Abstract
Abscisic acid (ABA) plays a key role in plant responses to abiotic stress, particularly drought stress. A wide number of ABA-hypersensitive mutants is known, however, only a few of them resist/avoid drought stress. In this work we have generated ABA-hypersensitive drought-avoidant mutants by simultaneous inactivation of two negative regulators of ABA signaling, i.e. the protein phosphatases type 2C (PP2Cs) ABA-INSENSITIVE1 (ABI1) and HYPERSENSITIVE TO ABA1 (HAB1). Two new recessive loss-of-function alleles of ABI1, abi1-2 and abi1-3, were identified in an Arabidopsis (Arabidopsis thaliana) T-DNA collection. These mutants showed enhanced responses to ABA both in seed and vegetative tissues, but only a limited effect on plant drought avoidance. In contrast, generation of double hab1-1 abi1-2 and hab1-1 abi1-3 mutants strongly increased plant responsiveness to ABA. Thus, both hab1-1 abi1-2 and hab1-1 abi1-3 were particularly sensitive to ABA-mediated inhibition of seed germination. Additionally, vegetative responses to ABA were reinforced in the double mutants, which showed a strong hypersensitivity to ABA in growth assays, stomatal closure, and induction of ABA-responsive genes. Transpirational water loss under drought conditions was noticeably reduced in the double mutants as compared to single parental mutants, which resulted in reduced water consumption of whole plants. Taken together, these results reveal cooperative negative regulation of ABA signaling by ABI1 and HAB1 and suggest that fine tuning of ABA signaling can be attained through combined action of PP2Cs. Finally, these results suggest that combined inactivation of specific PP2Cs involved in ABA signaling could provide an approach for improving crop performance under drought stress conditions.
The plant hormone abscisic acid (ABA) plays a crucial role in plant responses to several abiotic stresses such as drought, salt, and cold, as well as plant growth and development. In vegetative tissues, water stress produced by drought or high osmoticum treatment boosts ABA biosynthesis, leading to a variety of adaptive ABA-mediated responses such as stomatal closure and differential gene expression (Finkelstein et al., 2002; Nambara and Marion-Poll, 2005). ABA signaling in guard cells leads to stomatal closure, which occurs through rapid changes of ion fluxes and osmoregulation (Schroeder et al., 2001; Hetherington and Woodward, 2003). ABA regulation of the transpiration flow through stomatal pores is a crucial response of the plant to water deficit, as exemplified by the wilty phenotype of both ABA-deficient and ABA-insensitive mutants (Zeevaart and Creelman, 1988). Additionally, the ABA-dependent signaling pathway regulates stress-inducible gene expression, leading to a coordinated remodeling of gene expression that affects more than 1,000 genes of the plant transcriptome (Hoth et al., 2002; Seki et al., 2002; Takahashi et al., 2004).
Biochemical and genetic analyses have resulted in the identification of many elements of the ABA signal transduction pathway, although important pieces are still lacking. Recently, the RNA-binding protein FCA has been identified as an ABA-binding receptor with a singular role in flowering control, however, key responses to ABA such as inhibition of seed germination or stomatal response were not affected in the fca-1 mutant (Razem et al., 2006). Accordingly, FCA appears to be an ABA receptor involved in controlling flowering time but additional ABA receptors must perform ABA perception. Putative candidates might be some plasma membrane receptors, such as RPK1, which is known to be involved in ABA signaling (Osakabe et al., 2005). Furthermore, in guard cells several studies have indicated the presence of intracellular ABA receptors (Allan et al., 1994; Schwartz et al., 1994; Schwarz and Schroeder, 1998; Levchenko et al., 2005).
It is well known that a variety of second messengers contribute to the transmission of the ABA signal, which includes Ca2+, cADP-Rib, reactive oxygen species, nitric oxide, phosphoinositides, phosphatidic acid, and sphingosine 1-P (Schroeder and Hagiwara, 1989; Gilroy et al., 1990; McAinsh et al., 1990; Wu et al., 1997; Leckie et al., 1998; Jacob et al., 1999; Lemtiri-Chlieh et al., 2000; Pei et al., 2000; Allen et al., 2001; Ng et al., 2001; Neill et al., 2002; Guo et al., 2003). It is also known that phosphorylation/dephosphorylation events play a crucial role in ABA signaling, which involves a complex network of protein kinases and phosphatases as well as other signal transducers (for review, see Finkelstein et al., 2002). Finally, many transcriptional factors (TFs) of ABA-inducible genes are known. The TFs comprise ABA-responsive element (ABRE)-binding proteins (ABA-INSENSITIVE5 [ABI5]/ABF/AREB/AtbZIP family), ABI3/VP1/B3, ABI4/APETALA2, MYC, MYB, and HD-ZIP domain proteins (Giraudat et al., 1992; Suzuki et al., 1997; Finkelstein et al., 1998; Choi et al., 2000; Finkelstein and Lynch, 2000; Uno et al., 2000; Bensmihen et al., 2002; Himmelbach et al., 2002; Abe et al., 2003). Most of these TFs play a positive role in ABA signaling, but some of them function as repressors of ABA response (Himmelbach et al., 2002; Pandey et al., 2005; Song et al., 2005).
Genetic analyses of ABA signal transduction have identified both negative and positive regulators of ABA signaling (McCourt, 1999; Finkelstein et al., 2002). For instance, recessive mutations leading to ABA hypersensitivity were found in the era1 (Cutler et al., 1996), abh1 (Hugouvieux et al., 2001), fry1 (Xiong et al., 2001b), hypersensitive to ABA1 (hab1; Leonhardt et al., 2004; Saez et al., 2004), sad1 (Xiong et al., 2001a), and gcr1 (Pandey and Assmann, 2004) mutants. The intragenic revertants of abi1-1 and abi1-1R1 to R7 also carry recessive mutations that lead to enhanced responsiveness to ABA (Gosti et al., 1999). Loss-of-function mutants generated by RNA interference for the SOS3-like calcium-binding protein 5 and its interacting protein kinase 3 were also hypersensitive to ABA (Guo et al., 2002). As loss of function of the above-mentioned genes leads to enhanced ABA responsiveness, their corresponding gene products must represent negative regulators of ABA signaling. On the other hand, recessive mutations leading to reduced ABA sensitivity have been identified in the abi3 (Giraudat et al., 1992), abi4 (Finkelstein et al., 1998), abi5 (Finkelstein and Lynch, 2000), ost1 (Mustilli et al., 2002), rcn1 (Kwak et al., 2002), rpk1 (Osakabe et al., 2005), and the rbohD/F double mutants (Kwak et al., 2003). Therefore, these loci point out to positive regulators of ABA signal transduction.
