Abstract
Plant nucleotide binding and leucine-rich repeat (NB-LRR) proteins contain a region of homology known as the ARC domain located between the NB and LRR domains. Structural modeling suggests that the ARC region can be subdivided into ARC1 and ARC2 domains. We have used the potato (Solanum tuberosum) Rx protein, which confers resistance to Potato virus X (PVX), to investigate the function of the ARC region. We demonstrate that the ARC1 domain is required for binding of the Rx N terminus to the LRR domain. Domain-swap experiments with Rx and a homologous disease resistance gene, Gpa2, showed that PVX recognition localized to the C-terminal half of the LRR domain. However, inappropriate pairings of LRR and ARC2 domains resulted in autoactive molecules. Thus, the ARC2 domain is required to condition an autoinhibited state in the absence of elicitor as well as for the subsequent elicitor-induced activation. Our data suggest that the ARC region, through its interaction with the LRR, translates elicitor-induced modulations of the C terminus into a signal initiation event. Furthermore, we demonstrate that physical disruption of the LRR–ARC interaction is not required for signal initiation. We propose instead that this activity can lead to multiple rounds of elicitor recognition, providing a means of signal amplification.
INTRODUCTION
Race-specific, or gene-for-gene, resistance is a robust plant defense response whose initiation is dependent on the genotypes of both the host and the pathogen. The products of plant disease resistance (R) genes, known as R proteins, initiate this response when the appropriate elicitors, pathogen-encoded avirulence (Avr) gene products, are present (Flor, 1971; Keen, 1990). This response is often associated with a type of programmed cell death referred to as the hypersensitive response (HR) (Dangl et al., 1996; Hammond-Kosack and Jones, 1996). The type of R genes most prevalent in plant genomes encode proteins referred to as NB-LRR or NBS-LRR proteins because they contain a central nucleotide binding (NB) domain as well as a C-terminal leucine-rich repeat (LRR) domain. The NB domains of two NB-LRR proteins have been shown in vitro to bind and hydrolyze ATP (Tameling et al., 2002). The LRR domains of R proteins are highly divergent both in primary structure and number of repeats, appear to have undergone diversifying selection, and have been shown to be the region of the protein that confers recognition specificity (Meyers et al., 1998; Noel et al., 1999; Mondragon-Palomino et al., 2002).
Between the NB and LRR domains is a well-conserved region of homology whose function is poorly understood. This region has been defined as the ARC domain because of its presence in Apaf-1, R proteins, and CED-4 (van der Biezen and Jones, 1998) and is found in members of the apoptotic ATPase family of STAND (for signal transduction ATPases with numerous domains) NTPases (Leipe et al., 2004). Given that the NB and ARC domains are contiguous, these domains are often referred to as the NB-ARC domain. Recent molecular modeling of the ARC domain of plant NB-LRR proteins based on the crystal structure of Apaf-1 suggests that this domain is composed of two separate structural units: an N-terminal helical bundle and a C-terminal winged helix domain, referred to as the ARC1 and ARC2 subdomains, respectively (Albrecht and Takken, 2006; McHale et al., 2006).
Plant NB-LRR proteins can be divided into two classes based on the putative signaling domain present at the N terminus: those with an N-terminal TIR (for Toll and Interleukin-1 Receptor) homology domain and those without. The latter are identified by canonical motifs in the NB-ARC domain, and their N termini are often predicted to encode coiled-coil domains; thus, they are referred to as the CC class of NB-LRR proteins (Meyers et al., 1999; Cannon et al., 2002).
The potato (Solanum tuberosum) protein Rx confers resistance to Potato virus X (PVX), with the PVX coat protein (CP) acting as the Avr determinant (Bendahmane et al., 1995). Rx is a typical CC-NB-LRR protein and, of characterized R proteins, is most closely related to the potato proteins Rx2 and Gpa2 and the pepper (Capsicum annuum) protein Bs2, which confer resistance to PVX, the nematode Globodera pallida, and the bacterium Xanthomonas campestris, respectively (Tai et al., 1999; Bendahmane et al., 2000; van der Vossen et al., 2000).
Studies of the Rx and Bs2 proteins have demonstrated that expression of protein fragments consisting of either CC-NB-ARC plus LRR or CC plus NB-ARC-LRR reconstitutes the function of the full-length molecule in generating an elicitor-specific HR and that these same fragments undergo physical intramolecular interactions (Moffett et al., 2002; Leister et al., 2005). In the case of Rx, both of these interactions are disrupted in the presence of PVX CP. These disruptions likely play a role in R protein function; however, activation cannot be a simple matter of relieving a negative regulatory interaction, as physical removal of any of the domains does not lead to constitutive activation of the protein (Moffett et al., 2002).
A number of studies have described amino acid substitutions in NB-LRR proteins that result in constitutive activation of resistance responses and/or programmed cell death in the absence of elicitor (Li et al., 2001; Bendahmane et al., 2002; Shirano et al., 2002; Noutoshi et al., 2005). These constitutive gain-of-function mutants are referred to as autoactivators. Autoactivating proteins have also been obtained by swapping amino acid sequence between closely related paralogues or alleles of the Rp-1, L, and Mi-1 NB-LRR proteins (Hwang et al., 2000; Sun et al., 2001; Howles et al., 2005). This autoactivation can occur by the inappropriate pairing of several different regions of these proteins, although it has not been well defined which regions must be intercompatible.
