Abstract
The Dof (DNA binding with one finger) transcriptional activator rice (Oryza sativa) prolamin box binding factor (RPBF), which is involved in gene regulation of rice seed storage proteins, has been isolated from rice cDNA expressed sequence tag clones containing the conserved Dof. RPBF is found as a single gene per haploid genome. Comparison of RPBF genomic and cDNA sequences revealed that the genomic copy is interrupted by one long intron of 1,892 bp in the 5′ noncoding region. We demonstrated by transient expression in rice callus protoplasts that the isolated RPBF trans-activated several storage protein genes via an AAAG target sequence located within their promoters, and with methylation interference experiments the additional AAAG-like sequences in promoters of genes expressed in maturing seeds were recognized by the RPBF protein. Binding was sequence specific, since mutation of the AAAG motif or its derivatives decreased both binding and trans-activation by RPBF. Synergism between RPBF and RISBZ1 recognizing the GCN4 motif [TGA(G/C)TCA] was observed in the expression of many storage protein genes. Overexpression of both transcription factors gave rise to much higher levels of expression than the sum of individual activities elicited by either RPBF or RISBZ1 alone. Furthermore, mutation of recognition sites suppressed reciprocal trans-activation ability, indicating that there are mutual interactions between RISBZ1 and RPBF. The RPBF gene is predominantly expressed in maturing endosperm and coordinately expressed with seed storage protein genes, and is involved in the quantitative regulation of genes expressed in the endosperm in cooperation with RISBZ1.
Most plant species accumulate either globulins or prolamins in their seed storage tissues. Rice (Oryza sativa) and oat (Avena sativa) are unique among cereal crops in that they accumulate significant amounts (about 65%–85%) of glutelin and globulin as storage proteins in the endosperm. This is in contrast to other cereals that accumulate prolamins as their primary reserves. Rice glutelin and oat 12S globulins are homologous to leguminous 11S globulins, suggesting that these storage proteins are originated from a common ancestral gene (Takaiwa et al., 1999). In other cereals, most of the prolamins contain conserved sequences in their regions designated A, B, and C domains, irrespective of divergence in Mr and their overall sequences, constituting the superprolamin family (Shewry and Tatham, 1999). It should be noted that these storage proteins are specifically synthesized and deposited to protein bodies in the endosperm in spite of the difference in their evolutional origins. Therefore, it is interesting to examine what molecular mechanisms are responsible for endosperm-specific expression, and to investigate whether there are common regulatory mechanisms between rice and other cereal storage protein genes.
The spatial- and temporal-specific expression of storage protein genes is primarily regulated at the transcriptional level. Cis-regulatory elements involved in the endosperm-specific regulation of cereal storage protein genes have been mainly characterized by producing stable transgenic plants or by transient expression assays using particle bombardment (Morton et al., 1995). A number of consensus sequences such as the prolamin box (P box), GCN4, AACA, and ACGT motifs, among others, have been identified as elements that are critical in determining the endosperm specificity of cereal seed storage protein genes (Zheng et al., 1993; Takaiwa et al., 1996; Washida et al., 1999; Wu et al., 2000).
A conserved element, referred to as the endosperm box, located 300 bp upstream of the transcriptional start site, has been found in many cereal prolamin genes (Forde et al., 1985). The endosperm box has a bifactorial motif that is composed of the P box (TGTAAAG) and the GCN4 motif [TGA(G/C)TCA], which are separated by less than 10 nucleotides (Hammond-Kosack et al., 1993; Müller and Knudsen, 1993). The GCN4 motif is recognized by endosperm-specific basic Leu zipper (bZIP) trans-activators such as maize (Zea mays) Opaque2 (O2), wheat (Triticum aestivum) SPA1, barley (Hordeum vulgare) BLZ2, and rice RISBZ1 (Albani et al., 1997; Vicente-Carbajosa et al., 1998; Conlan et al., 1999; Onate et al., 1999; Onodera et al., 2001). The P box has been reported to be recognized by a Dof (DNA binding with one finger)-type transcription factor (Vicente-Carbajosa et al., 1997). Dof proteins are a group of plant-specific transcription factors that share a single highly conserved zinc finger motif named the Dof domain and have been associated with many biological processes (for review, see Yanagisawa, 2002, 2004). It has been reported that the Dof protein P-box binding factor (PBF) is implicated in trans-activation of cereal seed storage protein genes (Vicente-Carbajosa et al., 1997). Similar types of PBFs have been isolated from barley and wheat, which were designated BPBF (Mena et al., 1998) and WPBF (Mena et al., 1998). Maize PBF has also been shown to specifically interact in vitro with the bZIP protein O2 by protein-protein interactions.
We have characterized cis-regulatory elements required for endosperm-specific expression of glutelin genes (Takaiwa et al., 1996; Yoshihara et al., 1996; Wu et al., 1998, 2000; Washida et al., 1999). Combinatorial interactions of at least three cis-elements containing a P box or ACGT motif, in addition to the GCN4 and AACA motifs as common essential elements, are required to confer endosperm-specific expression in the native promoter (Zheng et al., 1993; Takaiwa et al., 1996; Washida et al., 1999; Wu et al., 2000). These glutelin gene cis-element arrangements are different from those found in many cereal prolamin genes. No typical bifactorial endosperm box composed of the P box and GCN4 motif is found in the glutelin genes.
It has been demonstrated that the GCN4 motif acts as a key regulatory element for determining tissue-specific expression, since a multimer of the GCN4 motif (with the minimum being a trimer) can confer endosperm-specific expression, and introduction of a mutation into the GCN4 motif of the native promoter alters expression patterns (Wu et al., 1998). In addition, the location and number of GCN4 motifs in glutelin promoters are strictly conserved. The GCN4 motif is recognized by at least five bZIP-type transcription factors, designated RISBZ1 to 5, in maturing seeds (Onodera et al., 2001). RISBZ1 is the only functional transcription factor with activation abilities, whereas the others are suggested to be involved in specific regulation via heterodimerization with RISBZ1. A similar situation has been observed for the maize O2 and O2 hetrodimerizing protein1 (Pysh et al., 1993). A Pro-rich activation domain has recently been identified at the N terminus of the protein (Onodera et al., 2001). On the other hand, the number and location of the P box vary widely among glutelin promoters, suggesting that this element may not be critical for determining endosperm-specific expression.
In this work, a cDNA clone encoding a rice P-box binding factor (RPBF) was isolated by screening Rice Genome Project expressed sequence tag (EST) clones containing a conserved Dof domain. There are an estimated 30 Dof genes in rice (Lijavetzky et al., 2003). The functional RPBF recognizing the P box in rice storage protein gene promoters was identified by its capacity to activate the expression of the rice glutelin and prolamin genes. We show that the RPBF gene is coordinately expressed with many seed storage protein genes irrespective of their involvement in regulation. This expression pattern is in marked contrast with maize and barley, because MPBF and BPBF precede the expression of storage protein genes. Furthermore, we demonstrate here that RPBF is involved in regulation in cooperation with RISBZ1 for not only many seed storage proteins but also some metabolic enzymes that are expressed in developing seeds.
RESULTS
Identification of the RPBF cDNA Clone
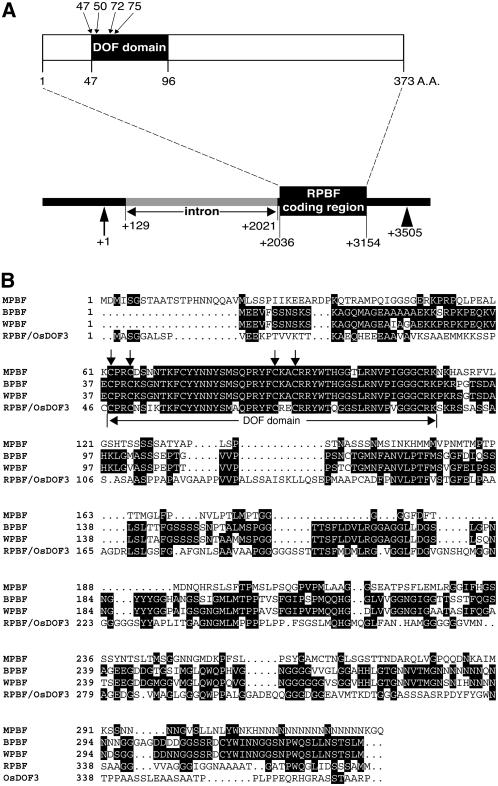
The rice seed EST databank was searched to isolate an endosperm-specific cDNA encoding the PBF. Only one out of an analyzed seven cDNA clones was identified as a candidate for the desired Dof class transcription factor, which was sequenced and designated as RPBF. The cDNA is 1,542 bp long, excluding the poly(A) tail, and contains an open reading frame encoding a potential protein of 373 amino acids with a predicted molecular mass of 36,843 D (Fig. 1A). A deduced Dof domain, responsible for DNA binding, is located between amino acids 47 and 96 of the RPBF protein (Fig. 1A). The Cys residues that are probably essential for formation of a zinc finger are located at positions 47, 50, 72, and 75 from the N terminus (Fig. 1A). The amino acid sequence deduced from the RPBF cDNA sequence is closely related to barley BPBF and wheat WPBF (Fig. 1B). The degree of overall amino acid sequence identities of the rice RPBF to barley BPBF, wheat WPBF, and maize PBF are 34.57%, 35.64%, and 26.23%, respectively, whereas the Dof domain regions are highly conserved (82.0%–84.0%; Fig. 1B). It is interesting to note that the nucleotide sequence of RPBF (accession no. AK107294) is nearly identical with that of rice OsDof3 (accession no. AB028131), except for a deletion of a single guanine in OsDof3, resulting in a frame shift. This indicates that RPBF may be the same gene to the rice OsDof3. As shown in Figure 1B, the amino acid sequences downstream from position 338 share very little homology. The accuracy of the RPBF sequence was confirmed by the rice whole genome sequence, which is now available.