Protein phosphatases type 2C (PP2Cs) were identified as components of ABA signaling pathway from pioneer work with the ABA-insensitive abi1-1 and abi2-1 mutants (Koornneef et al., 1984; Leung et al., 1994, 1997; Meyer et al., 1994; Rodriguez, et al., 1998). Currently, at least four Arabidopsis (Arabidopsis thaliana) PP2Cs, ABI1, ABI2, PP2CA, and HAB1 (formerly named AtP2C-HA), are known to regulate ABA signaling. Evidence on their role as negative regulators of ABA signaling has been provided by genetic approaches (Gosti et al., 1999; Merlot et al., 2001; Tahtiharju and Palva, 2001; Gonzalez-Garcia et al., 2003; Leonhardt et al., 2004; Saez et al., 2004; Kuhn, et al., 2006; Yoshida et al., 2006). For instance, the recessive T-DNA insertion mutant hab1-1 shows ABA-hypersensitive inhibition of seed germination and enhanced ABA-mediated stomatal closure (Leonhardt et al., 2004; Saez et al., 2004). HAB1 is broadly expressed in the plant and strongly induced by ABA (Leonhardt et al., 2004; Saez et al., 2004). Constitutive expression of HAB1 under a 35S promoter led to reduced ABA sensitivity both in seeds and vegetative tissues, compared to wild-type plants (Saez et al., 2004).
In the case of ABI1, recessive alleles were isolated as intragenic revertants of the originally dominant abi1-1 mutation, and named abi1-1R1 to R7 (Gosti et al., 1999). Therefore, these recessive alleles, in addition to the original Gly-180 Asp mutation, carry a second mutation that abolishes the dominant character of the abi1-1 mutation. The same approach was applied to the dominant mutant abi2-1, leading to the identification of the recessive abi2-1R1 allele (Merlot et al., 2001). It cannot be excluded that intragenic revertants of abi1-1 still retain some activity (not necessarily an enzymatic one) in the corresponding gene products, even though their in vitro protein phosphatase activity was shown to be negligible (Gosti et al., 1999). Thus, we were interested in the isolation of direct knockout alleles of ABI1, namely abi1-2 and abi1-3, to conclusively clarify its role in ABA signaling. Furthermore, double knockout mutants in PP2Cs have not yet been generated and we have analyzed hab1 abi1 double loss-of-function mutants here to determine whether these PP2Cs are strictly redundant or additive in their functions. Phenotypic analysis of abi1-2 and abi1-3 provided new data regarding the role of ABI1 in ABA-induced stomatal closure, transpiration, and ABA-mediated regulation of gene expression. The phenotypic effect on ABA signaling observed in single hab1-1, abi1-2, and abi1-3 mutants was notably reinforced in double mutants, which showed both enhanced responsiveness to ABA and drought avoidance. Thus, these results show a new biotechnological approach to increase plant drought avoidance, i.e. the combined inactivation of PP2Cs involved in ABA signaling.
RESULTS
Identification and Characterization of Knockout Alleles of ABI1
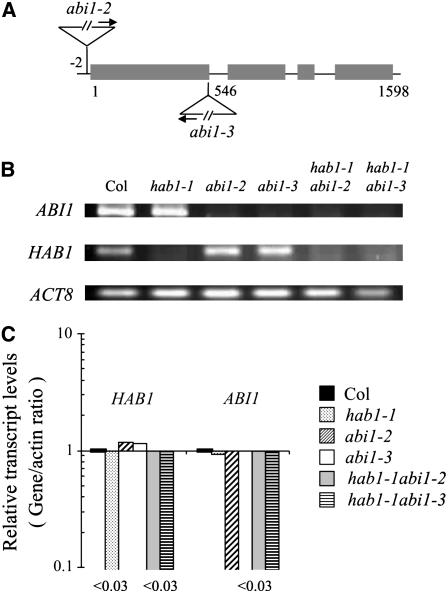
Two T-DNA insertion mutants of ABI1 were identified in the Salk collection (Columbia [Col] background), corresponding to donor stock numbers SALK_72009 and SALK_76309, and they were named abi1-2 and abi1-3, respectively. Homozygous individuals were identified by PCR and Southern-blot analyses (data not shown). Sequencing of the T-DNA flanking region in abi1-2 showed that the insertion was localized two nucleotides upstream of the ATG start codon (Fig. 1A). In the case of abi1-3, the T-DNA insert was localized 546 nucleotides downstream from the ATG start codon (Fig. 1A). Both T-DNA insertions severely impaired ABI1 expression, based on reverse transcription (RT)-PCR (Fig. 1B) and quantitative RT-PCR (qRT-PCR) analyses (Fig. 1C). Expression of HAB1 and ABI1 in wild type was quite similar to that in abi1-2/abi1-3 and hab1-1 mutant backgrounds, respectively (Fig. 1C).
Figure 1.
Map of abi1-2 and abi1-3 mutants. ABI1 and HAB1 transcript levels in wild type, hab1-1, abi1-2, abi1-3, and double hab1-1 abi1-2/hab1-1 abi1-3 mutants. A, Scheme of the ABI1 gene and localization of the T-DNA insertions in abi1-2 and abi1-3 mutants. The numbering begins at the ATG translation start codon. The T-DNA left border primer (LBpROK2) that was used to localize the T-DNA insertion is indicated by an arrow. B, RT-PCR analysis shows absence of full-length transcripts of ABI1 or HAB1 in genotypes containing either the abi1-2/abi1-3 or hab1-1 alleles, respectively. PCR reactions were performed as indicated in “Materials and Methods” and amplification of β-actin-8 was used as control. Samples were taken for analysis after 25 PCR cycles. C, Expression of HAB1 and ABI1 in wild type was similar to that in abi1-2/abi1-3 and hab1-1 mutants, respectively.
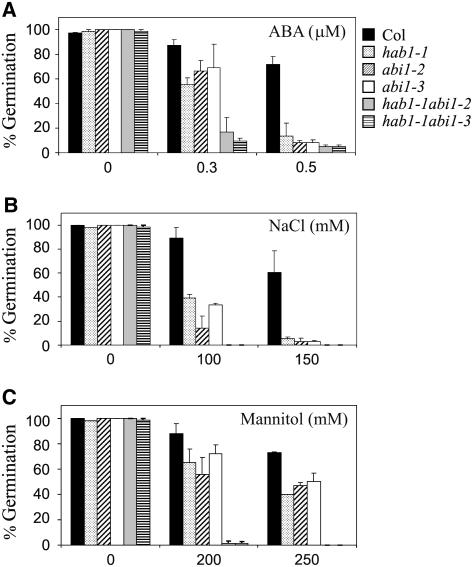
Progeny of both abi1-2 and abi1-3 homozygous individuals was harvested and different analyses to test their sensitivity to ABA were performed. First, the sensitivity of the mutants to inhibition of seed germination by ABA was analyzed (Fig. 2A). In the absence of exogenous ABA, abi1-2 and abi1-3 mutant seeds showed a germination ratio similar to wild type. However, in the presence of exogenous ABA, both the abi1-2 and abi1-3 mutants showed ABA-hypersensitive inhibition of seed germination (Fig. 2A; Supplemental Fig. 1). F1 seeds that were hemizygous for the T-DNA insertion present either in abi1-2 or abi1-3 showed wild-type germination on 0.5 μm ABA. In the next generation, F2 seeds showed an ABA-hypersensitive phenotype in approximately a 1:3 proportion (112 hypersensitive:313 wild type, χ2 = 0.42, P > 0.5 for abi1-2; 121 hypersensitive:319 wild type, χ2 = 1.4, P > 0.1 for abi1-3). Finally, F2 ABA-hypersensitive seedlings showed linkage between the ABA-hypersensitive phenotype and the presence of a homozygous T-DNA insertion in ABI1 as determined by PCR analysis (n = 40). Taken together, these data indicate that both the abi1-2 and abi1-3 mutations are recessive and segregate as a single nuclear locus linked to the T-DNA insertion present in the ABI1 gene. The ABA inhibitory concentration to achieve 50% inhibition (IC50) of seed germination was approximately 2-fold lower for abi1-2 and abi1-3 than for the wild type (0.35, 0.37, and 0.67 μm ABA, respectively; Supplemental Fig. 1).