We have investigated the role of the ARC domain in the regulation of the Rx protein. We show that the ARC1 subdomain plays a critical role in physically recruiting the LRR to the CC-NB-ARC. Domain-swap experiments between Rx and GPA2 demonstrate that pairing of at least two different regions of the Rx LRR with the GPA2 ARC2 subdomain resulted in autoactivation, suggesting that interplay between ARC2 and the LRR regulates the molecule's transition from an inactive to an active state. We suggest a model wherein recognition represents any event that results in a change at the interface between the ARC and LRR allowing this transition to take place. In addition, Rx/GPA2 swaps demonstrate that CP recognition specificity maps to the C terminus of the Rx LRR domain. We also present evidence that disruption of the interaction between the ARC and LRR by the CP is subsequent to, or coincident with, signal initiation and suggest that this disruption is required for multiple rounds of recognition, resulting in signal amplification.
RESULTS
Molecular Dissection of the Rx NB-ARC Domain
Previous studies have shown that the P-loop/Kinase 1 (PL) and Kinase 2 (K2) motifs of the Rx NB domain, as well as the ARC motif GxP (often referred to as GLPL), are critical to Rx function (Bendahmane et al., 2002). To determine the importance of the various conserved motifs within the ARC domain of Rx, we replaced a number of residues with Ala, as indicated in red in Figure 1. We chose conserved residues, as identified by van der Biezen and Jones (1998), including two highly conserved motifs, the RNBS-D and MHDV motifs, wherein autoactivating mutations have been identified previously (Bendahmane et al., 2002). Because preliminary experiments with L301A, F307A, and E318A showed no effect on Rx function, we generated combinations of these substitutions to test whether they had a cumulative effect.
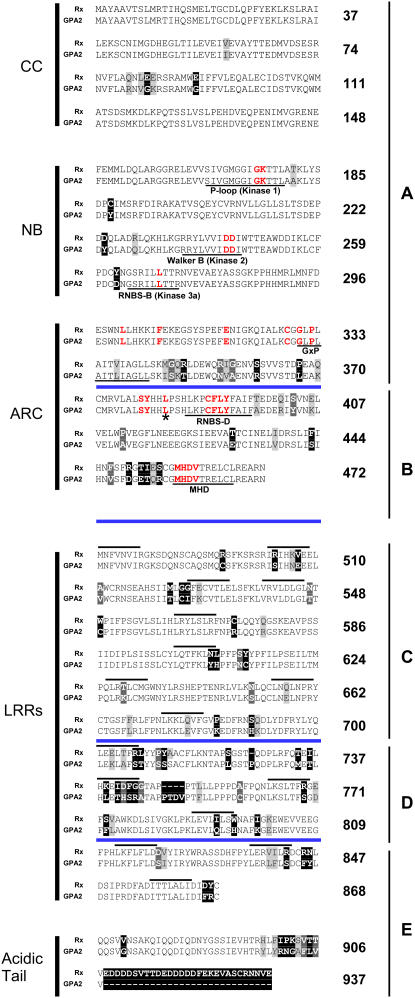
Figure 1.
Alignment of Rx and GPA2.
NB-ARC residues mutated in this work are shown in red, and conserved NB-ARC motifs are underlined. Blue lines delineate Rx/GPA2 regions A to E analyzed by sequence exchange in Figure 4. The asterisk marks the site of the largest (amino acids 1 to 382) C-terminal deletion still capable of binding the LRR domain (see Figure 2). Boxed residues highlight sequence differences between Rx and GPA2, with lighter boxes indicating more conservative substitutions. LRRs as originally annotated (Bendahmane et al., 1999) are overlined. Numbering refers to Rx residues.
Full-length Rx variants were transiently expressed under the control of the Rx genomic promoter (PRx) via agroinfiltration in Nicotiana benthamiana leaves together with either green fluorescent protein (GFP) or CP (Figure 2A). As reported previously (Bendahmane et al., 2002), mutations in the K2 (DD244AA) and GxP (GLP330ALA) motifs eliminated the ability of Rx to induce a CP-dependent HR. Substitution of RNBS-D (CFLY389AAAA) could not be investigated in this assay, as the full-length version of this protein was not stably expressed (data not shown). Surprisingly, all other ARC mutants initiated a CP-dependent HR, although SY378AA and MHDV458AAAA were consistently delayed compared with wild-type Rx (Figure 2A). The same substitutions were introduced into an Rx CC-NB-ARC construct driven from the 35S promoter to determine whether these mutations might have a greater effect in a protein fragment complementation assay. Despite the use of a stronger promoter in this assay, we found that, in addition to DD244AA (K2) and GLP330ALA (GxP), the CFLY389AAAA and MHDV458AAAA substitutions also abrogated the HR, whereas the SY378AA substitution resulted in a delayed HR (Figure 2A).
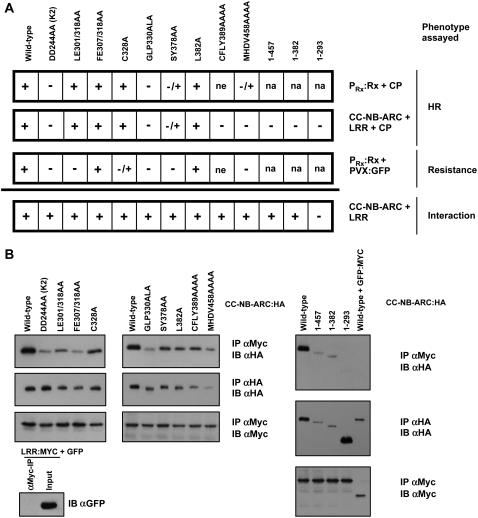
Figure 2.
Analysis of the Rx NB-ARC Domain.