Figure 1.
A, Organization of the RPBF protein and the RPBF locus. Schematic diagram of the primary structure of the RPBF protein (top section). The deduced length of the amino acid sequence encoded by the RPBF cDNA is shown. The shaded region represents the Dof domain. The location of the Cys residues is probably essential for formation of a zinc finger where indicated by arrows. Genomic structure of the RPBF gene (bottom section). Numbers indicate the positions from the putative transcription initiation site. The gray bar and the black box represent an intron and a coding region. The vertical arrow indicates the transcription initiation site as determined by primer extension analysis. The triangle indicates the putative transcription termination site. B, Comparison of the PBF protein primary sequences and Dof class proteins. Shaded boxes indicate identical amino acids. The vertical arrows indicate locations of the Cys residues likely to be essential for formation of a zinc finger. BPBF is from barley, WPBF is from wheat (Mena et al., 1998), MPBF is from maize (Vicente-Carbajosa et al., 1997), and OsDOF3 is from rice.
To identify orthologs of PBF-class Dof genes in noncereal plants, we did a BLAST search with RPBF and MPBF using the region downstream of the Dof domain as the query sequence. No related genes could be detected in dicot species, thus indicating that the PBF-class Dof proteins are cereal-specific transcription factors.
Genomic Structure of the RPBF Gene
Southern-blot analysis of rice genomic DNA showed that RPBF is encoded by a single gene (data not shown). It is now clear that RPBF is located on chromosome 2. The RPBF genomic clone was isolated by screening a rice bacterial artificial chromosome genomic library using the region of the RPBF cDNA between positions 572 and 1,597 as a hybridization probe. The site of transcription initiation was determined by primer extension analysis using poly(A) mRNA from maturing seeds. The initiation site was mapped at position −2,036 from the ATG start codon (data not shown). There was no typical TATA box [TATA(A/T) A(A/T)] found near the initiation site of transcription. However, the TATTAAA sequence between positions −26 and −32 from the transcription initiation site may function as a recognition site for the TATA box binding factor sequence.
Based on a comparison of the RPBF cDNA and genomic DNA sequences, a single intron (1,896 bp) is located in the 5′ noncoding region of the RPBF mRNA between positions +130 and +2,025 from the transcriptional start site (Fig. 1A). This intron is 15 bp longer than the one (1,881 bp) reported in the nucleotide database (accession no. AP005510), which may be because the genes are from different rice varieties (Nipponbare versus Shimokita) used for analysis.
Tissue Specificity of RPBF mRNA
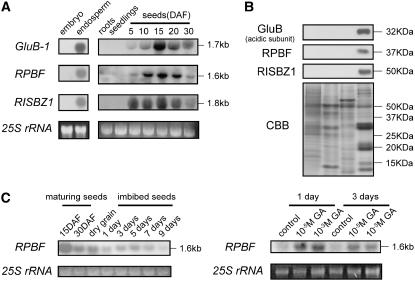
Total RNA and protein were extracted from embryo and endosperm of maturing seed (15 d after flowering [DAF]), roots, and seedlings, and subjected to northern-blot and western-blot analyses to examine the expression of RPBF. As shown in Figure 2, A and B, RPBF is specifically expressed in the endosperm of maturing seed and is not detected in the other tissues. This endosperm-specific expression pattern is also seen with RISBZ1, a transcriptional activator that determines the endosperm specificity of storage protein genes (Onodera et al., 2001), and the rice storage protein glutelin gene (GluB-1 mRNA/glutelin proteins). It was estimated by quantitative PCR that the RPBF transcript level is more than 10-fold higher than that of RISBZ1 (data not shown).
Figure 2.
Expression pattern of RPBF. A, Total RNA (30 μg) from roots, seedlings, and seeds during seed development (5, 10, 15, 20, and 30 DAF) was analyzed by northern blot using gene-specific sequences downstream of the DOF domain. To compare expression patterns of the RPBF gene with GluB-1 and RISBZ1 (Onodera et al., 2001) genes, the GluB-1 coding sequence and the RISBZ1-specific sequence corresponding to the region downstream of the Leu zipper were also used as probes. 25S rRNAs are shown as a loading control. B, Total protein from embryo and endosperm of 15 DAF seeds, roots, and seedlings was analyzed by western blot with anti-RPBF, anti-RISBZ1, and anti-glutelin GluB. Protein samples were separated on a 12.5% SDS-PAGE gel and stained with Coomassie Brilliant Blue (CBB). C, Total RNAs (7.5 μg) from imbibed seeds (top section) and from deembryonated half seeds incubated in the presence or absence of GA3 (bottom section) were subjected to northern-blot analysis. The blot was probed with the RPBF-specific sequence. Times of imbibition and GA treatment are at the top of the section.
The level of RPBF transcription during seed maturation reaches a maximum level at 15 DAF, and then drops off toward seed maturation (Fig. 2A). This profile of temporal expression parallels was observed for the glutelin gene (GluB-1), but is in remarkable contrast with the expression of the RISBZ1, which was detected before RPBF and glutelin (Fig. 2A). Thus, the temporal expression patterns of the GluB-1 and the RPBF genes differ slightly from RISBZ1.
We then examined whether the expression of RPBF in seed is induced following germination and whether its action is related to gibberellin (GA) synthesis, as is the case with barley BPBF (Mena et al., 2002). As shown in Figure 2B, RPBF transcript levels rapidly increased after imbibition, reached a plateau 5 d later, and then declined. When embryoless seeds were treated with either 10−3 or 10−5 m gibberellic acid (GA3), RPBF transcripts were induced (Fig. 2C). These results suggest that expression of the RPBF gene was induced by GA in the endosperm during early germination.
RPBF and RISBZ1 Proteins Can Activate Transcription from the Promoters of Rice Seed Storage Protein Genes
To examine the ability of the RISBZ1 and RPBF proteins to activate transcription of storage protein genes, we performed transient assays using rice callus protoplasts. In general, an internal standard has been used in such transient assays to normalize the transfection efficiency and to precisely evaluate data. Therefore, we first carried out pilot experiments to examine whether there is virtual difference in results of transcriptional activation abilities when internal standard gene (ubiquitin/luciferase) was cotransfected or protein concentrations were used as normalization standard. These results virtually showed little difference between them (data not shown). Therefore, each transfection was carried out using same number of protoplasts and normalized by their protein concentration in individual series of experiments. The RISBZ1 or RPBF coding regions were expressed under the control of a Cauliflower mosaic virus (CaMV) 35S promoter and used as the effector plasmid (35S/RISBZ1 or 35S/RPBF). Nine β-glucuronidase (GUS) reporter constructs directed by promoters from different storage protein genes were used: glutelin genes, GluA-1 (−827 to +11; Okita et al., 1989), GluA-2 (−682 to +11; Takaiwa et al., 1987a), GluA-3 (−897 to +11; Takaiwa and Oono, 1990a), GluB-1 (−199 to +23; Takaiwa et al., 1991); 16-kD prolamin gene (−931 to −1 from ATG; Qu and Takaiwa, 2004); 13-kD prolamin gene, NRP33 (−386 to −13 from ATG; Sha et al., 1996); 10-kD prolamin gene (−824 to −1 from ATG; Qu and Takaiwa, 2004); 26-kD α-globulin gene, Glb-1 (−340 to +73; Nakase et al., 1996); and 16-kD allergen (Alubmin) gene, RAG-1 (−472 to +26; Adachi et al., 1993). Protoplasts derived from rice calli were transfected with reporters or cotransfected with reporters and effectors by electroporation.
As shown in Table I, both the RISBZ1 and RPBF effectors activated transcription from storage protein promoters except for induction of the RAG-1 promoter by RISBZ1. It should be noted that RISBZ1 gave rise to higher trans-activation from GluA-1, GluA-2, GluB-1, and the 16-kD prolamin promoter than RPBF, whereas higher trans-activation was observed from the GluA-3, NRP33, 10-kD prolamin and Glb-1 promoters by the RPBF effector.
Table I.