Figure 2.
ABA-hypersensitive germination inhibition of hab1-1, abi1-2, abi1-3, and double hab1-1 abi1-2/hab1-1 abi1-3 mutants as compared to wild-type seeds. A to C, Percentage of seeds that germinated and developed green cotyledons in the presence of the indicated concentrations of ABA, NaCl, and mannitol. Approximately 200 seeds of each genotype were sowed on each plate and scored for germination and early growth 10 d later. Values are averages ± sd for three independent experiments.
ABA plays a critical role promoting inhibition of both seed germination and early seedling growth under high osmoticum (Gonzalez-Guzman et al., 2002). Thus, whereas ABA-hypersensitive mutants are generally more sensitive than wild type to the inhibition of seed germination promoted by osmotic stress (Saez et al., 2004), both ABA-deficient and ABA-insensitive mutants are more tolerant to osmotic stress at this stage (Leon-Kloosterziel et al., 1996; Gonzalez-Guzman et al., 2002). Dose-response analyses of germination and early growth in media supplemented with increasing concentrations of NaCl or mannitol were performed for abi1-2 and abi1-3 (Fig. 2, B and C). Both abi1-2 and abi1-3 mutants showed higher inhibition of germination and early growth by osmotic stress than wild-type seeds.
Generation and Analysis of hab1-1 abi1-2 and hab1-1 abi1-3 Double Mutants
Sequence similarity analysis of the Arabidopsis PP2C gene family reveals a branch composed by four members: ABI1, ABI2, HAB1, and HAB2 (Saez et al., 2004). ABI1 and HAB1 appear to play a predominant role over ABI2 and HAB2, respectively, according to their mRNA expression levels and mutant phenotype (Merlot et al., 2001; Leonhardt et al., 2004; Saez et al., 2004; Kuhn, et al., 2006; A. Saez, N. Robert, J. I. Schroeder, and P. L. Rodriguez, unpublished data). Double loss-of-function phenotypes in plant PP2Cs have not yet been analyzed in knockout mutants. To unravel a possible functional redundancy between ABI1 and HAB1, we decided to generate double mutant lines that contained knockout alleles of both genes. To this end we crossed the previously described hab1-1 mutant with either abi1-2 or abi1-3. PCR (data not shown) and RT-PCR analyses (Fig. 1B) of the resulting F2 population allowed the identification of hab1-1 abi1-2 and hab1-1 abi1-3 double mutants, and their response to ABA was analyzed in germination, growth, and transpiration assays.
Analysis of germination and early seedling growth in media supplemented with 0.3 μm ABA indicated an enhanced responsiveness to ABA of the double mutants as compared to the single parental mutants (Fig. 2A; Supplemental Fig. 1). Thus, the IC50 of ABA in seed germination was 0.18 μm for the double mutants versus 0.35 and 0.37 μm for abi1-2 and abi1-3, respectively. In agreement with this result, the double mutants were particularly sensitive to inhibition of germination and early growth promoted by both NaCl and mannitol (Fig. 2, B and C). Thus, a concentration of 100 mm NaCl practically abolished germination of the double mutants, whereas 15% to 40% germination was still observed in the single parental mutants (Fig. 2B). Likewise, 200 mm mannitol leads to almost complete inhibition of germination for the double mutants, whereas more than 50% germination is still observed in the single parental mutants (Fig. 2C).
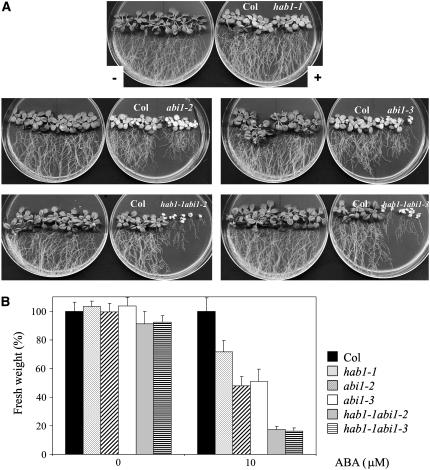
ABA has an inhibitory effect on plant growth when the medium is supplemented with micromolar concentrations of the hormone. For instance, the ABA-insensitive mutants abi1-1 and abi2-1 and 35S:HAB1 plants show ABA-resistant growth compared to wild-type plants (Leung et al., 1994, 1997; Meyer et al., 1994; Rodriguez, et al., 1998; Saez et al., 2004). In contrast, the recessive abi1-1R1 to R7 alleles were more sensitive to ABA inhibition of root growth than Landsberg erecta wild type (Gosti et al., 1999). Figure 3 shows that both abi1-2 and abi1-3 displayed enhanced sensitivity to ABA-mediated growth inhibition than wild-type plants. After 10 d in 10 μm ABA, both abi1-2 and abi1-3 plants showed yellowing and impaired growth of both leaves and roots. Under these conditions, the hab1-1 mutant also showed reduced growth as compared to wild-type plants, although growth was inhibited less in hab1-1 than in abi1-2 and abi1-3 mutants (Fig. 3). Finally, both double mutants showed a dramatic growth inhibition in medium supplemented with 10 μm ABA, and they were markedly more sensitive to ABA than the single parental mutants (Fig. 3).
Figure 3.
ABA-hypersensitive growth inhibition of hab1-1, abi1-2, abi1-3, and double hab1-1 abi1-2/hab1-1 abi1-3 mutants as compared to wild-type plants. A, Growth of the different mutants and wild type in medium supplemented (+) or not (−) with 10 μm ABA. The photographs were taken after 12 d of the transfer of 5-d-old seedlings from Murashige and Skoog medium to plates lacking or containing 10 μm ABA. B, Percentage of fresh weight from the different mutants as compared to wild type. The percentage was calculated with respect to the fresh weight of wild type in Murashige and Skoog medium either lacking or containing 10 μm ABA. Fresh weight of wild type was reduced by 35% in plates supplemented with ABA as compared to medium lacking ABA. Values are averages ± sd (n = 30).