(A) Functional assays of NB-ARC variants. Indicated mutations were incorporated into constructs containing the full-length Rx cDNA driven by the Rx genomic promoter. These variants were expressed via agroinfiltration in N. benthamiana leaves with CP and assessed for HR induction over a period of 3 d. The same variants were incorporated into Rx CC-NB-ARC constructs. These variants were transiently expressed via agroinfiltration in N. benthamiana leaves with CP plus the Rx LRR domain, all regulated by P35S. In this assay, + indicates that an HR was observed within 2 d after infiltration, and – indicates no cell death. –/+ indicates that an HR was observed but was consistently delayed by ∼24 h compared with wild-type Rx. na, not applicable; ne, not expressed (HA-tagged protein was not detectable by protein gel blotting when driven from P35S). The PVX:GFP resistance assay consisted of agroinfiltrating Rx variants with an infectious PVX:GFP clone and monitoring GFP fluorescence. In this assay, + indicates that no GFP fluorescence was observed 5 d after infiltration, and – indicates no restriction of virus accumulation. –/+ indicates a modest accumulation of PVX:GFP. All HRs were CP-dependent, and all infiltrations were performed at least twice.
(B) Coimmunoprecipitation of CC-NB-ARC variants. N. benthamiana leaves were agroinfiltrated with Rx CC-NB-ARC:HA variants and LRR:MYC under P35S regulation. Two days after infiltration, protein extracts were subjected to immunoprecipitation (IP) with anti-HA (αHA) or anti-Myc (αMyc) antibody–conjugated beads and immunoblotted (IB) with the indicated antibody. This experiment was repeated three times with similar results.
Rx is able to condition extreme resistance to PVX in the absence of cell death (Adams et al., 1986). To assess each variant's ability to confer viral resistance, we agroinfiltrated the various PRx:Rx constructs together with an infectious PVX:GFP clone (Peart et al., 2002). Full-length Rx prevents PVX:GFP accumulation, as does coexpression of either CC plus NB-ARC-LRR or CC-NB-ARC plus LRR (see Supplemental Figure 1 online). Interestingly, although the LE301/318AA and C328A substitutions were not obviously compromised in their ability to initiate an HR, they were compromised in their ability to fully suppress PVX accumulation. The inability of LE301/318AA to contain PVX:GFP is a result of the L301A mutation alone, as this substitution was compromised in PVX resistance, whereas E318A was not (data not shown).
To assess whether the mutant CC-NB-ARC fragments were compromised in their ability to bind the Rx LRR, we performed coimmunoprecipitation experiments. We also included in this experiment three C-terminal deletions of CC-NB-ARC terminating at residues 457, 382, and 293, which correspond to deletion of the C terminus up to and including the MHD motif, the RNBS-D motif, and the entire ARC region, respectively. All of the CC-NB-ARC variants, tagged with a hemagglutinin (HA) epitope, were coexpressed in N. benthamiana leaves with 6XMyc-tagged Rx LRR (LRR:MYC). Upon immunoprecipitation with anti-Myc antibodies, we found that 1-457 and 1-382 plus all of the Ala-substituted CC-NB-ARC variants were able to bind the LRR, although the binding of many constructs appears to be somewhat compromised, possibly as a result of the reduced accumulation of these fragments (Figure 2B). The 1-293 protein accumulated to very high levels but did not show appreciable binding to the LRR. These data suggest that residues between amino acids 293 and 382 are critical for interaction with the LRR domain but that this physical interaction is not sufficient to allow Rx to be activated (Figure 2A). Residues 293 to 382 encompass the ARC1 subdomain as annotated previously (Albrecht and Takken, 2006) plus ∼10 amino acids; thus, we conclude that the ARC1 subdomain is necessary for LRR binding.
LRR–NB-ARC Physical Interactions Lack Specificity
Previously, it was demonstrated that the CC-NB-ARC and LRR fragments of Rx and Bs2 were not functionally compatible and that the Bs2 LRR did not bind to the Rx CC-NB-ARC (Moffett et al., 2002). To further investigate this specificity, we tested the ability of the LRR and CC-NB-ARC fragments of Rx, Bs2, and the Arabidopsis thaliana HRT protein (Cooley et al., 2000) to interact with each other. Although in this study the interaction between the Bs2 LRR and Rx CC-NB-ARC was very weak, the other combinations of LRR and CC-NB-ARC fragments were found to coimmunoprecipitate, demonstrating that the interaction specificity between LRR and CC-NB-ARC fragments is not as stringent as suggested by the initial Bs2 LRR–Rx CC-NB-ARC experiments (Figure 3). None of the CC-NB-ARC or LRR fragments showed any binding to GFP (Figure 3). Likewise, the Rx LRR does not bind Rx CC-NB (amino acids 1 to 293) (Figure 2B), RAR1, SGT1, or CP (data not shown), suggesting that the LRR binds specifically to molecules containing an ARC region. However, no heterologous combination of CC-NB-ARC plus LRR resulted in an HR in the presence of any of the Avr determinants recognized by Rx, Bs2, or HRT (Moffett et al., 2002; Leister et al., 2005) (data not shown). Likewise, whereas coexpression of the Rx LRR with Rx CC-NB-ARC containing the autoactivating mutation D460V (Moffett et al., 2002) leads to an HR, none of the heterologous LRR domains were able to do so (data not shown). Thus, physical interaction between the LRR and CC-NB-ARC fragments is not sufficient to reconstitute a functional molecule.
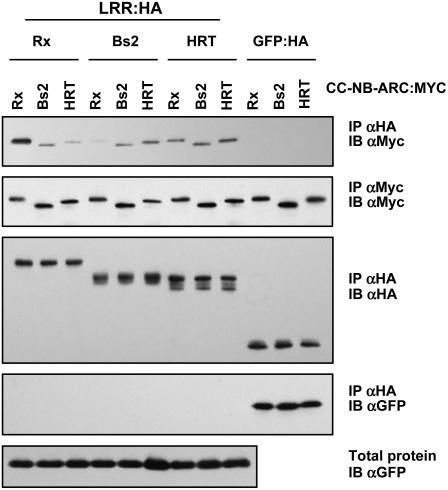
Figure 3.
