1994) and LEC2 (conditionally reproducing the development of somatic embryos in vegetative tissue described by Stone et al., 2001).
Plant lines that show inducible FUS3 expression illustrate the value of our gene expression system. In the absence of induction, transformants can be selected on antibiotic plates and their phenotype determined after induction in subsequent generations. Because ectopic expression of FUS3 using the constitutive and near-ubiquitous CaMV 35S promoter would lead to seed dormancy (as shown with the inducible system), primary transformants would be overlooked because they would be indistinguishable from nontransformants on a selection plate. Despite its strong effect on dormancy when ectopically expressed, FUS3 expression during seed development shows only marginal differences between strongly or moderately dormant Arabidopsis wild-type accessions (Baumbusch et al., 2004). This may reflect the sensitivity of seeds to the expression of FUS3. Significantly, FUS3 has been implicated as a positive regulator of abscisic acid synthesis (Gazzarrini et al., 2004) and abscisic acid is a hormone known to promote seed dormancy (Koornneef et al., 2002). The altered expression of such dormancy genes has the potential to resolve problems of preharvest sprouting in agriculturally important seeds (for review, see Gubler et al., 2005).
Modulated Gene Expression
A further advantage of an XVE-dependent system is that gene expression levels can be modulated using different concentrations of inducer (Zuo et al., 2002). In experiments with KNAT1 and FUS3 plant lines, different concentrations of 17-β-estradiol also modulated the severity of the phenotypes. This allows both weak and strong phenotypes to be examined in the same plant line—a real advantage. The method of inducer application can be adapted to suit requirements. We have developed a number of inducer application methods to enhance the flexibility of our system. Gene expression can be induced in roots, germinating seed, and juvenile plants by growing plants directly on Murashige and Skoog medium supplemented with 17-β-estradiol. Young seedlings can be induced after initial stages of development by transferring them to inductive media. In older plants, gene expression can be induced by allowing excised branches (inflorescences) to take up 17-β-estradiol by transpiration or, for reporter analysis, by submerging tissue in 17-β-estradiol solution. Alternatively 17-β-estradiol can be applied in 0.01% (v/v) Silwet 77 to plant sectors using an artist's paint brush. Less accessible cell types (e.g. the female gametophyte) can be induced by transpiration, painting, or dipping flowers in 17-β-estradiol solution containing 0.02% (v/v) Break thru S240 (Goldschmidt GmbH). This reduces surface tension and allows 17-β-estradiol to spread quickly over the hydrophobic cuticle. We do not recommend spraying 17-β-estradiol because, like glucocorticoid steroids, estrogens play a significant role in human physiology and should be used with caution.
In summary, we have shown that the two-component, Gateway-compatible XVE system can be used to generate faithful patterns of expression at high frequencies. The frequency with which these patterns are observed is largely independent of the position of both the activator and the responder in the genome. As the number of activator plant lines with cell type-specific activity rapidly grows and more Gateway-compatible, full-length cDNA libraries become available, this system will allow the inducible expression of any gene to be studied in any plant tissue type. This type of system will help to determine the cell type in which a gene's activity is required (i.e. for complementation studies). Furthermore, the system can be used to generate conditional mutant alleles to complement the early lethal effects of a mutation, revealing the effects of the same mutation at later stages of development. Similarly, mutations affecting early zygotic development could be conditionally complemented to generate seeds for second-site mutagenesis, revealing bypass mutants to the primary lesion in viable progeny. Conditional cell type-specific gene expression could further the development and analysis of new phenotypic traits, such as apomixis or dwarfism. Such analysis will be of particular relevance to the development of novel crop traits in which widespread transgene expression could impair plant viability or fertility (Curtis and Grossniklaus, 2006).
MATERIALS AND METHODS
Plasmid Construction
Standard gene-cloning methods (Sambrook and Russell, 2001) were used to make the constructs. An attR1-Cmr-ccdB-attR2 integration region from the Gateway cloning system (Invitrogen) was placed downstream of a promoter containing the LexA binding site (OlexA) and basal CaMV 35S promoter (TATA box; Zuo et al., 2000; Curtis and Grossniklaus, 2003). This responder cassette was subcloned into derivatives of the pCAMBIA (http://www.cambia.org; pMDC8) and derivatives of the pMoa vector series (Barrell et al., 2002; pMDC160, pMDC220, pMDC221, and pLB12), to produce responder constructs with a variety of selectable markers (see Fig. 1).