Enhanced ABA-Induced Stomatal Closing and Reduced Water Loss of the hab1-1 abi1-2 and hab1-1 abi1-3 Double Mutants
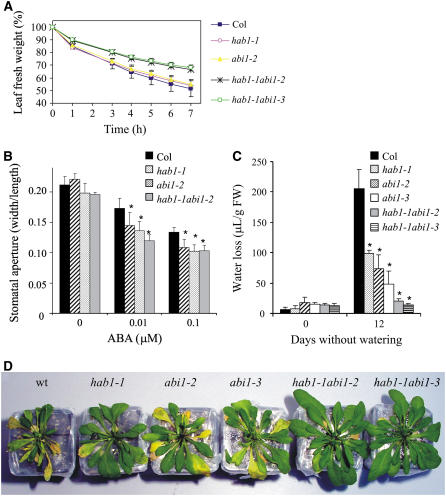
ABA signaling, by regulating stomatal aperture, plays a crucial role to reduce water loss under water shortage. Different analyses were performed to evaluate responses in wild type and the different mutant backgrounds (Fig. 4). Thus, short-term water-loss assays were performed by evaluating the decline in fresh weight of detached leaves (Verslues et al., 2006). The single loss-of-function abi1-2 and abi1-3 mutants, as well as hab1-1, did not exhibit significant differences in the transpiration rate of detached leaves compared to wild type (Fig. 4A). In contrast, combined inactivation of HAB1 and ABI1 resulted in a phenotype of reduced water loss in both double mutants (Fig. 4A).
Figure 4.
Reduced water loss of double hab1-1 abi1-2/hab1-1 abi1-3 mutants as compared to wild type or single parental mutants. A, Detached-leaves water-loss assays show reduced water loss in double hab1-1 abi1-2/hab1-1 abi1-3 mutants. Five leaves per individual at the same developmental stage and size from 21-d-old plants were excised and fresh weight was determined after submitting the leaves to the drying atmosphere of a flow laminar hood (n = 4). Results for abi1-2 and abi1-3 were almost identical. B, ABA-induced stomatal closing is ABA hypersensitive in hab1-1, abi1-2, and double mutant hab1-1 abi1-2 as compared to wild-type plants. Stomatal apertures were measured 2 h and 30 min after addition of 0.01 or 0.1 μm ABA. Data represent the average of three independent experiments ± sem (n = 30–40 stomata per experiment). C, Quantification of water loss in 5-week-old plants after 12 d without watering. Data shown are the average amounts of water loss measured in 10 leaves (μL/g fresh weight) collected from four different plants. Asterisks in B and C indicate P < 0.01 (Student's t test) when data was compared from mutant and wild type. D, Enhanced drought tolerance of double hab1-1abi1-2/hab1-1 abi1-3 mutants with respect to wild type or single parental mutants. Photograph was taken 14 d after water was withheld. Shoot was cut to better show the effect of drought on rosette leaves.
To further analyze stomatal responses to ABA in the mutants, direct measurements of stomatal closing were performed (Fig. 4B). ABA-induced stomatal closing was assayed in the single abi1-2 and hab1-1 mutants, as well as in the double mutant hab1-1 abi1-2 (Fig. 4B). Stomatal aperture measurements indicated that abi1-2, hab1-1, and double mutant hab1-1 abi1-2 were hypersensitive to ABA-induced stomatal closing in the range of 10 to 100 nm ABA. Moreover, the response of the double mutant hab1-1 abi1-2 to 10 nm ABA was more sensitive as compared to the single parental mutants (Fig. 4B). Similar results to those obtained for abi1-2 and double mutant hab1-1 abi1-2 were obtained for abi1-3 and double mutant hab1-1 abi1-3, respectively (Supplemental Fig. 2).
The era1, abh1, and gcr1 mutants display enhanced ABA-induced stomatal closing and reduced water loss as compared to wild-type plants (Pei et al., 1998; Hugouvieux et al., 2001; Pandey and Assmann, 2004). Therefore, we examined water loss of the different genetic backgrounds described here. Water-loss data were obtained, under greenhouse conditions, after exposing 21-d-old plants to drought stress by completely terminating irrigation and minimizing soil evaporation. Figure 4D shows that after 14 d without watering, wild-type plants wilted and many rosette leaves yellowed. In contrast, hab1-1 abi1-2 and hab1-1 abi1-3 double mutant plants did not show symptoms of wilting and they had turgid green rosette leaves. A limited improvement was observed under these conditions in single mutants (Fig. 4D), although far from the phenotype observed in the double mutants. Water loss was estimated by comparing fresh and turgid weight of rosette leaves after 12 d without watering (Fig. 4C). Under these experimental conditions, where the plants were submitted to a long period of drought, the single hab1-1, abi1-2, and abi1-3 mutants showed a reduced water loss as compared to wild type (Fig. 4C). Detached-leaf water-loss assays are likely not sensitive enough as to detect such variations (Kuhn et al., 2006), which are apparent after long periods of drought. Thus, whereas wild-type plants exhibited a marked water loss under these conditions, the ABA-hypersensitive mutants exhibited a reduced water loss, particularly in the case of the hab1-1 abi1-2 and hab1-1 abi1-3 double mutants.
Enhanced Expression of ABA-Inducible Genes in PP2C Mutants as Compared to Wild Type
The effect of the isolated single and double hab1 and abi1 loss-of-function mutations was analyzed on ABA-regulated gene expression. To this end, we used qRT-PCR to analyze the expression of the ABA- and drought-responsive RAB18, P5CS1, RD29B, KIN1, RD29A, and RD22 genes, in wild type, single, and double mutants. These gene markers have been widely used to monitor the ABA and stress response pathways in plants (Kurkela and Franck, 1990; Lang and Palva, 1992; Yamaguchi-Shinozaki and Shinozaki, 1994; Strizhov et al., 1997; Abe et al., 2003). In general, in the absence of ABA or stress treatments, these gene markers show a low expression, which is strongly up-regulated in response to the inductive signal.
Interestingly, in the absence of exogenous ABA treatment, the double hab1-1 abi1-2 and hab1-1 abi1-3 mutants showed approximately 2-fold higher mRNA levels of some gene markers (RAB18, RD29A, and RD29B) as compared to Col wild type (Table I). In the case of single mutants and under control conditions, only the RD29B marker was 2-fold up-regulated in all the single mutants. Upon ABA treatment, as a general trend, induction by ABA was higher in the mutants than in wild type. This enhanced response to ABA was particularly apparent in the double mutants for gene markers that contain ABRE but no typical drought-responsive element (DRE) at the promoter, such as RAB18, RD29B, and P5CS1 (between 4- and 8-fold higher expression level than wild type). Gene markers that contain both DRE and ABRE elements KIN1 and RD29A, were also hyperinduced by ABA in the double mutants, although to a lower level (2- to 3-fold). Finally, ABA-mediated induction of RD22, which lacks both ABRE and DRE consensus sequences at its promoter, was also up-regulated.