Interactions between Domains from Different R Proteins.
The indicated LRR:HA or GFP:HA proteins were transiently coexpressed in N. benthamiana leaves via agroinfiltration with the indicated CC-NB-ARC:MYC construct together with GFP, all under the control of P35S. Two days after infiltration, protein extracts were subjected to immunoprecipitation (IP) with either anti-HA (αHA) or anti-Myc (αMyc) antibodies and immunoblotted (IB) with the indicated antibody. This experiment was repeated three times with similar results.
Regions Required for LRR–CC-NB-ARC Compatibility and CP Recognition
Initial experiments investigating recognition specificity showed that certain domain swaps between Rx and GPA2 produced autoactive chimeras. We generated a series of swaps to identify the regions of Rx and GPA2 that are incompatible with each other. An alignment between Rx and GPA2 revealed two regions that appear to be the most variable: the ARC domain and the C-terminal 237 residues (Figure 1). We divided the Rx/GPA2 proteins into five segments spanning Rx residues 1 to 370 (region A), 371 to 472 (B), 473 to 700 (C), 701 to 809 (D), and 810 to 937 (E) (Figures 1 and 4) and generated a number of chimeras combining these segments. Each chimera was transiently expressed in N. benthamiana leaves with or without CP to assess whether the fusion protein was autoactive or could cause a CP-dependent HR (Figure 4).
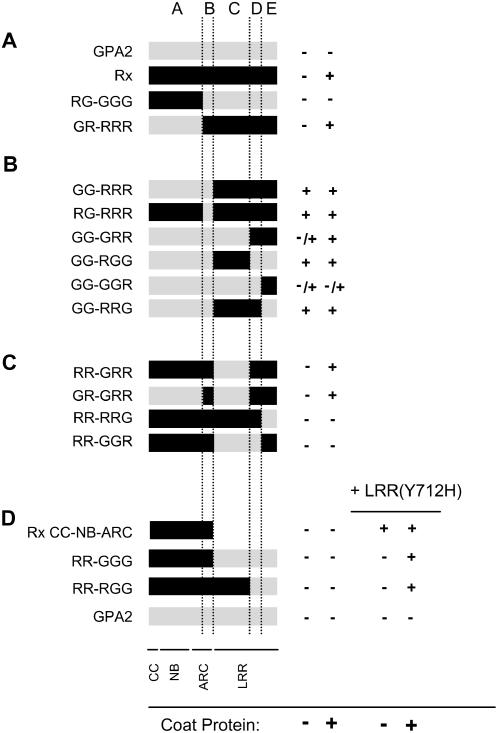
Figure 4.
Rx/GPA2 Chimera Analysis.
Chimeric proteins were generated from regions A to E as illustrated in Figure 1. + indicates that a strong HR was observed 2 d after infiltration, and – indicates no HR. –/+ indicates that an HR was observed but was consistently delayed by ∼24 h compared with wild-type Rx plus CP. Each experiment was repeated at least twice with similar results.
(A) to (C) Fusion proteins were transiently expressed in N. benthamiana leaves via agroinfiltration under P35S regulation with either P35S:GFP or P35S:CP.
(D) In addition to the experiments described above, these constructs were coexpressed with LRR(Y712H) in the presence or absence of CP. Identical results were obtained with wild-type Rx LRR. All constructs were expressed under the regulation of P35S.
We found that exchange of regions A of Rx and GPA2 did not alter the CP-dependent HR phenotype of either molecule (Figure 4A). However, chimeras that derived region B from GPA2 in combination with region C, CD, DE, or E of Rx generated an autoactive phenotype (Figure 4B). This finding demonstrates that at least two separate regions (C and E) of the Rx LRR must be compatible with region B to retain the molecule in an inactive state. All amino acid differences between regions B of Rx and GPA2 are within the region annotated as ARC2 (Albrecht and Takken, 2006). Interestingly, this ARC2-initiated autoactivation was not reciprocal in that Rx ARC2 did not cause autoactivation when paired with the GPA2 LRR (Figure 4D).
Region DE of Rx was sufficient to mediate a CP-dependent HR as long as the appropriate ARC2 subdomain was present to prevent autoactivation (Figure 4C, GR-GRR). At the same time, the HR induced by GG-GRR was greatly enhanced in the presence of CP (Figure 4B), suggesting that the initial recognition event mediated by the C terminus occurs regardless of which ARC2 is present. Attempts to further delimit a minimal recognition region by making exchanges at the D/E junction were unsuccessful. Neither region CD nor region E of Rx was able to confer CP recognition (Figure 4C).
It is possible that some of the chimeric molecules do not function because of a lack of intramolecular interaction between the CC-NB-ARC and LRR fragments of the protein. To test this notion, we used an Rx LRR fragment containing the Y712H mutation. This mutation in the LRR domain causes an autoactive phenotype when present in full-length Rx (Farnham, 2003) as well as in the LRR plus CC-NB-ARC complementation assay (Figure 4D). Full-length Rx is not activated in trans by LRR(Y712H), presumably because the NB-ARC domain is bound by its cis LRR. Coexpression of LRR(Y712H) did not transactivate any of the full-length chimeric molecules (Figure 4D), demonstrating that the LRR–NB-ARC cis interaction is intact. Unexpectedly, LRR(Y712H) or wild-type LRR did result in a CP-dependent HR when coexpressed with a number of full-length molecules (Figure 4D; see Supplemental Figure 2 online). This phenomenon was observed only in chimeras in which the entire CC-NB-ARC was derived from Rx. It is unclear at present why this combination of molecules results in an HR; however, this phenomenon further underlines the importance of compatibility between the LRR and N-terminal sequences. It is prudent to be cautious in evaluating evidence from nonactive chimeras; therefore, we have based the majority of our conclusions on molecules that initiate an HR in at least one assay. All nonautoactivating chimeric molecules were able to complement Rx NB-ARC-LRR for a CP-dependent HR, suggesting that at least the CC domain of these proteins is functional (see Supplemental Figure 2 online).