Trans-activation of reporter genes with storage protein promoters and metabolic enzyme promoters
The proximal GluA-1 (−827 to +11), GluA-2 (−682 to +11), GluA-3 (−897 to +11), GluB-1 (−199 to +23), 16 kD prolamin gene (−391 to −1 from ATG), NRP33 (−386 to −13), 10 kD prolamin gene (−824 to −1 from ATG), Glb-1 (−340 to +73), RAG-1 (−472 to +26), AlaAT (−930 to +81), and cytoplasmic ppdk (−738 to +67) promoters were fused to the GUS reporter gene. Each of the reporter genes was electroporated into protoplasts in the presence of RISBZ1, RPBF, or RISBZ1 + RPBF. The results shown are the mean ± sd.
| Promoter/GUS | Effector Plasmid GUS Activity
|
|||
|---|---|---|---|---|
| None | RISBZ1 | RPBF | RISBZI + RPBF | |
| pM 4-MU/min/mg | ||||
| Seed storage protein promoters | ||||
| Glutelin | ||||
| GluA-1 | 10.23 ± 2.09 (1.0) | 438.87 ± 80.65 (42.9) | 125.64 ± 11.15 (12.3) | 1,763.38 ± 219.47 (172.4) |
| GluA-2 | 10.56 ± 1.68 (1.0) | 641.46 ± 194.32 (60.7) | 106.05 ± 16.42 (10.0) | 2,318.26 ± 364.71 (219.5) |
| GluA-3 | 10.89 ± 2.82 (1.0) | 371.25 ± 59.39 (34.1) | 885.90 ± 241.58 (81.3) | 1,532.28 ± 375.48 (140.7) |
| GluB-1 | 8.61 ± 1.71 (1.0) | 128.33 ± 18.79 (14.9) | 41.63 ± 8.35 (4.8) | 465.10 ± 99.19 (54.0) |
| 16 kD Prolamin | 5.29 ± 0.03 (1.0) | 227.26 ± 9.93 (43.0) | 37.59 ± 3.73 (7.1) | 1,376.88 ± 58.32 (260.3) |
| 13 kD Prolamin | ||||
| NRP33 | 8.36 ± 1.80 (1.0) | 32.75 ± 7.62 (3.9) | 80.34 ± 36.13 (9.6) | 622.71 ± 164.74 (74.5) |
| 10 kD Prolamin | 5.62 ± 0.65 (1.0) | 135.44 ± 5.26 (24.1) | 707.30 ± 27.07 (125.9) | 1,396.76 ± 3.36 (248.5) |
| α Globulin | ||||
| Glb-1 | 14.08 ± 7.12 (1.0) | 101.94 ± 17.78 (7.2) | 146.79 ± 49.89 (10.4) | 616.54 ± 255.63 (43.8) |
| Alubmin | ||||
| RAG-1 | 8.45 ± 2.45 (1.0) | 8.34 ± 1.58 (1.0) | 26.67 ± 5.45 (3.2) | 72.39 ± 12.85 (8.6) |
| Metabolic enzyme promoters | ||||
| AlaAT | 5.18 ± 0.17 (1.0) | 55.84 ± 7.05 (10.8) | 53.67 ± 6.62 (10.4) | 338.14 ± 154.94 (65.3) |
| PPDK | 19.29 ± 1.44 (1.0) | 167.18 ± 6.27 (8.7) | 525.81 ± 20.37 (27.3) | 631.30 ± 265.87 (32.7) |
To examine whether the RISBZ1 and the RPBF effectors trans-activate the storage protein genes cooperatively, protoplasts were cotransfected with both effector plasmids. The activation level of the GluA-1 promoter directed by cotransfection with RISBZ1 and RPBF was higher than the sum of the trans-activation levels directed by RISBZ1 and the RPBF individually. Similarly, these transcription factors gave rise to synergistic trans-activation from the GluA-2, GluA-3, GluB-1, NRP33, and Glb-1 promoters as well as GluA-1. It should be noted that RISBZ1 + RPBF activated the RAG-1 promoter greater than RPBF alone, though RISBZ1 did not independently affect promoter activity.
It has been reported that maize cytoplasmic orthophosphate dikinase (ppdk) is a target of maize O2 and maize Dof activators (Maddaloni et al., 1996; Yanagisawa, 2000). We next examined whether metabolic enzyme gene promoters could be trans-activated by RISBZ1 and RPBF. When Ala amino transferase (AlaAT) (−930 to +81; Kikuchi et al., 1999) and cytoplasmic ppdk (−738 to +67; Imaizumi et al., 1997), which are highly expressed in the endosperm, were transfected with the RISBZ1 or RPBF effectors, activities were synergistically increased for the AlaAT promoter, whereas there was an additive effect with the ppdk promoter.
Identification of RPBF and RISBZ1 Protein Binding Sites
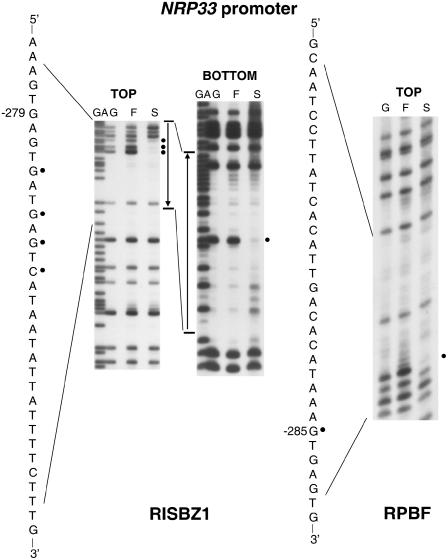
We have previously shown that RISBZ1 protein recognizes the GCN4 motif (TGAGTCA) between positions −165 and −159 of the GluB-1 promoter (Onodera et al., 2001). We determined the binding sites of RISBZ1 on the NRP33 and the Glb-1 promoters using methylation interference experiments. Two regions of the NRP33 promoter that contain the GCN4 motifs, between positions −279 and −272, and −158 and −152 upstream from the translation initiation site, were found to be protected by RISBZ1 (Figs. 3 and 4; Supplemental Fig. 1). It should be noted that a motif similar to GCN4 (TGACACA) between positions −296 and −290 was also partially protected (Fig. 4; Supplemental Fig. 1), indicating that RISBZ1 binds to canonic and variant GCN4 motifs with different affinities. The RISBZ1 protein also protects three regions of the Glb-1 promoter between positions −340 and +73, which are located at positions −254 to −250, −236 to −232, and −101 to −97 upstream of the transcription start site, and have been designated as GbR1, GbR2, and GbR3. These sites are not identical to the canonic GCN4 nor the ACGT core motifs. As candidate target sites for bZIP protein, however, all of them contain TG followed at a distance of up to 7 bp by CA (TGNx ≅ 7CA; Fig. 4; Supplemental Fig. 1). It is interesting to note that G residues flanked by the three protected sequences were partially protected by RISBZ1 (Fig. 4; Supplemental Fig. 1).
Figure 3.
Methylation interference experiments of RISBZ1 and RPBF binding to NRP33 promoter. DNA fragments corresponding to NRP33 (−386 to −276 and −295 to −172) were labeled on both strands (top and bottom). Both strands were partially methylated and incubated with GST-RISBZ1 or GST-RPBF. Free (F) and retarded (S) protein-DNA complex fragments were separated by PAGE. Fragments were eluted from the gel, chemically digested with piperidine, and separated on a sequencing gel in parallel with the sequencing ladder of this fragment.
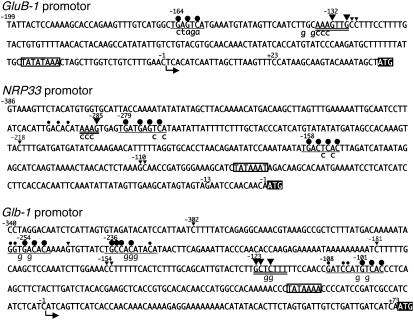
Figure 4.
Sequences of the proximal GluB-1, NRP33, and Glb-1 promoters, and summary of RISBZ1 and RPBF binding sites. Nucleotide positions relative to the transcriptional (GluB-1 and Glb-1) or translational (NRP33) start sites are indicated. The shaded boxes indicate ATG start codons. G residues, which are protected from methylation by RISBZ1 (larger circle) and RPBF (larger arrowhead), are indicated. G residues partially protected by RISBZ1 (smaller circle) and RPBF (smaller arrowhead) are also indicated. Mutant nucleotides introduced for loss-of-function analyses are shown in lowercase letters below each of the corresponding RISBZ1 and RPBF binding sites.
We then determined the binding sites of RPBF on these promoters. RPBF protein protects G residues between positions −132 and −127 of the GluB-1 promoter, whereas the P box [TG(T/A/C)AAAG] is found between positions −138 and −132 (Fig. 4; Supplemental Fig. 1). The two C residues 3′ to the P box were partially protected, indicating that the CTTT sequence motif adjacent to the P box may be recognized by RPBF (Fig. 4; Supplemental Fig. 1). The G residue 285 bp upstream of the NRP33 translation initiation site was protected by RPBF (Figs. 3 and 4). Although there was no typical P box around the G residue of NRP33 protected by RPBF, the G residue is present within the AAAG sequence motif that is a candidate target site for Dof-class proteins. The AAAG sequence motif between positions −288 and −285 of the NRP33 promoter is located only seven nucleotides upstream of the GCN4 motif, and thus was referred to as the P box. Together, the two motifs may form a bipartite motif corresponding to the endosperm box (Fig. 4). It is notable that RPBF partially protects the G residues at 218, 110, and 109 bp upstream of NRP33 translation initiation site when the AAAG sequence motif is present (Fig. 4; Supplemental Fig. 1). Similarly, RPBF protects the region containing the CTTT sequence motif (Fig. 4; Supplemental Fig. 1), between positions −123 and −120 of the Glb-1 promoter, and partially protects the G residues at positions −302, −245, −181, −154, and −153, all of which are adjacent to or within the AAAG/CTTT sequence motif.