The AtGL2 promoter was amplified from Arabidopsis (Arabidopsis thaliana) Columbia genomic DNA using PCR with Gateway adapter-GL2 promoter-specific primers (5′-AAAAAGCAGGCTAAGCTTTTGAATTGTAGATAAATCATCTGC-3′and 5′-AGAAAGCTGGGTGCTAGCTTCTTTGCTTAATTATGATCTCTTCCC-3′). This PCR fragment could not be further amplified with attB1 and attB2 adapter primers, as recommended by Invitrogen, and was, therefore, digested with EcoRI and NheI to yield a truncated fragment of 1.5 kb. This AtGL2-promoter fragment was cloned into the EcoRI and XbaI sites of the pBluescript vector (CLONTECH) and amplified using Gateway-compatible primers designed to anneal to the T7 and T3 primer sequences of the pBluescript vector. The forward primer contained the AttB1 tail T7 sequence (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGTAATACGACTCACTATAGGGC-3′) and the reverse primer contained the AttB2 tail T3 sequence (5′-GGGGACCACTTTGTACAAGAAAGCTGGCTAATTAACCCTCACTAAAGGG-3′).
The 35S promoter/enhancer region was amplified from the pCAMBIA 3300 plasmid (http://www.cambia.org) using PCR with the Gateway adapter-CaMV 35S promoter-specific primers 35S-F (5′-AAAAAGCAGGCTGTTTGCGTATTGGCTAGAGCAGCTTG-3′) and 35S-R (5′-AGAAAGCTGGGTGCGTCATCCCTTACGTCAGTGGAG-3′) and the AtEASE sequence was amplified from pWY-093.1 (Yang et al., 2005) using the Gateway adapter-AtEASE enhancer-specific primers EAFP (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCACGATGCAAATATATCG-3′) and EARP (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGCCTTAATATCATACGAAAG-3′). The plasmid pWY-093.1 contains four tandem repeats of AtEASE; however, due to these repeats, our amplified fragment fortuitously contained five tandem repeats. H. An and G. Coupland provided the AtSUC2, ROLC, and NtTobRB7 promoters as entry clones (An et al., 2004).
The Gateway-compatible PCR products AtGL2, CaMV 35S, and AtEASE were introduced into the Gateway pDONR207 (Invitrogen) vector using BP reactions to generate promoter entry clones. Different promoter fusions in the vectors pMDC163 (Curtis and Grossniklaus, 2003), pMDC150, and LB12 were produced using LR (Invitrogen) reactions.
The DT-A entry clone was generated by amplifying the DT-A chain from plasmid pIBI30-DT-A (Maxwell et al., 1986; Harrison et al., 1991), flanked by attB sites using the primers DIP-forward-attB1 (5′-AAAAAGCAGGCTATGGATCCTGATGATGTTGT-3′) and DIP-reverse-attB2 (5′-AGAAAGCTGGGTCACAAAGATCGCCTGACACG-3′). The amplified product was integrated into pDONR207 using BP clonase and subsequently integrated into pMDC221 using LR clonase.
The pYAT5G17430 entry clone (Gong et al., 2004), containing the full-length cDNA of BBM, was obtained from the Arabidopsis Biological Resource Center (ABRC) stock center. This was integrated into the vector pMDC221 using LR clonase.
The KNAT1 entry clone was generated by subcloning a KpnI-NotI fragment that contained the full-length cDNA from the clone U10690 (Yamada et al., 2003), obtained from the ABRC, into the KpnI-NotI sites of pENTR1A vector (Invitrogen). This fragment, flanked by attL1 and attL2 sites, was integrated by into the vector pMDC221 using LR clonase.