Disruption of the LRR–ARC Interaction Is Not Required for Signal Initiation
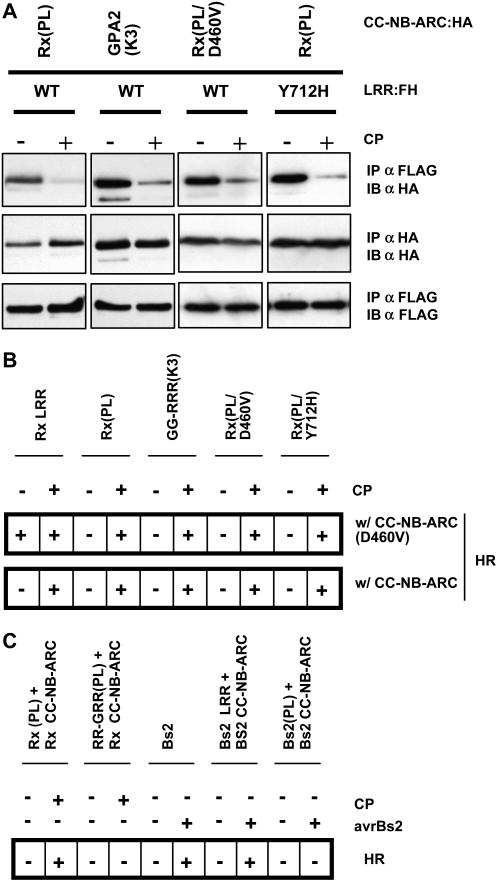
We demonstrated previously that the Rx LRR binds CC-NB-ARC in the absence, but not in the presence, of CP, suggesting that the release of the LRR is an important step in Rx signaling (Moffett et al., 2002). We thus predicted that autoactivating mutations in Rx would compromise the interaction between these fragments. To test this idea, we introduced the autoactivating D460V mutation (Bendahmane et al., 2002) into the CC-NB-ARC:HA construct and the Y712H mutation into an LRR construct with a FLAG epitope and 6×His tags (LRR:FH). We also tested the autoactivating combination of the GPA2 CC-NB-ARC:HA plus the Rx LRR:FH. To prevent an HR, we introduced inactivating mutations into the CC-NB-ARC versions of Rx and GPA2 in the P-loop (PL; GK175AA) and Kinase 3a motifs (K3; L270P), respectively. We found that LRR:FH coimmunoprecipitated CC-NB-ARC:HA regardless of whether or not one of the fragments contained an autoactivating mutation (Figure 5A) and that this interaction was attenuated in the presence of the CP. Although these results indicate that the interaction between LRR and CC-NB-ARC is not affected by the autoactivating mutations, it is possible that a small percentage of these molecules are in fact dissociated. We tested this possibility in a functional assay. The Rx LRR fragment can complement Rx CC-NB-ARC(D460V), whereas the LRR present in Rx(PL) can do so only in the presence of the CP. Consistent with this fact, the LRR of a full-length molecule cannot interact physically with another CC-NB-ARC fragment (Moffett et al., 2002). Thus, if there is a dissociation of some of the LRR domains from full-length autoactive variants, these should be available to complement Rx CC-NB-ARC(D460V). None of the full-length (PL or K3) molecules with autoactivating mutations was able to complement CC-NB-ARC(D460V) in the absence of CP (Figure 5B). These results suggest that the LRR–ARC interaction is strictly preferred in cis, even in molecules possessing autoactivating mutations. Thus, only the presence of CP releases the LRR domains of the full-length molecules from the intramolecular interaction, allowing them to associate in trans with a functional CC-NB-ARC.
Figure 5.
Effect of Autoactivating Mutations on LRR–ARC Interaction Dynamics.
(A) Physical interaction of autoactive variant domains. Wild-type LRR:FH or Rx LRR(Y712H):FH were expressed via agroinfiltration in N. benthamiana leaves with the indicated CC-NB-ARC:HA variants plus either GFP or CP. In combinations that normally would result in an HR, inactivating mutations were introduced into the P-loop (PL: GK175AA) and Kinase 3a motifs (K3: L270P) of Rx and GPA2, respectively. All constructs were regulated by P35S. Two days after infiltration, protein was extracted and subjected to immunoprecipitation (IP) with either anti-FLAG (αFLAG) or anti-HA (αHA) antibody–conjugated beads and immunoblotted (IB) with the indicated antibody. This experiment was repeated three times with similar results.
(B) Functional assays of Rx autoactive variants. The indicated variants were transiently expressed via agroinfiltration in N. benthamiana leaves with either CC-NB-ARC or CC-NB-ARC(D460V) in the presence or absence of CP. + indicates that an HR was observed 2 d after infiltration, and – indicates no HR.
(C) Lack of transactivation by the LRRs in some full-length molecules. Inactivating mutations were introduced into the P-loops of RR-GRR (GK175AA) and Bs2 (GK186AA) to create RR-GRR(PL) and Bs2(PL). The indicated constructs were expressed under the regulation of P35S with or without CP or avrBs2. + indicates that an HR was observed 2 d after infiltration, and – indicates a lack of HR.