Table I.
Enhanced expression of ABA-inducible genes in PP2C mutants with respect to wild type
Numbers indicate the induction level of the stress-responsive genes under mock or ABA treatment (10 μm for 3 h) in wild type and mutants. Values are the expression level reached in each mutant genotype with respect to the wild type (value 1). qRT-PCR analyses were made in triplicate on RNA samples obtained from mock-treated plants or plants treated once with 10 μm ABA.
| Genotype
|
||||||
|---|---|---|---|---|---|---|
| RAB18 | KIN1 | RD22 | P5CS1 | RD29a | RD29b | |
| Mock | ||||||
| Col | 1 | 1 | 1 | 1 | 1 | 1 |
| hab1-1 | 0.9 | 0.8 | 1.5 | 0.9 | 2.7 | 2.0 |
| abi1-2 | 1.1 | 1.0 | 1.4 | 1.1 | 1.3 | 2.6 |
| abi1-3 | 0.8 | 0.7 | 1.1 | 0.8 | 1.0 | 2.6 |
| hab1-1 abi1-2 | 2.1 | 1.6 | 2.1 | 1.7 | 2.0 | 2.5 |
| hab1-1 abi1-3 | 2.3 | 1.2 | 1.6 | 1.9 | 2.0 | 2.7 |
| ABA treatment | ||||||
| Col | 1 | 1 | 1 | 1 | 1 | 1 |
| hab1-1 | 2.6 | 1.7 | 2.5 | 3.0 | 3.5 | 2.1 |
| abi1-2 | 3.7 | 1.6 | 1.7 | 2.6 | 1.7 | 2.4 |
| abi1-3 | 2.7 | 1.5 | 1.5 | 2.0 | 1.5 | 1.3 |
| hab1-1 abi1-2 | 6.0 | 2.7 | 3.0 | 6.4 | 2.2 | 3.9 |
| hab1-1 abi1-3 | 8.6 | 3.6 | 3.1 | 6.0 | 2.2 | 4.9 |
DISCUSSION
In this work, we report the identification and characterization of two new ABI1 recessive alleles, abi1-2 and abi1-3, as well as hab1-1 abi1-2 and hab1-1 abi1-3 double mutants. The knockout abi1-2 and abi1-3 mutants (Col background) showed enhanced ABA sensitivity in germination and growth assays, which is in agreement with previous results reported for intragenic revertants of abi1-1 (Landsberg erecta background). ABA-induced stomatal closing was also ABA hypersensitive in abi1-2 and abi1-3 (Supplemental Fig. 2) in the range of 10 to 100 nm, in contrast to the recessive abi1-1R4 allele, which showed a wild-type response at 100 nm ABA (Merlot et al., 2001). This discrepancy might be due to the different genetic background of each mutant or might reflect that abi1-1R4 is not a knockout mutation. In spite of the enhanced response to ABA in stomatal closure assays, water-loss measurements in detached-leaf assays did not reveal significant differences with respect to wild type for single mutants. This may be due to the finding that detached-leaf water-loss assays to a degree reflect differences in stomatal apertures of wild type compared to a mutant at the beginning of drought experiments rather than later wilting-induced signaling events (Kuhn et al., 2006). In intact plants after a longer drought period, both abi1-2 and abi1-3 showed reduced water loss as compared to wild type (Fig. 4C). Finally, both abi1-2 and abi1-3 showed an enhanced up-regulation of some ABA- and drought-inducible genes compared to wild type, although to a modest level (1.5- to 3-fold). In general, a similarly enhanced response to ABA was observed in the hab1-1 mutant, except that ABA-mediated inhibition of growth was stronger in both abi1-2 and abi1-3 than hab1-1, indicating that ABI1 plays a predominant role in this particular response to ABA. Finally, these phenotypes conclusively indicate that ABI1 is a global negative regulator of ABA signaling. We speculate that the reduced sensitivity to ABA observed in the dominant abi1-1 allele might be due to the formation of an inactive complex between abi1-1 and one of its substrates (Gosti et al., 1999), which might be a master positive regulator of ABA signaling. In both abi1-2 and abi1-3 recessive mutants the putative target of ABI1 might be hyperactive in response to ABA; conversely, it would be inactivated by the effect of the abi1-1 dominant allele.
Previous studies have not analyzed double knockout mutants in plant PP2Cs. An abi1-1R4 abi2-1R1 double mutant was more responsive to ABA than the single parental mutants (Merlot et al., 2001). Combined inactivation of HAB1 and ABI1 in the hab1-1 abi1-2 and hab1-1 abi1-3 double mutants led to an additive ABA hypersensitivity compared to the single parental mutants. Thus, the IC50 for ABA-mediated inhibition of germination was 2-fold lower in the double mutants than in single parental mutants. The double mutants were also more sensitive than single parental mutants to inhibition of germination and early growth mediated by osmotic stress. Imposing osmotic stress at the seedling stage leads to increased ABA biosynthesis and consequently to early growth arrest (Lopez-Molina et al., 2001; Gonzalez-Guzman et al., 2004). Thus, whereas in adult plants ABA plays a crucial role to coordinate the various aspects of the low water potential response to allow plant survival, in seeds and seedlings ABA action is mainly focused to prevent germination and to arrest seedling growth. Interestingly, lowering the osmotic potential of the media by using 200 mm mannitol (−0.5 MPa) had a limited effect on wild type or single mutants, but practically abolished early growth of the double mutants (Fig. 2C). According to the dramatic effect of the combined loss-of-function phenotype, ABI1 and HAB1 must cooperate to negatively regulate ABA signaling at the seed and seedling stage. Another PP2C, PP2CA, was recently shown to strongly and negatively regulate ABA signaling during germination (Kuhn, et al., 2006; Yoshida et al., 2006). The ABA-mediated seed germination phenotype of pp2ca or hab1-1 abi1-2/hab1-1 abi1-3 mutants was apparent even though HAB1 and ABI1, or PP2CA, respectively, were functional. Therefore, at least two branches of ABA signaling (or not completely redundant functions of these proteins) appear to exist during seed germination, and the impairing of any of them leads to strong ABA hypersensitivity.
In addition to enhanced ABA-mediated inhibition of seed germination, vegetative responses to ABA were superinduced in the double mutant compared to single parental mutants. For instance, inhibition of growth upon prolonged culture in medium supplemented with ABA was particularly dramatic in hab1-1 abi1-2 and hab1-1 abi1-3 double mutants. Transpiration water loss was also noticeably reduced in the double mutants, either measured as detached-leaf assays or after a long period of drought. Finally, ABA-inducible gene expression was notably up-regulated in the double mutants compared to single parental mutants, particularly for those stress-responsive genes mostly regulated through an ABA-dependent pathway, such as RAB18, RD29B, and P5CS1. Taken together, these results indicate partially overlapping functions for HAB1 and ABI1 as negative regulators of ABA signaling, although a predominant role for ABI1 in growth control can be deduced from the ABA-mediated growth-inhibition phenotype observed in abi1-2 and abi1-3. Additionally, these results reveal fine modulation of ABA signaling through the combined action of HAB1 and ABI1 and suggest that different degrees of ABA sensitivity can be engineered in plants through PP2C modulation of the ABA signal transduction pathway.