The FUS3 and LEC2 entry clones were generated by amplifying full-length cDNAs kindly provided by Francois Parcy, flanked by attB sites using the primers FUS3-forward-attB1 (5′-AAAAAGCAGGCTATGGTTGATGAAAATGTGG-3′) and FUS3-reverse-attB2 (5′-AGAAAGCTGGGTCTAGTAGAAGTCATCGAGAG-3′), and LEC2-forward-attB1 (5′-AAAAAGCAGGCTATGGATAACTTCTTACCCTTTCC-3′) and LEC2-reverse-attB2 (5′-AGAAAGCTGGGTTCACCACCACTCAAAGTCG-3′). The amplified product was integrated into pDONR221 using BP clonase and subsequently integrated into pMDC221 using LR clonase.
Plant Materials, Growth Conditions, and Plant Transformation
Arabidopsis Landsberg erecta plants were used for plant transformation using the floral-dip method (Clough and Bent, 1998). Plants were grown under 14-h white light/10-h dark at 22°C on Murashige and Skoog agar (1× Murashige and Skoog salts, 3% Suc, 0.8% agar) or in the greenhouse for mature plants.
17-β-Estradiol Induction Methods
A stock of 20 mm 17-β-estradiol (Sigma-Aldrich) in 70% ethanol or 100% dimethylsulfoxide was made and stored at −20°C in small aliquots (17-β-estradiol is light sensitive and its activity slowly declines in a light intensity-dependent manner). The ethanol alone has no effect on transgene expression and at a concentration of ≤0.1% (v/v) in sterile culture media has no inhibitory effect on seed germination. Chemical treatments were carried out by either germinating seeds directly on Murashige and Skoog medium supplemented with 10, 5, or 2 μm 17-β-estradiol, or by transferring seedlings from a noninductive to an inductive medium. Alternative methods include 17-β-estradiol uptake through transpiration or by submerging tissue in 17-β-estradiol solution. Restricted regions of the plant can be induced by applying 17-β-estradiol solution, supplemented with 0.01% Silwet 77 with an artist's paint brush to sectors of the plant. When applying 17-β-estradiol to the exterior of the flower buds to induce expression in the egg apparatus deep within the carpels, 17-β-estradiol solution was supplemented with 0.02% Break thru S240 (Goldschmidt GmbH), which aids spreading.
GUS Staining
In situ GUS staining was carried out by vacuum infiltrating GUS staining solution (50 mm sodium phosphate buffer, pH 7.0, 1 mm EDTA, 0.5 mg/mL 5-bromo-4-chloro-3-indolyl β-d GlcUA [X-Gluc; Biosynth AG], 0.4% Triton X-100, 100 mg/mL chloramphenicol, and 5 mm each of potassium ferri/ferrocyanide), and incubating at 37°C for 24 h.
Acknowledgments
We thank Nam-Hai Chua (Rockefeller University) for kindly providing the vector PER8 and George Coupland and Hailong An (Max Planck Institut für Züchtungsforschung) for kindly providing the entry clones containing the promoters for NtTobRB7, AtSUC2, and RolC. We thank Ian Maxwell (University of Colorado) for the plasmid pIBI30-DT-A, the ABRC for distributing BBM entry clone PYAT5G17430 and the full-length cDNA clone U10690, and Francois Parcy for cDNA clones containing LEC2 and FUS3. We thank Valeria Gagliardini, Jana Schneider, and Brigitte Gabathuler for help with sequencing, Peter Kopf for technical assistance, and Urs Jauch for scanning electron microscopy. We are also grateful to Célia Baroux, Margaret Collinge, and Siân Curtis for critical reading of the manuscript.
This work was supported by the Swiss National Science Foundation (grant no. 3100A0–100281 to M.D.C. and grant no. 3100–064061 to U.G.), the University of Zürich, and the Forschungskredit of the University of Zürich (to M.D.C.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Mark D. Curtis (mcurtis@botinst.unizh.ch).
The online version of this article contains Web-only data.