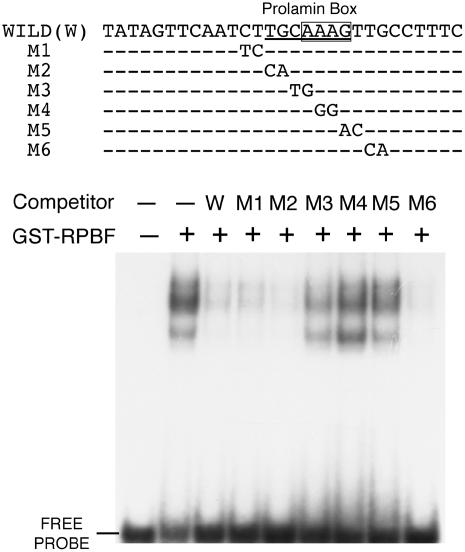
It has been reported that the maize PBF and barley BPBF proteins specifically interact with an AAAG motif (P box) within the endosperm box (Vicente-Carbajosa et al., 1997; Mena et al., 1998). To examine whether the AAAG motif in the endosperm box is specifically recognized by the RPBF protein, electrophoretic mobility shift assay (EMSA) was performed for the GluB-1 promoter. Binding of the glutathione S-transferase (GST)-RPBF fusion protein to the 29 bp fragment containing the AAAG motif was detected as a retarded band (Fig. 5). As shown in Figure 5, a 29 bp sequence containing the AAAG motif was sequentially mutagenized by two bases, which were used as 100-fold molar excess competitor. The retarded band was abolished by the presence of 100-fold molar excess wild-type fragment. Mutagenized nucleotides introduced into any position of the AAAG motif as a competitor had little or no effect on binding of the native fragment, whereas introduction of mutations outside of the AAAG sequence resulted in the loss of the retarded band (Fig. 5). Thus, mutations within the AAAG sequence motif were critical for the binding of RPBF, indicating that RPBF interacts with AAAG in a sequence-specific manner.
Figure 5.
EMSA of RPBF protein with a P box in the GluB-1 promoter. Nucleotide sequences of oligonucleotides used as probe and competitors are depicted. WILD, 29 bp sequence containing a P box from the GluB-1 promoter (−152 to −123); M1 to M6, 29 bp sequence with successive dinucleotide mutations. The P-box and AAAG motif are underlined and boxed, respectively. The GST-RPBF fusion protein was used for EMSA with the 29 bp sequence containing the P box. Competitors were added in 100-fold molar excess. Lane 1, No protein; lane 2, no competitor; lanes 3 to 9, with competitors (wild type [W] and M1 to M6).
Contribution of RPBF and RISBZ1 Binding Sites to Trans-Activation of Rice Seed Storage Protein Gene Promoters
The ability of RPBF to activate expression from the P box was examined by transient assays in rice callus protoplasts. P-box trimers in the GluB-1 (TCGAGTTCAATCTTGCAAAGTTGCCTTTC) and NRP33 (ACATAAAGT) promoters were fused to a −46 CaMV/GUS reporter gene. When trans-activated by RPBF, expression levels of the 3× P-box (GluB-1)/−46 CaMV/GUS and 3× P-box (NRP33)/−46 CaMV/GUS genes resulted in 3.1- and 1.6-fold increases, respectively (Table II). These results indicate that RPBF is able to at least partially activate the reporter genes through binding to the P box.
Table II.
Transient expression assay of P-box motif/−46 CaMV/GUS reporter genes in rice callus protoplasts
Trimers of the P box in GluB-1 and NRP33 were fused to GUS genes and were electroporated into protoplasts in the presence of RPBF. The results shown are the mean ± sd.
| Reporter | Effector (Fold Induction)
|
|---|---|
| RPBF | |
| 3× P box (GluB-1)/−46 CaMV | 6.61 ± 0.41 |
| 3× P box (NRP33)/−46 CaMV | 1.72 ± 0.38 |
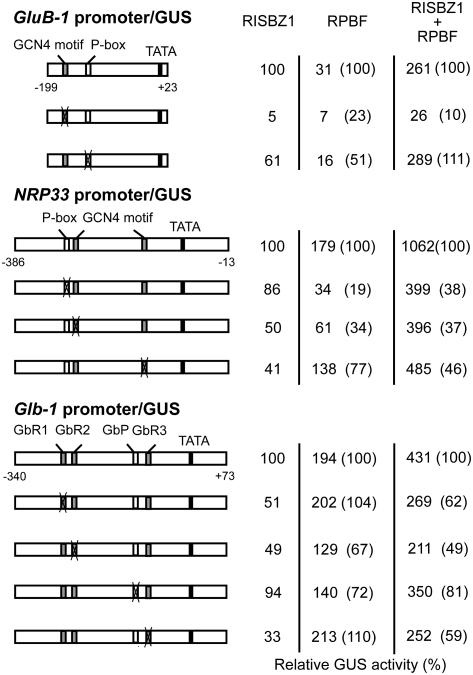
To investigate the extent to which RISBZ1 and RPBF binding sites are responsible for transcriptional activation, site-directed mutations were introduced into the RISBZ1 and RPBF binding sites of the GluB-1, NRP33, and Glb-1 promoters. As shown in Figures 4 and 6, the GCN4 motifs and P boxes of GluB-1 and NRP33, and GbR1, GbR2, GbR3, and GbP of Glb-1 were eliminated by base substitutions and then mutagenized fragments were transcriptionally fused to the GUS reporter gene. Transcriptional activation of these reporter genes driven by the RISBZ1 and RPBF activators was assayed by transient expression. As shown in Figure 6, a mutation in the GCN4 motif of the GluB-1 promoter resulted in about a 20-fold reduction of the activation level by the RISBZ1 activator. This mutation also caused a reduction in the level of trans-activation of the GluB-1 promoter by RPBF (23% of the intact promoter; Fig. 6). Elimination of the P box reduced the trans-activation level of the GluB-1 promoter by both RPBF (51% of the intact promoter) and RISBZ1 (61% of the intact promoter; Fig. 6). A mutation of the distal or proximal GCN4 motif of the NRP33 promoter reduced expression to 50% and 41% of the intact region, respectively, when driven by RISBZ1 (Fig. 6). These mutations also reduced trans-activation of the NRP33 promoter by RPBF (34% and 77% of the intact promoter, respectively; Fig. 6). Alterations in the P box of the NRP33 promoter reduced activation by both RPBF (19% of the intact promoter) and RISBZ1 (86% of the intact promoter; Fig. 6). Introduction of mutations into the GbR1, GbR2, or GbR3 motifs caused a 50% to 70% reduction in trans-activation of the Glb-1 promoter by RISBZ1 (Fig. 6). Mutation of the GbR2 motif also reduced trans-activation of the Glb-1 promoter by RPBF (67% of the intact promoter), though mutations of the GbR1 and GbR3 motifs failed to significantly affect activation (Fig. 6). Mutation of the GbP motif resulted in about a 30% reduction in the trans-activation of the Glb-1 promoter by RPBF, but did not significantly affect the RISBZ1 activation level (Fig. 6).
Figure 6.
Functional analysis of RISBZ1 and RPBF binding sites using a transient expression assay. Schematic diagrams of the proximal GluB-1 (−199 to +23), NRP33 (−386 to −13), and Glb-1 (−340 to +73) promoters are shown. The mutagenized promoters, each carrying one mutated site (marked with an X), were designed for loss-of-function assays. These promoters were linked to the GUS reporter gene and electroporated into protoplasts in the presence of RISBZ1, RPBF, or RISBZ1 + RPBF. Expression of a GUS reporter gene in the presence of the RISBZ1 was used as a control (100%). Relative GUS activities with reference to each wild-type promoter activity are shown in parentheses. RISBZ1 binding sites are the GCN4 motif, GbR1, GbR2, and GbR3. RPBF binding sites are the P box and GbP.
The possibility of synergistic activation by RISBZ1 and RPBF co-trans-activation was also examined with the introduction of mutations into activator binding sites in the GluB-1, NRP33, and Glb-1 promoters. As shown in Figure 6, when the GCN4 motif in the GluB-1 promoter was mutagenized, about a 10-fold reduction in trans-activation was observed. Even with such high suppression, this activation level (0.26) was much higher than the additive level of the individual RISBZ1 and RPBF activation levels (0.12), indicating a synergistic effect between the two proteins. However, when a mutation was introduced into the P box, trans-activation by both effectors was enhanced to about the same level as intact promoter. This may be because the flanking CTTT RPBF binding site between positions −127 and −124 substituted for the mutagenized P box. Mutations in either the P box or GCN4 motifs in the NRP33 promoter depressed transcriptional activation by RISBZ1 + RPBF effectors (37%–46% of the intact promoter). However, a synergistic effect directed by RISBZ1 + RPBF was not abolished by any of these mutations. When mutations were introduced into the GbR1, GbR2, and GbR3 motif RISBZ1 binding sites of the Glb-1 promoter, trans-activation levels by RISBZ1 + RPBF resulted in levels of about 50% to 60% of the intact promoter, which were almost equal to the additive levels. A mutation in GbP of the Glb-1 promoter caused a 20% reduction in transcriptional activation by the RISBZ1 + RPBF effectors, which was higher than the additive level.
To verify whether RISBZ1 and RPBF binding sites are sufficient to cause transcriptional activation synergism by RISBZ1 + RPBF, the endosperm box (wild type: ACATAAAGTGAGTGATGAGTCATAATA) of NRP33 was fused to the −46 CaMV/GUS reporter gene. As a negative control reporter gene, the P box and GCN4 motif in endosperm box were mutagenized (M1: ACATAcccTGAGTGATGAGTCATAATA, or M2: ACATAAAGTGAGTcccccccccTAATA), and then fused to the reporter gene. The M1 endosperm box and M2 endosperm box lack a P box and GCN4 motif, respectively. As shown in Table III, expression of the wild-type endosperm box/−46 CaMV/GUS gene was activated by RISBZ1 (15.2-fold), as well as by RPBF (1.3-fold). Activation levels of this reporter gene were further enhanced by cotransfection of the RISBZ1 and RPBF effectors, which produced much higher levels (62.6-fold) than the sum (16.4-fold) of activation levels obtained independently by RISBZ1 and RPBF (Table III). When a mutation was introduced into the P box, trans-activation levels were decreased to about 46% of the intact construct for RISBZ1 and RPBF, although little effect on activation level was directed by RPBF alone. On the other hand, a mutation in the GCN4 motif reduced RISBZ1-dependent expression levels, but failed to affect RPBF-dependent levels (Table III). Either of these two mutations highly suppressed RISBZ1 + RPBF synergism in transcriptional activation (Table III).