ABA biosynthetic and signaling pathways can be considered as potential targets to improve plant performance under drought. Thus, it has been demonstrated that transgenic plants producing high levels of ABA display improved growth under drought stress than wild type (Iuchi et al., 2001; Qin and Zeevaart, 2002). Priming of ABA biosynthesis can be obtained by direct overexpression of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in the biosynthetic pathway (Iuchi et al., 2001; Qin and Zeevaart, 2002), or through the use of chemicals that accelerate ABA accumulation (Jakab et al., 2005). Alternatively, mutants affected in ABA signal transduction might also show an enhanced ABA response leading to stress-tolerant phenotypes.
Many examples of ABA-hypersensitive mutants have been reported (Finkelstein et al., 2002); however, in spite of the critical role of ABA to coordinate plant response to drought, a general correlation between enhanced response to ABA and drought tolerance has not been well established. Thus, although some mutants (i.e. era1, abh1, and gcr1) with enhanced response to ABA have been shown to cause reduced water consumption (Pei et al., 1998; Hugouvieux et al., 2001; Pandey and Assmann, 2004), many examples of mutants that do not match this assertion are known. For instance, the fry1 and sad1 mutants, which show ABA-hypersensitive inhibition of seed germination and superinduction of ABA-responsive genes, have compromised tolerance to drought stress (Xiong et al., 2001a, 2001b). Likewise, the calcineurin B-like 9, the calcineurin B-like-interacting protein kinase, and the APETALA2-like ABA repressor 1 mutants, which display ABA hypersensitivity and enhanced expression of ABA signaling genes, do not correlate with stress-tolerance phenotypes (Kim et al., 2003; Pandey et al., 2004, 2005). Therefore, superinduction of ABA- and stress-inducible genes in ABA-hypersensitive mutants does not appear to be sufficient to induce drought avoidance. A differential feature of the era1, abh1, and gcr1, as well as hab1-1 abi1-2/hab1-1 abi1-3 double mutants is an enhanced response to ABA in stomata and reduced water loss. Thus, an important consideration for engineering drought avoidance by enhancing ABA responses may include amplifying the molecular mechanisms through which ABA closes stomata. Prospecting of fully or partially sequenced plant genomes from other plants than Arabidopsis reveals the presence of gene products that are likely orthologous to the PP2Cs involved in ABA signaling in Arabidopsis, such as ABI1 and HAB1. Therefore, based on the results presented here, we suggest that silencing in crop plants of genes encoding PP2Cs with similar roles to ABI1 and HAB1 may provide a new biotechnological approach to enhance drought avoidance mechanisms.
A major advance in the study of ABA effect on stomatal closure and opening has been recently reported by Mishra et al. (2006). This work shows that ABA signaling bifurcates at ABI1 and the heterotrimeric G-protein α-subunit GPA1 to regulate ABA-mediated stomatal closure and inhibition of stomatal opening. In this work, an abi1 knockout line (abi1-ko, corresponding to SALK_076309, here named abi1-3) was used to show a genetic interaction with the phospholipase Dα1 mutant (pldα1). Thus, whereas the single mutant pldα1 abolished both ABA promotion of stomatal closure and ABA inhibition of stomatal closure, the double mutant pldα1 abi1-ko remained insensitive to ABA in the ABA inhibition of stomatal closing response, but was sensitive to ABA for promotion of stomatal closure. This result suggests that inhibition of stomatal opening by ABA is not governed through ABI1, whereas ABI1 inhibits ABA promotion of stomatal closure. The results further suggest that PLDα1 is not needed for ABA-induced stomatal closing when ABI1 is deleted. These findings are interesting in light of these and other recent findings that several PP2Cs function as negative regulators of ABA signaling (Leonhardt et al., 2004; Saez et al., 2004; Kuhn et al., 2006; Yoshida et al., 2006), but deletion of the ABI1 PP2C is sufficient to restore PLDα1-independent ABA-induced stomatal closing in pldα1 (Mishra et al., 2006). Finally, we show here that the abi1-2 and abi1-3 knockout lines show enhanced ABA-induced stomatal closing. The fact that the abi1-3 line reported by Mishra et al. (2006) did not show an ABA-hypersensitive phenotype in the stomatal closure response can likely be explained because a high dose (50 μm) of ABA was assayed.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were routinely grown under greenhouse conditions in pots containing a 1:3 vermiculite-soil mixture. For in vitro culture, seeds were surface sterilized by treatment with 70% ethanol for 20 min, followed by commercial bleach (2.5% sodium hypochlorite) containing 0.05% Triton X-100 for 10 min, and finally, four washes with sterile distilled water. Stratification of the seeds was conducted in the dark at 4°C for 3 d. Then, seeds were sowed on Murashige and Skoog (1962) plates composed of Murashige and Skoog basal salts, 0.1% 2-[N-morpholino]ethanesulfonic acid, 1% agar, and 1% Suc. The pH was adjusted to 5.7 with potassium hydroxide before autoclaving. Plates were sealed and incubated in a controlled environment growth chamber at 22°C under a 16-h light, 8-h dark photoperiod at 80 to 100 μE m−2 s−1.
Mutant Identification by PCR Screening
Two lines containing a single T-DNA insertion in ABI1 were identified in the SALK T-DNA collection (SALK_72009 and SALK_76309; Alonso et al., 2003) and obtained from the Nottingham Arabidopsis Stock Center (http://nasc.nott.ac.uk). To identify individuals homozygous for the T-DNA insertion, genomic DNA was obtained from kanamycin-resistant seedlings and submitted to PCR genotyping using the following ABI1 primers: line SALK_72009, 5′-AGGAAACCCTTATTGAAATTC and 5′-CTCTGTTCTGCTGATCATCT; line SALK_76309, 5′-CCGGCCCTCGAGATGATCAGCAGAACAGAGAGT and 5′-CCGGCCCTCGAGTCAGTTCAAGGGTTTGCT. As T-DNA left border primer of the pROK2 vector, we used LBpROK2 (5′-GCCGATTTCGGAACCACCATC).
To generate the hab1-1 abi1-2 and hab1-1 abi1-3 double mutants, we transferred pollen of either abi1-2 or abi1-3 to the stigmas of emasculated flowers of hab1-1. The resulting F2 individuals were genotyped by PCR for the presence of homozygous hab1-1 (Saez et al., 2004), abi1-2, and abi1-3 alleles (see above).
Germination Assays
To measure ABA sensitivity, seeds were plated on solid medium composed of Murashige and Skoog basal salts, 1% Suc, and increasing concentrations of ABA. To determine sensitivity to inhibition of germination by high osmoticum the medium was supplemented with increasing concentrations of either sodium chloride or mannitol, respectively. To score seed germination, the percentage of seeds that had germinated and developed fully green expanded cotyledons was determined.