References
- An H, Roussot C, Suarez-Lopez P, Corbesier L, Vincent C, Pineiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, et al (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Baroux C, Blanvillain R, Betts H, Batoko H, Craft J, Martinez A, Gallois P, Moore I (2005) Predictable activation of tissue-specific expression from a single gene locus using the pOp/LhG4 transactivation system in Arabidopsis. Plant Biotechnol J 3: 91–101 [DOI] [PubMed] [Google Scholar]
- Barrell PJ, Yongjin S, Cooper PA, Conner AJ (2002) Alternative selectable markers for potato transformation using minimal T-DNA vectors. Plant Cell Tissue Organ Cult 70: 61–68 [Google Scholar]
- Baumbusch LO, Hughes DW, Galau GA, Jakobsen KS (2004) LEC1, FUS3, ABI3 and Em expression reveals no correlation with dormancy in Arabidopsis. J Exp Bot 55: 77–87 [DOI] [PubMed] [Google Scholar]
- Blanc G, Barakat A, Guyot R, Cooke R, Delseny M (2000) Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12: 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker J, Chatfield S, Leyser O (2003) Auxin acts in xylem-associated or medullary cells to mediate apical dominance. Plant Cell 15: 495–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu CM, van Lammeren AA, Miki BL, et al (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14: 1737–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collier RJ (1967) Effect of diphtheria toxin on protein synthesis: inactivation of one of the transfer factors. J Mol Biol 25: 83–98 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2005) Thale cress (Arabidopsis thaliana) genome. In RA Meyers, ed, Encyclopedia of Molecular Cell Biology and Molecular Medicine. Wiley-VCH, Weinheim, Germany, pp 245–282
- Curtis MD, Grossniklaus U (2006) Conditional gene expression in plants. In JA Teixeira da Silva, ed, Floriculture, Ornamental and Plant Biotechnology: Advances and Topical Issues, Ed 1, Vol 2. Global Science Books, London, pp 77–87
- De Veylder L, Beeckman T, Van Montagu M, Inze D (2000) Increased leakiness of the tetracycline-inducible Triple-Op promoter in dividing cells renders it unsuitable for high inducible levels of a dominant negative CDC2aAt gene. J Exp Bot 51: 1647–1653 [DOI] [PubMed] [Google Scholar]
- Deveaux Y, Peaucelle A, Roberts GR, Coen E, Simon R, Mizukami Y, Traas J, Murray JA, Doonan JH, Laufs P (2003) The ethanol switch: a tool for tissue-specific gene induction during plant development. Plant J 36: 918–930 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL (2001) Establishment of polarity in lateral organs of plants. Curr Biol 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P (2004) The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell 7: 373–385 [DOI] [PubMed] [Google Scholar]
- Gong W, Shen YP, Ma LG, Pan Y, Du YL, Wang DH, Yang JY, Hu LD, Liu XF, Dong CX, et al (2004) Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol 135: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Hardt R, Lenhard M, Laux T (2002) WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev 16: 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Millar AA, Jacobsen JV (2005) Dormancy release, ABA and pre-harvest sprouting. Curr Opin Plant Biol 8: 183–187 [DOI] [PubMed] [Google Scholar]
- Harrison GS, Maxwell F, Long CJ, Rosen CA, Glode LM, Maxwell IH (1991) Activation of a diphtheria toxin A gene by expression of human immunodeficiency virus-1 Tat and Rev proteins in transfected cells. Hum Gene Ther 2: 53–60 [DOI] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA (2000) DNA cloning using in vitro site-specific recombination. Genome Res 10: 1788–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CY, Lin Y, Zhang M, Pollock S, Marks MD, Schiefelbein J (1998) A common position-dependent mechanism controls cell-type patterning and GLABRA2 regulation in the root and hypocotyl epidermis of Arabidopsis. Plant Physiol 117: 73–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T, Fox GL, Meyerowitz EM (1994) Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76: 703–716 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H (2002) Seed dormancy and germination. Curr Opin Plant Biol 5: 33–36 [DOI] [PubMed] [Google Scholar]