Table III.
Transient expression assay of endosperm-box motif/−46 CaMV/GUS reporter genes in rice callus protoplasts
The endosperm box (WT: ACATAAAGTGAGTGATGAGTCATAATA) of NRP33 was fused to a −46 CaMV/GUS reporter gene. An endosperm box containing a mutation either in the P-box (M1: ACATAcccTGAGTGATGAGTCATAATA) or GCN4 motif (M2: ACATAAAGTGAGTcccccccccTAATA) was used as a negative control. Each of the reporter genes was electroporated into protoplasts in the presence of RISBZ1, RPBF, or RISBZ1 + RPBF. The results shown are the mean ± sd.
| Reporter | Effector (Fold Induction)
|
||
|---|---|---|---|
| RISBZ1 | RPBF | RISBZ1 + RPBF | |
| WT endosperm box/−46 CaMV | 15.18 ± 3.32 | 1.26 ± 0.23 | 62.63 ± 9.32 |
| M1 endosperm box/−46 CaMV | 16.33 ± 4.03 | 1.11 ± 0.07 | 28.91 ± 11.52 |
| M2 endosperm box/−46 CaMV | 2.35 ± 0.65 | 0.92 ± 0.14 | 6.77 ± 0.71 |
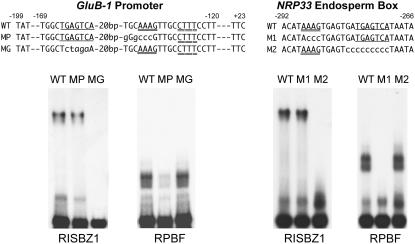
Mutations introduced into RPBF binding sites caused a reduction of transcriptional activation by overexpressed RISBZ1 protein in the GluB-1 and NRP33 promoters. Similarly, mutations in RISBZ1 binding sites, i.e. GCN4 and GR2 motifs, also reduced transcriptional activation by RPBF (Fig. 6; Table III). We performed EMSA assays to test whether a mutation in either the RPBF or RISBZ1 binding sites caused a reduction in trans-activation by either RISBZ1 or RPBF as a result of lower affinities for target sites by the effector proteins. The intact (wild type) and mutagenized GluB-1 promoters (positions −199 and +23) containing a mutation in the GCN4 motif (MG) or P-box motif (MP), were used as probes. As shown in Figure 7, wild-type and MP GluB-1 promoters were bound by RISBZ1 with similar affinities based on the retardation of fragments, but MG promoters were not recognized. RPBF was able to bind not only to wild type and MG, but its binding to the MP GluB-1 promoter was significantly reduced. Binding to the MP promoter may be accounted for if the CTTT motif flanked by a mutagenized P box is used as a target site by RPBF. Similar work was carried out with the endosperm box in the NRP33 promoter. As shown in Figure 7, the intact endosperm box in the NRP33 promoter (wild type) and the corresponding mutagenized endosperm boxes containing mutations in the P box (M1) or GCN4 motif (M2) were used as probes. RISBZ1 bound to wild-type and M1 endosperm boxes at almost equal affinities, but not to the M2 endosperm box. RPBF binds to wild-type endosperm and M2 endosperm boxes, but not to M1 endosperm. These results suggest that mutations in RPBF or RISBZ1 binding sites may not lead to a reduction in the affinity of the endosperm box for RISBZ1 or RPBF.
Figure 7.
EMSA of RISBZ1 and RPBF with the GluB-1 promoter and endosperm box in the NRP33 promoter. DNA fragments, corresponding to the intact GluB-1 promoter (positions −199 and +23; WT) and to the GluB-1 promoter containing a mutation in either the GCN4 motif or P box (MG or MP), were used as probes. The oligonucleotides, corresponding to the intact endosperm box in the NRP33 promoter (WT) and to the endosperm box containing a mutation in either P-box (M1) or GCN4 motif (M2), were used as probes. The 6xHis tagged RISBZ1 and 6xHis tagged RPBF fusion proteins were used for EMSA with the NRP33 promoter.
DISCUSSION
We have characterized the rice PBF Dof zinc finger transcription factor that activates the expression of a number of storage protein genes through binding to the AAAG or CTTT sequences in their promoters. The ability of RPBF to trans-activate and bind to specific sites in several seed genes is implicated in seed-specific expression not only of storage protein genes, but also of some metabolic enzyme genes such as AlaAT and ppdk. Rice PBF is an ortholog of maize PBF and barley BPBF Dof proteins, an endosperm-specific protein which binds to the P box (TGTAAAG) and its derivatives. Furthermore, it has been proposed that RPBF may be identical to the OsDof3 characterized as an activator of GA regulation, because their cDNA sequences are nearly identical, altered at only one position, which results in a different C terminus due to a frame shift. The difference between RPBF and OsDof3 primary sequences might be due to a sequencing error of OsDof3 or a nucleotide polymorphism between rice varieties (Shimokita/Nipponbare versus Yukihikari), because Southern-blot analysis of rice genomic DNA using RPBF/OsDof3 specific sequence showed that RPBF/OsDof3 is encoded by a single gene (data not shown). An OsDof3 cDNA clone was isolated from the aleurone cells of germinating seeds and has been reported to be involved in up-regulation of hydrolase-encoding genes such as carboxypeptidase 3 or α-amylase promoters expressed in aleurone cells following germination. The binding site of OsDOF3 is a pyrimidine box in GA-responsive elements (Washio, 2001, 2003). A similar induction of OsDof3 transcripts in GA-treated seeds was observed from RPBF (Fig. 2). Although the involvement of OsDof3 in seed protein gene regulation has, to our knowledge, not yet been reported, it can be concluded that RPBF corresponds to OsDof3. Therefore, there is a possibility that GA is implicated in the quality control of rice seed development via the RPBF function.
The RPBF gene is specifically expressed in endosperm tissue and its mRNA is not detected in leaf, roots, stem, pericarp, nor young spikes before flowering. It is noteworthy that the temporal expression pattern of RPBF during seed development is very similar to that of the storage protein glutelin gene. This expression pattern is slightly different from those of maize and barley PBFs, which precedes the expression of storage protein genes (Vicente-Carbajosa et al., 1997; Mena et al., 1998). Therefore, these results indicate that RPBF is a unique transcription factor among cereal seed PBFs. The coordinated expression with storage protein genes suggests that RPBF may not be a critical transcription factor controlling the expression of storage protein genes, but may act as an accessory activator that is involved in seed-specific expression of storage protein genes by interaction with other central transcription factors regulating the expression of seed storage genes.
It has been reported that the PBFs from maize and barley recognize either an AAAAG or a TAAAG motif in the highly conserved P box of the endosperm box as the target sequence in the promoters of seed storage protein genes (Vicente-Carbajosa et al., 1997; Yanagisawa and Schmidt, 1999; Onodera et al., 2001). We have shown by methylation interference experiments that G residues of both strands covering the 5′ (T/A)AAAG 3′ or 5′ CTTT(T/A) 3′ motifs at multiple sites in the GluB-1, Glb-1, and NRP33 promoters were specifically protected by the RPBF protein. Base-specific recognition by RPBF was also confirmed by competition experiments using a 100-fold molar excess of mutagenized fragments. When mutations were introduced into the AAAG motif, binding was abolished. It has been reported that the flanking sequences of the binding site core motif (AAAG) of many types of Dof zinc finger transcription factors determine specificity of expression (Yanagisawa and Schmidt, 1999). DNA-binding selection experiments for maize PBF revealed that the (A/T)AAAG core sequence was preferentially recognized (Yanagisawa and Schmidt, 1999). When the binding sites of RPBF were identified within the GluB-1, NRP33, and Glb-1 promoters, it was determined that most of the binding sites were (A/T)AAAG, except for the sites (CAAAG and CTTTC) between positions −136 and −132 and between positions −127 and −123 of the GluB-1 promoter. We previously reported that the binding activity of nuclear proteins from maturing endosperm, interacting with the sequence between −130 and −120 of the glutelin GluA-2 promoter, was closely correlated to the levels of glutelin mRNA during seed maturation (Takaiwa and Oono, 1990b). Levels of transcription factors gradually increased and reached a peak at 15 DAF, and then declined toward seed maturation. The CTTTC sequence was found in this sequence, suggesting that this site may be recognized by RPBF. It should be noted that such binding activity changes during seed maturation were very similar to the temporal pattern of RPBF mRNA levels during seed maturation. Furthermore, it was demonstrated that the region between −437 and −317 has a TAAAG RPBF target site that is responsible for quantitative regulation of the glutelin GluA-3 gene (Yoshihara and Takaiwa, 1996). Endosperm-specific expression was observed when this region was fused to a −90 truncated CaMV promoter. We previously showed that this region contains general positive and negative elements, and is involved in suppressing expression in nonendospermic tissues. It was suggested that the TAAAG and AACA elements act as general positive and specific negative regulators during seed maturation, respectively. These results suggest that binding of the RPBF protein to the AAAG target site may be involved in quantitative regulation of seed-specific expression of storage protein genes.