Growth and Stomatal Aperture Assays
The ABA-resistant growth was scored by weighting whole plants after 12 d of the transfer of 5-d-old seedlings onto Murashige and Skoog plates supplemented with 10 μm ABA. Data were obtained for three independent experiments, each done with 15 plants. For assays of ABA-induced stomatal closing, leaves of 5- to 6-week-old plants were used. Measurements were performed on epidermal peels, which were first incubated for 2 h and 30 min in stomatal opening buffer containing 10 mm KCl, 7.5 mm iminodiacetic acid, and 10 mm MES/Tris, pH 6.2, at 20°C. Then, they were incubated for 2 h and 30 min in the same buffer supplemented or not with 10 and 100 nm ABA. Data were expressed as the average of four experiments where 30 to 40 stomata were measured for each one.
Drought Stress and Water-Loss Assays
Two different water-loss assays were performed. Short-term assays were performed in detached leaves at the same developmental stage and size from 21-d-old plants. Five leaves per individual were excised and fresh weight was determined after submitting the leaves to the drying atmosphere of a flow laminar hood. Kinetics analysis of water loss was performed and represented as the percentage of initial fresh weight at each time point.
Long-term assays were performed after removing watering in plants maintained under greenhouse conditions. To this end, plants (10 individuals per experiment, three independent experiments) were grown under normal watering conditions for 21 d and then subjected to drought stress by completely terminating irrigation and minimizing soil evaporation by covering pots with plastic Saran Wrap film. Ten leaves from each plant were removed at the time points indicated. Subsequently, leaves were weighted, incubated in demineralized water for 3 h, and weighed again. The difference in weight was considered as water loss.
RNA Analyses
Plants were grown on Murashige and Skoog plates supplemented with 1% Suc. After 7 d, approximately 30 to 40 seedlings were either mock or 10 μm ABA treated. After 3 h, plant material was collected and frozen in liquid nitrogen. Total RNA was extracted using a Qiagen RNeasy plant mini kit and 1 μg of the RNA solution obtained was reverse transcribed using 0.1 μg oligo(dT)15 primer and Moloney murine leukemia virus reverse transcriptase (Roche) to finally obtain a 40 μL cDNA solution. qRT-PCR amplifications and measurements were performed using an ABI PRISM 7000 sequence detection system (Perkin-Elmer Applied Biosystems). The sequences of the primers used for PCR amplifications were the following ones: for HAB1 (At1g72770), forward 5′-AACTGCTGTTGTTGCCTTG and reverse 5′-GGTTCTGGTCTTGAACTTTCT; for ABI1 (At4g26080), forward 5′-ATGATCAGCAGAACAGAGAGT and reverse 5′-TCAGTTCAAGGGTTTGCT; for KIN1 (At5g15960), forward 5′-GCTGGCAAAGCTGAGGAGAA and reverse 5′-TTCCCGCCTGTTGTGCTC; for RD29A (At5g52310), forward 5′-GTCCAAAGTTAC-TGATCCCAC and reverse 5′-CTTCATATCAAAATCATGACT; for P5CS1 (At2g39800), forward 5′-TTTATGGTGCTATAGATCACA and reverse 5′-GAATGTCCTGATGGGTGTAAAC; for RAB18 (At5g66400), forward 5′-ATG GCG TCT TACCAGAACCGT and reverse 5′-CCAGATCCGGAGCGGTGAAGC; for RD29B (At5g52300), forward 5′-ATG GAG TCA CAG TTG ACA CGT CC and reverse 5′-GAG ATA GTC ATC TTC ACC ACC AGG; for RD22 (At5g25610), forward 5′-ATG GCG ATT CGG CTT CCT CTG ATC and reverse 5′-GAC ATT CAT TTC TTT CCC GCG AAC; and for β-actin-8 (At1g49420), forward 5′-AGTGGTCGTACAACCGGTATTGT and reverse 5′-GAGGATAGCATGTGGAAGTGAGAA.
qRT-PCR amplifications were monitored using the Eva-Green fluorescent stain (Biotium). Relative quantification of gene expression data was carried out using the 2T−ΔΔC or comparative CT method (Livak and Schmittgen, 2001). Expression levels were normalized using the CT values obtained for the β-actin-8 gene. The presence of a single PCR product was further verified by dissociation analysis in all amplifications. All quantifications were made in triplicate on RNA samples obtained from plants treated once with ABA.
Supplementary Material
Acknowledgments
We thank Joseph Ecker and the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants, and the Arabidopsis Biological Resource Center/Nottingham Arabidopsis Stock Center for distributing these seeds.
This work was supported by the Ministerio de Educación y Ciencia and Fondo Europeo de Desarrollo Regional (grant nos. BIO2002–03090 and BIO2005–01760 to P.L.R.) and by the National Institutes of Health (grant no. R01GM060396 to J.I.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Pedro L. Rodriguez (prodriguez@ibmcp.upv.es).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.081018.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan AC, Fricker MD, Ward JL, Beale MH, Trewavas AJ (1994) Two transduction pathways mediate rapid effects of abscisic acid in Commelina guard cells. Plant Cell 6: 1319–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GJ, Chu SP, Harrington CL, Schumacher K, Hoffmann T, Tang YY, Grill E, Schroeder JI (2001) A defined range of guard cell calcium oscillation parameters encodes stomatal movements. Nature 411: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, Granier F, Giraudat J, Parcy F (2002) The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14: 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P (1996) A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science 273: 1239–1241 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy S, Read ND, Trewavas AJ (1990) Elevation of cytoplasmic calcium by caged calcium or caged inositol triphosphate initiates stomatal closure. Nature 346: 769–771 [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Garcia MP, Rodriguez D, Nicolas C, Rodriguez PL, Nicolas G, Lorenzo O (2003) Negative regulation of abscisic acid signaling by the Fagus sylvatica FsPP2C1 plays a role in seed dormancy regulation and promotion of seed germination. Plant Physiol 133: 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Abia D, Salinas J, Serrano R, Rodriguez PL (2004) Two new alleles of the abscisic aldehyde oxidase 3 gene reveal its role in abscisic acid biosynthesis in seeds. Plant Physiol 135: 325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Apostolova N, Belles JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodriguez PL (2002) The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14: 1833–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo FQ, Okamoto M, Crawford NM (2003) Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302: 100–103 [DOI] [PubMed] [Google Scholar]
- Guo Y, Xiong L, Song CP, Gong D, Halfter U, Zhu JK (2002) A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev Cell 3: 233–244 [DOI] [PubMed] [Google Scholar]
- Hetherington AM, Woodward FI (2003) The role of stomata in sensing and driving environmental change. Nature 424: 901–908 [DOI] [PubMed] [Google Scholar]
- Himmelbach A, Hoffmann T, Leube M, Hohener B, Grill E (2002) Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J 21: 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoth S, Morgante M, Sanchez JP, Hanafey MK, Tingey SV, Chua NH (2002) Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J Cell Sci 115: 4891–4900 [DOI] [PubMed] [Google Scholar]
- Hugouvieux V, Kwak JM, Schroeder JI (2001) An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106: 477–487 [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27: 325–333 [DOI] [PubMed] [Google Scholar]