Additional evidence that LRR disruption is not required for signal initiation is provided by the RR-GRR construct. The RR-GRR chimera has the ability to confer a CP-dependent HR (Figure 4C), but a P-loop mutant variant of this molecule fails to transactivate CC-NB-ARC (Figure 5C), suggesting that the chimeric LRR is either not displaced by CP or cannot reassociate with another CC-NB-ARC fragment in trans. Unfortunately, these possibilities could not be tested because chimeric LRR domains were not stable when expressed as the LRR fragment (data not shown). The RR-GRR(PL) and GG-GRR(PL) constructs are expressed at a level comparable to Rx(PL), excluding the possibility that its inability to transactivate CC-NB-ARC is a result of less accumulation of RR-GRR(PL) protein (see Supplemental Figure 3 online). Similarly, recent work with Bs2 demonstrated that the LRR–CC-NB-ARC physical interaction is not disrupted in the presence of the avrBs2 elicitor (Leister et al., 2005). Consistent with this finding, a P-loop mutant (GK186AA) of Bs2 is unable to transactivate Bs2 CC-NB-ARC in the presence of avrBs2 (Figure 5C). Bs2(PL) is expressed at a level comparable to Bs2 (see Supplemental Figure 3 online). These results suggest that Rx and Bs2 differ with respect to elicitor-induced disruption of the interaction between ARC and LRR and that this disruption is not required for signal initiation.
Molecular Dissection of the LRR Domain
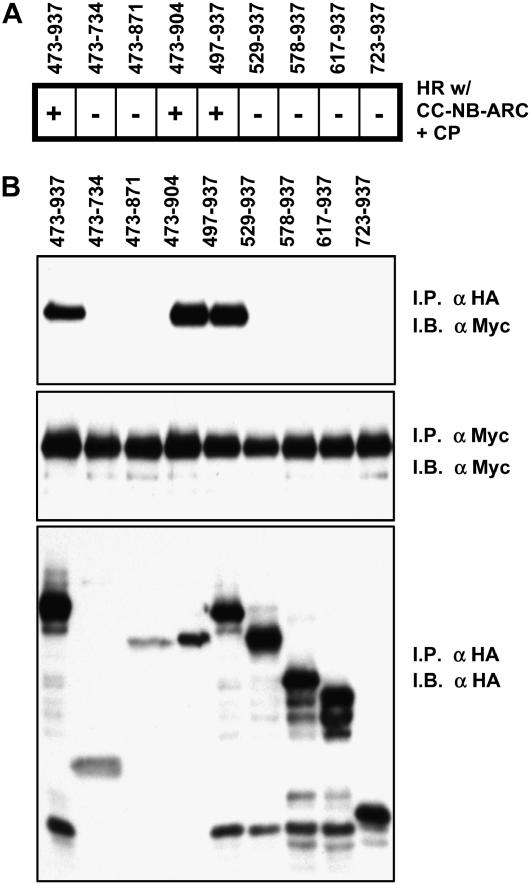
The Rx LRR domain has been shown to bind CC-NB-ARC and to be essential for the HR elicited either by CP or by autoactivating mutations (Moffett et al., 2002). To determine whether we could delimit regions of the LRR domain necessary or sufficient for CC-NB-ARC binding and/or signaling, we generated HA-tagged LRR constructs that were deleted from either the N or C terminus of the domain. C-terminal deletions were generated that terminated at residues 904, 871, and 734, corresponding to removal of the acidic tail, all sequence C terminal to the last predicted LRR, and all sequence C terminal to LRR 11, respectively, as annotated previously (Bendahmane et al., 1999). N-terminal deletions begin at residue 497 (LRR 2), 529 (LRR 4), 578 (LRR 6), 617 (LRR 7), and 723 (LRR 12). Constructs lacking either the acidic tail or the first LRR were functional in complementing CC-NB-ARC (Figure 6A) and were able to coimmunoprecipitate CC-NB-ARC (Figure 6B). However, all other deletions completely abrogated both physical and functional interaction with CC-NB-ARC. These data demonstrate that the LRR functions as a single domain and is not easily dissected into a minimal ARC binding component.
Figure 6.
Deletion Analysis of the Rx LRR Domain.
(A) Functional assays of LRR variants. The indicated Rx LRR:HA fragments were transiently expressed via agroinfiltration in N. benthamiana leaves with Rx CC-NB-ARC and CP, all under P35S regulation. + indicates that an HR was observed 2 d after infiltration, and – indicates no HR.
(B) Coimmunoprecipitation of CC-NB-ARC by LRR variants. N. benthamiana leaves were agroinfiltrated with Rx CC-NB-ARC:MYC and LRR:HA variants under P35S regulation. Protein was extracted 2 d after infiltration, immunoprecipitated (I.P.) with anti-Myc (αMyc) or anti-HA (αHA) antibodies, and immunoblotted (I.B.) with the indicated antibody. These experiments were repeated at least three times with similar results.
DISCUSSION
Molecular Functions of the ARC Region of Homology
Motifs within the ARC region of homology are well conserved among plant NB-LRR proteins as well as other members of the apoptotic-ATPase clade of STAND NTPases, including Apaf-1 and CED-4 (van der Biezen and Jones, 1998; Leipe et al., 2004). Recent modeling based on Apaf-1 suggests that this region actually contains two structural subdomains termed ARC1 and ARC2 (Albrecht and Takken, 2006; McHale et al., 2006). The junction between ARC1 and ARC2 corresponds roughly to the region immediately N terminal to the highly conserved SY motif (or motif 2) (van der Biezen and Jones, 1998) of Rx and thus corresponds well with the two regions that we have defined functionally. We show that the ARC1 subdomain of Rx is necessary for the physical interaction between the N terminus of the protein and the LRR domain (Figure 2B). Because the ARC1 domain is unstable on its own (data not shown), we do not know whether this region is sufficient for LRR binding and thus do not rule out the possibility of additional contacts between the LRR and other regions of the N terminus, particularly given the functional interaction between the LRR and the ARC2 region. However, mutation of the P-loop motif, which is critical for NTP binding (Tameling et al., 2002), does not abrogate LRR binding (Moffett et al., 2002), nor does mutation of the Kinase 2 or Kinase 3a motif (Figures 2B and 5A); thus, NTP binding is not required for this initial interaction. In addition, an ARC-LRR construct is capable of activating CC-NB-ARC(D460V) only in the presence of CP (Moffett et al., 2002), suggesting that the ARC domain is sufficient to sequester the LRR domain in an intramolecular interaction.