The function of the RPBF protein has been suggested by loss-of-function analysis of its binding site using homologous stable transgenic rice. Mutation of the P box in the minimal region (from −199 to +23) of the glutelin GluB-1 promoter, which confers endosperm-specific expression, suppressed its promoter activities without altering endosperm-specific expression (Wu et al., 2000). This result also indicates that interaction of the RPBF and the P box is involved in controlling the quantitative level of seed storage gene expression. Seed storage protein promoters contain multiple copies of AAAG or CTTT motifs. It is interesting to note that trans-activation levels driven by RPBF may be related to the number of these binding sites. Trans-activation levels by RPBF in the NRP33 and Glb-1 promoters were higher than that in the GluB-1 promoter. One major and two or three minor binding sites were detected within these promoters by methylation interference experiments (Figs. 3 and 4; Supplemental Fig. 1). Trans-activation levels by the RPBF effector were examined for 2.3, 1.3, and 0.2 kb GluB-1 promoters, which included 26, 18, and four tentative AAAG motifs, respectively, and it was shown that transcription levels were reduced depending on the number of RPBF binding sites (data not shown). It is notable that only one GCN4 motif is present irrespective of differences in the length of the promoter used. These results suggest that binding of RPBF to the AAAG motif, in a dose-dependent manner, may be responsible for quantitative regulation of seed-specific genes.
In many cereal prolamin promoters, the P box (or endosperm motif) and the GCN4 motif are linked, and are then designated as the endosperm box. The GCN4 motif is the binding sequence of bZIP proteins such as maize O2, wheat SPA 1, barley Blz2, and rice RISBZ1. The close linking of two motifs (bifactorial motifs) is limited to cereal prolamin genes, including the rice 13 and 16 kD prolamin promoters, and is not found in many other rice seed storage protein promoters containing glutelin and 26 kD globulin genes. In the case of the GluB-1 promoter, these motifs are separated from each another by about 25 bp, which is in contrast to the less than 10 bp of prolamin promoters. Typical pairing of the two motifs has not been found in the Glb-1 promoter.
Interactions between O2-like bZIP and PBF-like Dof proteins, which recognize the GCN4 motif and P box, respectively, have been reported in some cereal storage protein genes. Combinatorial interactions between these two activators in the transcriptional apparatus have been demonstrated to confer a synergistic effect on the expression of storage protein genes. When the RISBZ1 and RPBF activators were cotransfected with the storage promoter/GUS reporter gene, trans-activation abilities were much higher than the additive increase provided individually by RISBZ1 and RPBF, thus indicating that there is a synergistic interaction between RISBZ1 and RPBF. A 1.3- to 5.5-fold increase in trans-activation activity over additive levels directed by individual RPBF and RISBZ1 activators was observed for the seed storage protein promoters (Table I). The highest synergistic enhancement was detected in the NRP33 promoter and the lowest was in the glutelin GluA-3 promoter. Such synergistic interactions may be related to the number of potential binding sites for RPBF and RISBZ1, and distance between a potent GCN4 motif and P box. As shown in Figures 4 and 6, there is only one endosperm box in which the GCN4 motif is separated from the P box by 25 bp, and an additional P box in the 0.2 kb GluB-1 promoter. On the other hand, the NRP33 promoter has one typical and two truncated P box, and two typical and one truncated GCN4 motifs, respectively. These two typical motifs between positions −288 and −271 form a bipartite endosperm box linked to each another by 7 bp. In the case of the 0.4 kb, 16 kD prolamin promoter, one bipartite endosperm box linked by 7 bp and three additional P boxes are present, whereas the 0.8 kb, 10 kD prolamin promoter has one endosperm box composed of the GCN4 motif and P box linked by 37 bp and 18 members of the P box. It should be noted that much higher synergistic interactions (5.2- and 5.5-fold) than additive levels were observed for the NRP33 and 16 kD prolamin promoters, whereas the GluB-1 and 10 kD promoters exhibited lower synergistic interactions (1.6- and 2.7-fold). These results suggest that the distance between two motifs may be related to the interactions of bZIP and PBF transcription factors leading to synergistic activation, rather than to their number.
Full activation capacity may depend on the presence of an intact P box in the vicinity of the target site of the GCN4 motif. The Dof domain is known to be a bifunctional domain that mediates not only DNA binding but also protein-protein interactions. The Dof domain and bZIP of the bZIP protein may be responsible for these interactions. Protein-protein interactions between the bZIP O2 and PBF of maize were examined by pull-down assays (Vicente-Carbajosa et al., 1997). Although we do not yet have evidence confirming this type of protein-protein interaction between RISBZ1 and RPBF by EMSA (data not shown), functional analysis of their binding sites, in which mutations introduced into them failed to abolish trans-activation synergism by the RISBZ1 + RPBF complex, indicated the presence of direct interaction in vivo. In addition, there remains a possibility that such interaction may give rise to more efficient DNA binding, resulting in their synergism. It has been reported that barley PBF interacts not only with bZIP BLZ2, but also with the GAMYB transcription factor (Diaz et al., 2002). The AACA motif conserved in all glutelin promoters is one key regulatory motif controlling the endosperm-specific expression, which may be the target site of GAMYB. Therefore, combinatorial interactions between RPBF and GAMYB were examined by cotransfection of RPBF and GAMYB with the 0.2 kb GluB-1 promoter linked to GUS, which did not reveal any significant synergistic trans-activation, indicating little, if any, interaction between them (data not shown). Because rice GAMYB mRNA levels are negligible during seed maturation (data not shown), synergistic interactions of transcription factors regulating seed-specific expression in rice may be different from other cereals.
It has been demonstrated in transgenic rice that the GCN4 motif is the key regulatory element determining endosperm-specific expression, since trimers of the GCN4 motif in the GluB-1 minimal region are capable of conferring endosperm-specific expression (Wu et al., 1998). On the other hand, multimers of the P box are not sufficient to confer seed-specific expression in stable transgenic rice (Wu et al., 2000). Thus, the RISBZ1 protein, recognizing the GCN4 motif, may act as a key transcription factor in determining endosperm-specific expression. Furthermore, the timing of RISBZ1 transcript appearance, coming before storage protein mRNAs, is a reasonable clue to what might be a critical activator in controlling seed storage protein genes, and is consistent with the possibility that RISBZ1 may be implicated in regulation of RPBF via protein-protein interactions and direct binding to target sites of seed storage protein promoters.
Present results suggest that the newly characterized RPBF plays an important role as an activator involved in quantitative regulation by combinatorial interactions with RISBZ1 in determining endosperm-specific expression. Rice RPBF is different from other cereal PBF proteins in several regards, such as temporal regulation and interactions with other transcription factors such as GAMYB. Further work on uncovering the underlying mechanisms of protein-protein interactions and characterizing its activation domain will be required to understand the synergistic interactions between transcription factors.
Yanagisawa et al. (2004) have recently demonstrated that expression of the Dof1 transcription factor, which is involved in the expression of multiple genes associated with organic acids, enhanced nitrogen assimilation and exhibited better growth under low nitrogen conditions in transgenic plants. This result suggests that the RPBF transcription factor could be used as a metabolic engineering tool for the improvement of rice grain quality and quantity. Further work on transgenic rice lines that overexpress RPBF will be required to assess this possibility.
MATERIALS AND METHODS
Plant Materials
Rice (Oryza sativa cv Mangetsumochi) was germinated in tap water; 14-d-old leaves and roots were frozen in liquid nitrogen and stored at −80°C until use for RNA extraction. Developing seeds were harvested from field-grown rice.
GA Treatment
To examine the effect of GAs on RPBF transcription, seeds (cv kitaake) were dehusked and deembryonated and incubated in 20 mm CaCl2 and 10−3 or 10−5 m GA3 in the dark at 30°C.
Identification of the RPBF cDNA Clone
cDNA clones containing the Dof domain were screened from rice ESTs of the National Institute of Agrobiological Sciences (NAIS) DNA bank using the BLAST algorithm. Seven cDNA clones were identified and provided by T. Sasaki (NAIS).
Screening of a Rice Genomic Library
To determine the genomic structure coding for full-length RPBF, a rice bacterial artificial chromosome genomic library was screened using the region coding for the C-terminal portion downstream of the Dof domain of the RPBF cDNA clone. Labeling of the DNA fragment and hybridization were carried out using an ECL direct nucleic acid labeling and detection system (Amersham-Biosciences).
Northern-Blot Analysis
Total RNA from roots, seedlings, and seeds was isolated as described (Takaiwa et al., 1987b). Blots were probed using the DNA fragment encoding the C-terminal portion (between positions 511 and 1,218) downstream of the Dof domain of the RPBF cDNA clone (accession no. AK107294). Hybridization was carried out at 45°C in 50% formamide, 5× SSC, 0.1% SDS, and 5× Denhardt's solution and then washed twice at 55°C in 0.1× SSC and 0.1% SDS for 30 min each.
Western-Blot Analysis
Total proteins were extracted from a fine frozen powder of tissues with SDS/urea buffer (4% SDS, 8 m urea, 5% 2-mercaptoethanol, 50 mm Tris-HCl pH 6.8, 20% (v/v) glycerol; 10 mg powder per 100 μL) as described (Tada et al., 2003). Proteins were fractionated by SDS-PAGE (Laemmli, 1970) and electroblotted onto Immobilon-P (Millipore) using a Mini Trans-Blot Cell (Bio-Rad). The membrane was blocked by incubation in Tris-buffered saline plus Tween 20 (TBST; 25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.05% Tween 20) containing 5% nonfat dried milk for 1 h at room temperature, and then incubated with blocking buffer containing antiserum (1 μg/mL) for 2 h at room temperature. After three washes with TBST for 10 min each, the membrane was incubated with the secondary antibody (a 1:3,000 dilution of horseradish peroxidase-conjugated anti-rabbit IgG; Cell Signaling Technology) in blocking buffer for 1 h at room temperature, and washed three times in TBST for 10 min each. Signal bands were detected using the ECL detection system (Amersham-Biosciences).