- Jacob T, Ritchie S, Assmann SM, Gilroy S (1999) Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc Natl Acad Sci USA 96: 12192–12197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakab G, Ton J, Flors V, Zimmerli L, Metraux JP, Mauch-Mani B (2005) Enhancing Arabidopsis salt and drought stress tolerance by chemical priming for its abscisic acid responses. Plant Physiol 139: 267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KN, Cheong YH, Grant JJ, Pandey GK, Luan S (2003) CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15: 411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant 61: 377–383 [Google Scholar]
- Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI (2006) The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol 140: 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkela S, Franck M (1990) Cloning and characterization of a cold- and ABA-inducible Arabidopsis gene. Plant Mol Biol 15: 137–144 [DOI] [PubMed] [Google Scholar]
- Kwak JM, Moon JH, Murata Y, Kuchitsu K, Leonhardt N, DeLong A, Schroeder JI (2002) Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 14: 2849–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JD, Schroeder JI (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang V, Palva ET (1992) The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 20: 951–962 [DOI] [PubMed] [Google Scholar]
- Leckie CP, McAinsh MR, Allen GJ, Sanders D, Hetherington AM (1998) Abscisic acid-induced stomatal closure mediated by cyclic ADP-ribose. Proc Natl Acad Sci USA 95: 15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F, MacRobbie EA, Brearley CA (2000) Inositol hexakisphosphate is a physiological signal regulating the K+-inward rectifying conductance in guard cells. Proc Natl Acad Sci USA 97: 8687–8692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10: 655–661 [DOI] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264: 1448–1452 [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko V, Konrad KR, Dietrich P, Roelfsema MR, Hedrich R (2005) Cytosolic abscisic acid activates guard cell anion channels without preceding Ca2+ signals. Proc Natl Acad Sci USA 102: 4203–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh MR, Brownlee C, Hetherington AM (1990) Abscisic acid-induced elevation of guard cell cytosolic Ca2+ precedes stomatal closure. Nature 343: 186–188 [Google Scholar]
- McCourt P (1999) Genetic analysis of hormone signaling. Annu Rev Plant Physiol Plant Mol Biol 50: 219–243 [DOI] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signaling pathway. Plant J 25: 295–303 [DOI] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Mishra G, Zhang W, Deng F, Zhao J, Wang X (2006) A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 312: 264–266 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Neill SJ, Desikan R, Clarke A, Hancock JT (2002) Nitric oxide is a novel component of abscisic acid signaling in stomatal guard cells. Plant Physiol 128: 13–16 [PMC free article] [PubMed] [Google Scholar]
- Ng CK, Carr K, McAinsh MR, Powell B, Hetherington AM (2001) Drought-induced guard cell signal transduction involves sphingosine-1-phosphate. Nature 410: 596–599 [DOI] [PubMed] [Google Scholar]
- Osakabe Y, Maruyama K, Seki M, Satou M, Shinozaki K, Yamaguchi-Shinozaki K (2005) Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell 17: 1105–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GK, Cheong YH, Kim KN, Grant JJ, Li L, Hung W, D'Angelo C, Weinl S, Kudla J, Luan S (2004) The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 16: 1912–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GK, Grant JJ, Cheong YH, Kim BG, Li L, Luan S (2005) ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol 139: 1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Assmann SM (2004) The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein alpha subunit GPA1 and regulates abscisic acid signaling. Plant Cell 16: 1616–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei ZM, Ghassemian M, Kwak CM, McCourt P, Schroeder JI (1998) Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 282: 287–290 [DOI] [PubMed] [Google Scholar]
- Pei ZM, Murata Y, Benning G, Thomine S, Klusener B, Allen GJ, Grill E, Schroeder JI (2000) Calcium channels activated by hydrogen peroxide mediate abscisic acid signaling in guard cells. Nature 406: 731–734 [DOI] [PubMed] [Google Scholar]
- Qin X, Zeevaart JA (2002) Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol 128: 544–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razem FA, El Kereamy A, Abrams SR, Hill RD (2006) The RNA-binding protein FCA is an abscisic acid receptor. Nature 439: 290–294 [DOI] [PubMed] [Google Scholar]
- Rodriguez PL, Benning G, Grill E (1998) ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett 421: 185–190 [DOI] [PubMed] [Google Scholar]
- Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL (2004) Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signaling. Plant J 37: 354–369 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D (2001) Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol 52: 627–658 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Hagiwara S (1989) Cytosolic calcium regulates ion channels in the plasma membrane of Vicia faba guard cells. Nature 338: 427–430 [Google Scholar]
- Schwartz A, Wu WH, Tucker EB, Assmann SM (1994) Inhibition of inward K+ channels and stomatal response by abscisic acid: an intracellular locus of phytohormone action. Proc Natl Acad Sci USA 91: 4019–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz M, Schroeder JI (1998) Abscisic acid maintains S-type anion channel activity in ATP-depleted Vicia faba guard cells. FEBS Lett 428: 177–182 [DOI] [PubMed] [Google Scholar]
- Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002) Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct Integr Genomics 2: 282–291 [DOI] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17: 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strizhov N, Abraham E, Okresz L, Blickling S, Zilberstein A, Schell J, Koncz C, Szabados L (1997) Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J 12: 557–569 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, McCarty DR (1997) The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9: 799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahtiharju S, Palva T (2001) Antisense inhibition of protein phosphatase 2C accelerates cold acclimation in Arabidopsis thaliana. Plant J 26: 461–470 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Seki M, Ishida J, Satou M, Sakurai T, Narusaka M, Kamiya A, Nakajima M, Enju A, Akiyama K, et al (2004) Monitoring the expression profiles of genes induced by hyperosmotic, high salinity, and oxidative stress and abscisic acid treatment in Arabidopsis cell culture using a full-length cDNA microarray. Plant Mol Biol 56: 29–55 [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45: 523–539 [DOI] [PubMed] [Google Scholar]
- Wu Y, Kuzma J, Marechal E, Graeff R, Lee HC, Foster R, Chua NH (1997) Abscisic acid signaling through cyclic ADP-ribose in plants. Science 278: 2126–2130 [DOI] [PubMed] [Google Scholar]
- Xiong L, Gong Z, Rock CD, Subramanian S, Guo Y, Xu W, Galbraith D, Zhu JK (2001. a) Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev Cell 1: 771–781 [DOI] [PubMed] [Google Scholar]
- Xiong L, Lee B, Ishitani M, Lee H, Zhang C, Zhu JK (2001. b) FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev 15: 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T (2006) ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol 140: 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JA, Creelman RA (1988) Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol Plant Mol Biol 39: 439–473 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.