For the most part, mutation of even the most conserved residues within the ARC domain appears to have only a quantitative effect on LRR binding (Figure 2B). It is worth noting, however, that although Rx CC-NB-ARC(FE307/318AA) shows lower binding efficiency to the LRR (Figure 2B), this mutant is not compromised in any of the functional assays (Figure 2A). Therefore, we conclude that the binding strength of the interaction between CC-NB-ARC and LRR is not likely responsible for the observed functional attenuation of these Rx mutants. Rather, these mutants appear to be compromised in their ability to become activated. Binding between LRRs and CC-NB-ARCs of different R proteins (Figure 3) suggests that LRR binding is a general property of ARC1 subdomains. Deletion analysis of the Rx LRR revealed that essentially the entire LRR structure is necessary to bind to the Rx CC-NB-ARC fragment (Figure 6B). Thus, the interface between these two domains may consist of multiple contact points, allowing the interaction to be maintained despite the high degree of variability seen in NB-LRR proteins.
The physical binding between the LRR and ARC domains of distantly related R proteins does not result in functional molecules (Moffett et al., 2002; Leister et al., 2005). At the same time, this and other studies (Hwang et al., 2000; Hwang and Williamson, 2003; Howles et al., 2005) illustrate that compatibility between protein domains is often necessary to retain chimeras of closely related R proteins in an inactive state. Combining the GPA2 region B (ARC2 subdomain) with region C, DE, or E of the Rx LRR resulted in a constitutively active molecule. The ARC2 subdomain contains the MHDV motif, and mutation of the highly conserved Asp in this motif in several NB-LRR proteins results in a strong autoactivation phenotype (Bendahmane et al., 2002; de la Fuente van Bentem et al., 2005; Howles et al., 2005). Deletion of the last 15 amino acids of the CC-NB-ARC, up to and including the MHDV motif, results in an inactive molecule, and Ala substitution of this motif compromises Rx function (Figure 2A), illustrating the requirement of this motif for activation. Furthermore, structural studies with Apaf-1 suggest that the MHDV motif acts as a sensor domain, in which the highly conserved His interacts directly with the bound nucleotide along with Ser-422, which appears to correspond to Rx Ser-440 (Riedl et al., 2005). The ARC2 region also contains the RNBS-D motif, in which inactivating (Figure 2A) and autoactivating mutations have also been identified (Bendahmane et al., 2002). Thus, given its requirement for activation and autoinhibition of Rx, we suggest that the ARC2 subdomain acts as a switch that relays recognition events into changes in the NB-ARC domain that result in activation of the protein.
Domain-swap experiments with Mi1.2 and Mi1.1 demonstrated autoactivating incompatibilities between the LRRs of Mi1.2 and a region of 751 residues encompassing the entire NB-ARC domain (Hwang et al., 2000). This allows for the possibility that it is the Mi1.1 ARC2 subdomain that conditions inappropriate signaling in the presence of the Mi1.2 LRRs. An apparent autoactivating phenotype is also correlated with LRR exchange between homologs at the maize (Zea mays) Rp1 locus (Sun et al., 2001). The authors of that study note that RP1 homologs possess a hypervariable region immediately preceding the MHDV motif that can also be seen in the Rx/Rx2/GPA2 family and the RPP8/HRT/RCY-1 locus (Bendahmane et al., 2000; Takahashi et al., 2002).
In the crystal structure of Apaf-1, the P-loop, Kinase 2, Kinase 3a, and GxP motifs are all located in the same ADP binding pocket (Riedl et al., 2005), and mutations in any of these motifs caused complete loss of function of Rx (or GPA2 in the case of Kinase 3a). Surprisingly, with the exception of the RNBS-D substitution, mutation of other highly conserved residues within the ARC domain of full-length Rx did not fully compromise the CP-dependent HR (Figure 2A). However, each of the mutants tested, except FE307/318AA and L382A, was compromised in at least one assay, indicating that these residues have at least a quantitative effect on the ability of Rx to translate a recognition event into a signal initiation event.
Some mutants, such as LE301/318AA and C328A, were able elicit a CP-dependent HR indistinguishable from wild-type Rx but were compromised in conferring extreme resistance to PVX:GFP in a transient assay (Figure 2A; see Supplemental Figure 1A online). At the same time, although the CP-mediated HR conditioned by either CC plus NB-ARC-LRR or CC-NB-ARC plus LRR fragments (Moffett et al., 2002) is slightly delayed compared with full-length Rx, the outcomes in the transient PVX:GFP resistance assay are the same (see Supplemental Figure 2 online). Thus, although a protein may be competent to elicit a rapid HR, it is not necessarily able to mediate effective viral resistance.