Preparation of Antisera
The C-terminal 113 amino acids of the predicted RISBZ1 protein and the C-terminal 277 amino acids of the predicted RPBF were expressed as GST fusion proteins in Escherichia coli, and purified as described (Suzuki et al., 1998). Immunization of rabbits and purification of antisera were performed according to standard methods (Sambrook et al., 1989). The antiserum against RISBZ1 was immunoadsorbed by passing it over GST-coupled CNBr-activated Sepharose 4B (Amersham-Biosciences), and then affinity purified using antigens coupled to CNBr-activated Sepharose 4B according to the manufacturer's recommendations. The anti-RPBF antiserum was immunoadsorbed to GST Sepharose, and then affinity purified using the antigen based on a western-blotting technique (Mishkind et al., 1987).
Primer Extension Analysis
Primer extension analysis was carried out as described by Sambrook et al. (1989). A 30-mer antisense oligonucleotide (5′-RPBF1: 5′-CTGTTGGTGAGAGATCTAGCTTGCTTGAAA-3′) was labeled at the 5′ end with T4 polynucleotide kinase and [γ32P] ATP with a MegaPrime kit (Amersham-Biosciences) and then used as the primer of a reverse transcriptase reaction with poly(A) mRNA with a Supertranscriptase kit (GIBCO BRL).
Expression of the Fusion Protein in E. coli
The coding sequence for RPBF and RISBZ1 cDNAs were inserted into the vector pGET4T-1 (Amersham-Biosciences). GST-fusion proteins were expressed and isolated as described (Suzuki et al., 1998). For the production of 6xHis tagged proteins, the coding sequences for RPBF and RISBZ1 cDNAs were ligated to the cold-shock expression vector pCold II DNA (TAKARA BIO). 6xHis tagged proteins were induced according to the manufacturer's instructions and purified using a nickel-nitrilotriacetic acid agarose spin column (Qiagen). After purification by affinity chromatography, fusion proteins were dialyzed overnight against 20 mm HEPES-KOH (pH 7.9), 50 mm KCl, 1 mm EDTA, and 10% glycerol.
EMSA
The four oligonucleotide probes described in Figures 5 and 7 were generated by annealing a complementary single-stranded oligonucleotide that created a four base (TCGA) overhang at the 5′ end. The SalI to BamHI fragment (−199 and +23) of GluB-1 5′ flanking region and its mutagenized derivatives described in Figure 7 were also used as probes. These probes were end labeled with [α32P]dCTP by fill-in reaction with Klenow fragment and fractionated by electrophoresis in a 5% polyacrylamide native gel. For the mutant competitors (M1–M6), 29 bp complementary oligonucleotides with successive mutations by three bases were synthesized and annealed. Gel-retardation assays were performed by the addition of GST-RPBF, 6xHis tagged RISBZ1, or 6xHis tagged RPBF fusion proteins as described previously (Onodera et al., 2001).
The radioactive probe was incubated with 0.5 μg fusion proteins in binding buffer (Onodera et al., 2001) for 20 min at room temperature. For competition assays, 100-fold molar excess of unlabeled competitor DNA was added to the binding reaction. The mixture was loaded onto a 5% native polyacrylamide gel in 0.25× Tris-borate/EDTA buffer at room temperature.
Methylation Interference Experiments
Methylation interference experiments were carried out as described by Weinberger et al. (1986). The 5′ flanking regions of glutelin GluB-1 (−199 to −155, −140 to −80, and −110 to −42), NRP33 (−386 to −276, −295 to −172, and −188 to −91), and Glb-1 (−340 to −228, −245 to −122, and −137 to −29) were amplified by PCR and cloned into the TA cloning vector (pCR 2.1). Cloned DNA was digested with restriction enzymes and end labeled by fill-in reaction with Klenow fragment and [α32P]dCTP, and then methylated by dimethylsulphate. The end-labeled fragments were incubated with GST-RPBF or GST-RISBZ1 and fractionated by electrophoresis on a 5% native acrylamide gel. After free and protein-bound DNA fragments were eluted and purified on a DEAE-Sephacel column, fragments were electrophoresed through a 6% sequencing gel containing 7 m urea in parallel with sequencing ladders (G and G/A specific).
Construction of an Effector Plasmid
35S/RISBZ1 plasmid (Onodera et al., 2001) was used as an effector. For the construction of a 35S/RPBF plasmid, the regions encoding the entire RPBF protein were amplified by PCR using forward (AGCTAGATCTCTCACCAACAGGAGGTG) and reverse (GGTTGATTTGATTGAGCTTACATCATGGCCGAGCTGCT) primers with overhanging XhoI and BamHI recognition sites. After digestion, these cDNAs were inserted into the corresponding sites of pRT100 (Töpfer et al., 1987).
Construction of a Reporter Plasmid
A GluB-1 (−199 to +23) promoter/GUS reporter gene, constructed as described (Wu et al., 2000), was used as a reporter. For the construction of NRP33 and Glb-1 gene promoter/GUS reporters, 5′ flanking regions of NRP33 (between positions −386 and −13) and Glb-1 (between positions −340 and +73) were amplified by PCR using forward and reverse primers containing overhanging PstI and BamHI recognition sites, respectively. PCR fragments were digested with PstI and BamHI and introduced into the corresponding sites of pBI201. Mutagenized GluB-1 (−199 to +23) promoters were constructed as described (Hammond-Kosack et al., 1993; Wu et al., 2000). Base substitutions in the NRP33 and Glb-1 promoters described in Figure 4 were introduced by the method of Mikaelian and Sergeant (1992).
Three times P-box (GluB-1)/−46 CaMV/GUS reporter genes were constructed as described (Wu et al., 2000). To construct trimers of the P-box sequence (ACATAAAGT) in NRP33, 27 b complementary oligonucleotide pairs containing the sticky ACGT sequence at the 5′ end of the sense strand were annealed. For the construction of the monomer-normal endosperm box (ACATAAAGTGAGTGATGAGTCATAATA) of NRP33 and mutant endosperm boxes (ACATAcccTGAGTGATGAGTCATAATA and ACATAAAGTGAGTcccccccccTAATA), double-stranded oligonucleotides were generated by annealing the 27 bp complementary oligonucleotide pairs containing the sticky ACGT sequence at the 5′ end of the sense strand. These double-stranded oligonucleotides were inserted into the SalI and StuI sites of the −46 CaMV/GUS reporter gene.
Transient Expression and Enzyme Assays
Transient expression assays in rice callus protoplasts were performed by electroporation as described previously (Wu et al., 1998). GUS activities were measured by fluorometric quantification of 4-methylumbelliferone produced from the glucuronide precusor according to Jefferson (1987). Protein concentrations were determined with a Bio-Rad kit using bovine serum albumin as standard. The means and sds are presented for at least three assays performed with independent transfections.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AK107294 (RPBF cDNA), AP005510 (RPBF genome), and AB028131 (OsDof3 cDNA).
Supplementary Material
This work was supported by research grants from the Ministry of Agriculture, Forestry and Fisheries of Japan (Functional analysis of genes relevant to agriculturally important traits in rice genome: IP2001) and the Research and Development Program for New Bio-Industry Initiatives of the Bio-Oriented Technology Research Advancement Institution (to F.T.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Fumio Takaiwa (takaiwa@nias.affrc.go.jp).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.082826.