- Laufs P, Coen E, Kronenberger J, Traas J, Doonan J (2003) Separable roles of UFO during floral development revealed by conditional restoration of gene function. Development 130: 785–796 [DOI] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S (1994) A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6: 1859–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel A, Weigel D (2004) Temporally and spatially controlled induction of gene expression in Arabidopsis thaliana. Plant J 38: 164–171 [DOI] [PubMed] [Google Scholar]
- Martinez A, Sparks C, Hart CA, Thompson J, Jepson I (1999) Ecdysone agonist inducible transcription in transgenic tobacco plants. Plant J 19: 97–106 [DOI] [PubMed] [Google Scholar]
- Matsuhara S, Jingu F, Takahashi T, Komeda Y (2000) Heat-shock tagging: a simple method for expression and isolation of plant genome DNA flanked by T-DNA insertions. Plant J 22: 79–86 [DOI] [PubMed] [Google Scholar]
- Maxwell IH, Maxwell F, Glode LM (1986) Regulated expression of a diphtheria toxin A-chain gene transfected into human cells: possible strategy for inducing cancer cell suicide. Cancer Res 46: 4660–4664 [PubMed] [Google Scholar]
- Mizukami Y, Ma H (1992) Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71: 119–131 [DOI] [PubMed] [Google Scholar]
- Nye AC, Rajendran RR, Stenoien DL, Mancini MA, Katzenellenbogen BS, Belmont AS (2002) Alteration of large-scale chromatin structure by estrogen receptor. Mol Cell Biol 22: 3437–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell JT, Nagy F, Chua NH (1985) Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313: 810–812 [DOI] [PubMed] [Google Scholar]
- Pien S, Wyrzykowska J, McQueen-Mason S, Smart C, Fleming A (2001) Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc Natl Acad Sci USA 98: 11812–11817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz R, Bergervoet JHW, Koornneef M (2001) Sequential steps for the developmental arrest in Arabidopsis seeds. Development 128: 243–252 [DOI] [PubMed] [Google Scholar]
- Roslan HA, Salter MG, Wood CD, White MR, Croft KP, Robson F, Coupland G, Doonan J, Laufs P, Tomsett AB, et al (2001) Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J 28: 225–235 [DOI] [PubMed] [Google Scholar]
- Salter MG, Paine JA, Riddell KV, Jepson I, Greenland AJ, Caddick MX, Tomsett AB (1998) Characterisation of the ethanol-inducible alc gene expression system for transgenic plants. Plant J 16: 127–132 [Google Scholar]
- Sambrook J, Russell D (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski DB, Jilk RA, Pollock SM, Marks MD (1998) Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125: 1161–1171 [DOI] [PubMed] [Google Scholar]
- Truernit E, Sauer N (1995) The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of β-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta 196: 564–570 [DOI] [PubMed] [Google Scholar]
- Tumbar T, Sudlow G, Belmont AS (1999) Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J Cell Biol 145: 1341–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali M, Steger DJ, Neely KE, Workman JL (2000) Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J 19: 2629–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Van Hamburg JP, Van Rijn E, Hooykaas PJ, Offringa R (2003) Diphtheria toxin-mediated cell ablation reveals interregional communication during Arabidopsis seed development. Plant Physiol 133: 1882–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Yamamoto YT, Taylor CG, Acedo GN, Cheng CL, Conkling MA (1991) Characterization of cis-acting sequences regulating root-specific gene expression in tobacco. Plant Cell 3: 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Jefferson RA, Huttner E, Moore JM, Gagliano WB, Grossniklaus U (2005) An egg apparatus-specific enhancer of Arabidopsis, identified by enhancer detection. Plant Physiol 139: 1421–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SY, Bomblies K, Yoo SK, Yang JW, Choi MS, Lee JS, Weigel D, Ahn JH (2005) The 35S promoter used in a selectable marker gene of a plant transformation vector affects the expression of the transgene. Planta 221: 523–530 [DOI] [PubMed] [Google Scholar]
- Zuo J, Hare PD, Chua NH (2006) Applications of chemical-inducible expression systems in functional genomics and biotechnology. In J Salinas, JJ Sanchez-Serrano, eds, Methods in Molecular Biology-Arabidopsis Protocols. Humana Press, Totowa, NJ [DOI] [PubMed]
- Zuo J, Niu QW, Chua NH (2000) Technical advance: an estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Frugis G, Chua NH (2002) The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J 30: 349–359 [DOI] [PubMed] [Google Scholar]