References
- Adachi T, Izumi H, Yamada T, Tanaka K, Takeuchi S, Nakamura R, Matsuda T (1993) Gene structure and expression of rice seed allergenic proteins belonging to the alpha-amylase/trypsin inhibitor family. Plant Mol Biol 21: 239–248 [DOI] [PubMed] [Google Scholar]
- Albani D, Hammond-Kosack MC, Smith C, Conlan S, Colot V, Holdsworth M, Bevan MW (1997) The wheat transcriptional activator SPA: a seed-specific bZIP protein that recognizes the GCN4-like motif in the bifactorial endosperm box of prolamin genes. Plant Cell 9: 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan RS, Hammond-Kosack M, Bevan M (1999) Transcription activation mediated by the bZIP factor SPA on the endosperm box is modulated by ESBF-1 in vitro. Plant J 19: 173–181 [DOI] [PubMed] [Google Scholar]
- Diaz I, Vicente-Carbajosa J, Abraham Z, Martinez M, Isabel-LaMoneda I, Carbonero P (2002) The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J 29: 453–464 [DOI] [PubMed] [Google Scholar]
- Forde BG, Heyworth A, Pywell AH, Kreis M (1985) Nucleotide sequence of a B1 hordein gene and the identification of possible upstream regulatory elements in endosperm storage protein genes from barley, wheat and maize. Nucleic Acids Res 13: 7327–7339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack MC, Holdsworth MJ, Bevan MW (1993) In vivo footprinting of a low molecular weight glutenin gene (LMWG-1D1) in wheat endosperm. EMBO J 12: 545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi N, Ku MSB, Ishihara K, Samejima M, Matsuoka M (1997) Characterization of the gene for pyruvate, orthophosphate dikinase from rice, a C3 plant, and a comparison of structure and expression between C3 and C4 genes for this protein. Plant Mol Biol 34: 701–716 [DOI] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 [Google Scholar]
- Kikuchi H, Hirose S, Toki S, Akama K, Takaiwa F (1999) Molecular characterization of a gene for alanine aminotransferase from rice (Oryza sativa). Plant Mol Biol 39: 149–159 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lijavetzky D, Carbonero P, Vicente-Carbajosa J (2003) Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol Biol 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddaloni M, Donini G, Balconi C, Rizzi E, Gallusci P, Forlani F, Lohmer S, Thompson R, Salamini F, Motto M (1996) The transcriptional activator Opaque-2 controls the expression of a cytosolic form of pyruvate orthophosphate dikinase-1 in maize endosperms. Mol Gen Genet 250: 647–54 [DOI] [PubMed] [Google Scholar]
- Mena M, Cejudo FJ, Isabel-Lamoneda I, Carbonero P (2002) A role for the DOF transcription factor BPBF in the regulation of gibberellin-responsive genes in barley aleurone. Plant Physiol 130: 111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena M, Vicente-Carbajosa J, Schmidt RJ, Carbonero P (1998) An endosperm-specific DOF protein from barley, highly conserved in wheat, binds to and activates transcription from the prolamin-box of a native B-hordein promoter in barley endosperm. Plant J 16: 53–62 [DOI] [PubMed] [Google Scholar]
- Mikaelian I, Sergeant A (1992) A general and fast method to generate multiple site directed mutations. Nucleic Acids Res 20: 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishkind ML, Plumley FG, Raikhel NV (1987) Immunochemical analysis of plant tissue. In Vaughn KC, ed, CRC Handbook of Plant Cytochemistry, Vol 2. CRC Press, Boca Raton, FL, pp 65–119
- Morton RL, Quiggin D, Higgins TJV (1995) Regulation of seed storage protein gene expression. In J Kigel, G Galili, eds, Seed Development and Germination. Marcel Dekker, NY, pp 103–138
- Müller M, Knudsen S (1993) The nitrogen response of a barley C-hordein promoter is controlled by positive and negative regulation of the GCN4 and endosperm box. Plant J 6: 343–355 [DOI] [PubMed] [Google Scholar]
- Nakase M, Hotta H, Adachi T, Alvarez AM, Aoki N, Nakamura R, Masumura T, Tanaka K, Matsuda T (1996) Cloning of the rice seed alpha-globulin-encoding gene: sequence similarity of the 5′-flanking region to those of the genes encoding wheat high-molecular-weight glutenin and barley D hordein. Gene 170: 223–226 [DOI] [PubMed] [Google Scholar]
- Okita TW, Hwang YS, Hnilo J, Kim WT, Aryan AP, Larson R, Krishnan HB (1989) Structure and expression of the rice glutelin multigene family. J Biol Chem 264: 12573–12581 [PubMed] [Google Scholar]
- Onate L, Vicente-Carbajosa J, Lara P, Diaz I, Carbonero P (1999) Barley BLZ2, a seed-specific bZIP protein that interacts with BLZ1 in vivo and activates transcription from the GCN4-like motif of B-hordein promoters in barley endosperm. J Biol Chem 274: 9175–9182 [DOI] [PubMed] [Google Scholar]
- Onodera Y, Suzuki A, Wu CY, Washida H, Takaiwa F (2001) A rice functional transcriptional activator, RISBZ1, responsible for endosperm-specific expression of storage protein genes through GCN4 motif. J Biol Chem 276: 14139–14152 [DOI] [PubMed] [Google Scholar]
- Pysh LD, Aukerman MJ, Schmidt RJ (1993) OHP1: a maize basic domain/leucine zipper protein that interacts with opaque2. Plant Cell 5: 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu LQ, Takaiwa F (2004) Evaluation of tissue specificity and expression strength of rice seed component gene promoters in transgenic rice. Plant Biotechnol J 2: 113–125 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sha S, Sugiyama Y, Mitsukawa N, Masumura T, Tanaka K (1996) Cloning and sequencing of a rice gene encoding the 13-kDa prolamin polypeptide. Biosci Biotechnol Biochem 60: 335–337 [DOI] [PubMed] [Google Scholar]
- Shewry PR, Tatham AS (1999) The characteristics, structures and evolutionary relationships pf prolamins. In PR Shewry, R Casey, eds, Seed Proteins. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 11–33
- Suzuki A, Wu CY, Washida H, Takaiwa F (1998) Rice MYB protein OSMYB5 specifically binds to the AACA motif conserved among promoters of genes for storage protein glutelin. Plant Cell Physiol 39: 555–559 [DOI] [PubMed] [Google Scholar]
- Tada Y, Utsumi S, Takaiwa F (2003) Foreign gene products can be enhanced by introduction into low storage protein mutants. Plant Biotechnol J 1: 411–422 [DOI] [PubMed] [Google Scholar]
- Takaiwa F, Ebinuma H, Kikuchi S, Oono K (1987. a) Nucleotide sequence of a rice glutelin gene. FEBS Lett 221: 43–47 [Google Scholar]
- Takaiwa F, Kikuchi S, Oono K (1987. b) A rice glutelin family-A major type of glutelin mRNAs can be divided into two classes. Mol Gen Genet 208: 15–22 [Google Scholar]
- Takaiwa F, Ogawa M, Okita TW (1999) Rice glutelins. In PR Shewry, R Casey, eds, Seed Proteins. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 401–425
- Takaiwa F, Oono K (1990. a) Genomic DNA sequences of two new genes for new storage protein glutelin in rice. Jpn J Genet 66: 161–171 [DOI] [PubMed] [Google Scholar]
- Takaiwa F, Oono K (1990. b) Interaction of an immature seed-specific trans-acting factor with the 5′ upstream region of a rice glutelin gene. Mol Gen Genet 224: 289–293 [DOI] [PubMed] [Google Scholar]
- Takaiwa F, Oono K, Wing D, Kato A (1991) Sequence of three members and expression of a new major subfamily of glutelin genes from rice. Plant Mol Biol 17: 875–885 [DOI] [PubMed] [Google Scholar]
- Takaiwa F, Yamanouchi U, Yoshihara T, Washida H, Tanabe F, Kato A, Yamada K (1996) Characterization of common cis-regulatory elements responsible for the endosperm-specific expression of members of the rice glutelin multigene family. Plant Mol Biol 30: 1207–1221 [DOI] [PubMed] [Google Scholar]
- Töpfer R, Matzeit V, Gronenborn B, Schell J, Steinbiss HH (1987) A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Res 15: 5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Moose SP, Parsons RL, Schmidt RJ (1997) A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc Natl Acad Sci USA 94: 7685–7690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Onate L, Lara P, Diaz I, Carbonero P (1998) Barley BLZ1: a bZIP transcriptional activator that interacts with endosperm-specific gene promoters. Plant J 13: 629–640 [DOI] [PubMed] [Google Scholar]
- Washida H, Wu CY, Suzuki A, Yamanouchi U, Akihama T, Harada K, Takaiwa F (1999) Identification of cis-regulatory elements required for endosperm expression of the rice storage protein glutelin gene GluB-1. Plant Mol Biol 40: 1–12 [DOI] [PubMed] [Google Scholar]
- Washio K (2001) Identification of Dof proteins with implication in the gibberellin-regulated expression of a peptidase gene following the germination of rice grains. Biochim Biophys Acta 1520: 54–62 [DOI] [PubMed] [Google Scholar]
- Washio K (2003) Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol 133: 850–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger J, Baltimore D, Sharp PA (1986) Distinct factors bind to apparently homologous sequences in the immunoglobulin heavy-chain enhancer. Nature 322: 846–848 [DOI] [PubMed] [Google Scholar]
- Wu CY, Suzuki A, Washida H, Takaiwa F (1998) The GCN4 motif in a rice glutelin gene is essential for endosperm-specific gene expression and is activated by Opaque-2 in transgenic rice plants. Plant J 14: 673–683 [DOI] [PubMed] [Google Scholar]
- Wu CY, Washida H, Onodera Y, Harada K, Takaiwa F (2000) Quantitative nature of the Prolamin-box, ACGT and AACA motifs in a rice glutelin gene promoter: minimal cis-element requirements for endosperm-specific gene expression. Plant J 23: 415–421 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S (2000) Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J 21: 281–288 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S (2002) The Dof family of plant transcription factors. Trends Plant Sci 7: 555–560 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S (2004) Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol 45: 386–391 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Akiyama A, Kisaka H, Uchimiya H, Miwa T (2004) Metabolic engineering with Dof1 transcription factor in plants: improved nitrogen assimilation and growth under low-nitrogen conditions. Proc Natl Acad Sci USA 101: 7833–7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Schmidt RJ (1999) Diversity and similarity among recognition sequences of Dof transcription factors. Plant J 17: 209–214 [DOI] [PubMed] [Google Scholar]
- Yoshihara T, Takaiwa F (1996) Cis-regulatory elements responsible for quantitative regulation of the rice seed storage protein glutelin GluA-3 gene. Plant Cell Physiol 37: 107–111 [DOI] [PubMed] [Google Scholar]
- Yoshihara T, Washida H, Takaiwa F (1996) A 45-bp proximal region containing AACA and GCN4 motif is sufficient to confer endosperm-specific expression of the rice storage protein glutelin gene, GluA-3. FEBS Lett 383: 213–218 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Kawagoe Y, Xiao S, Li Z, Okita TW, Hau TL, Lin A, Murai N (1993) 5′ distal and proximal cis-acting regulator elements are required for developmental control of a rice seed storage protein glutelin gene. Plant J 4: 357–366 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.