Role of CP-Mediated Disruption of the LRR–ARC Interaction
We have shown that binding of CC-NB-ARC by the LRR is not affected by autoactivating mutations, nor is the CP-dependent disruption of this binding (Figure 5). These data suggest that the disruption of LRR binding to CC-NB-ARC is not required for signaling but likely occurs coincident with, or subsequent to, CP recognition and signal initiation. If this activity is not required for activation, what role might it play in Rx function? The assay presented in Figure 5B suggests one possibility. The LRR present in full-length Rx(PL) can complement CC-NB-ARC when it is activated by CP but not when activated by autoactivating mutations. We suggest that, in this assay, the Rx LRR dissociates from its cognate ARC domain in the presence of CP and can subsequently associate with another ARC domain in trans followed by another recognition event that activates signaling through the wild-type CC-NB-ARC fragment. If this scenario can occur in trans, then it should also be possible, and even preferred, in cis. As such, CP recognition would cause activation of a wild-type Rx molecule; the LRR would dissociate, reassociate, and undergo a further round of recognition and activation. Therefore, although disruption of the LRR–ARC interaction may not be required for the initial activation of the protein, it may provide a mechanistic advantage in that multiple rounds of recognition and activation would allow for amplification of the resistance response. We suggest that the autoactivating mutations (or domain swaps) are sufficient to induce a signal initiation event but not for resetting the protein for multiple rounds of recognition. That the dissociation of the LRR from the NB-ARC plays a role other than signal initiation is supported by data showing that the Bs2 LRR–CC-NB-ARC interaction is not disrupted by its elicitor, avrBs2 (Leister et al., 2005) (Figure 5C). Furthermore, the RR-GRR chimera is able to initiate a CP-dependent HR, but its P-loop variant is unable to complement Rx CC-NB-ARC (Figure 5C). It is also interesting that whereas the CP-mediated HR responses of wild-type Rx and RR-GRR do not differ, RR-GRR cannot mediate extreme resistance in a transient virus resistance assay (see Supplemental Figure 2 online). Signal amplification through an iterative mechanism might explain the ability of some R genes, including Rx, to mediate extreme resistance to viruses (Adams et al., 1986; Kang et al., 2005). Given the differences observed between Bs2, Rx, and Rx/GPA2 chimeras, this characteristic may vary among R proteins, attributable either to intrinsic differences in the R proteins themselves or to differences in the strength of Avr recognition.
Our results do not directly address the dynamics of ATP binding or hydrolysis. However, as the CP-mediated disruption of the interaction between ARC and LRR takes place in the presence of a P-loop mutation (Moffett et al., 2002), the recognition might occur independent of nucleotide binding and downstream signaling. We propose that the cycle of disruption and reassociation would allow the protein to return to its initial state after a signal initiation event and undergo further rounds of recognition. This, in turn, would allow for multiple rounds of activation and the associated events that take place within the nucleotide binding pocket. The bacterially produced NB-ARC domains of I-2 and Mi-1 have been shown to hydrolyze ATP in vitro (Tameling et al., 2002). The proteins used in the latter study lacked LRR domains, and given the importance of the LRR domain for activity as well as recognition (Figure 5B), it is unclear whether the full-length molecules in planta would also undergo constitutive ATP hydrolysis. Importantly, however, subsequent studies demonstrated that upon ATP hydrolysis, the dissociation of bound ADP from the I-2 NB-ARC was extremely slow (Tameling et al., 2006), suggesting that other factors may be required for nucleotide exchange and further rounds of hydrolysis. Thus, it is conceivable that a single round of nucleotide hydrolysis and/or exchange is sufficient to initiate signaling events leading to an HR. However, multiple rounds of activation and signaling may require multiple recognition events.
The Rx C Terminus Conditions Recognition Specificity
Sequence swaps between Rx and GPA2 also allowed us to identify the region of Rx that determines recognition specificity. Because much of the variation between the Rx and GPA2 proteins is found in the ARC domain and in the LRR C terminus, we hypothesized that the recognition determinants would lie in one of these two regions. Accordingly, constructs containing Rx region DE (amino acids 701 to 937) condition CP-dependent HR similar to Rx. Region B (ARC2) did not appear to contribute to CP recognition but was required to avoid autoactivation in the presence of Rx LRR sequences (Figure 4C). Similarly, sequence-swap experiments have shown that recognition specificity of MLA1 and MLA6 lies within regions containing the C-terminal half of their LRR domains plus a C-terminal extension (Shen et al., 2003).
Implications for Recognition and Activation
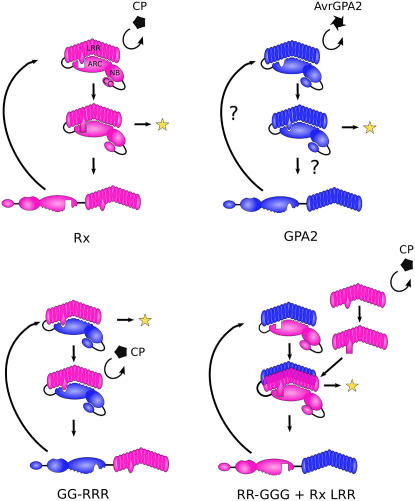
It is tempting to speculate that the many autoactivating point mutations found in NB-LRR proteins disrupt negative regulatory regions of the protein. However, in domain-swap experiments, autoactivation occurs when wild-type domains are placed in an inappropriate context. Furthermore, autoactivating fusions can be obtained by exchanging different, nonoverlapping Rx LRR sequences for the equivalent region of GPA2. Autoactivating mutations found in human NOD-LRR proteins are numerous and map throughout the proteins in both conserved and nonconserved residues (Tanabe et al., 2004; Ting and Davis, 2005; Rairdan and Moffett, 2006). We propose that NB-LRR proteins are normally present in the cell in a conformation that is autoinhibited from progressing to an active state by virtue of a perfect fit between the ARC and LRR domains but that this inactive state is a hair trigger that is highly sensitive to perturbation. At least one mechanism to overcome this inactive state is to alter the interface between the ARC and LRR domains through an elicitor-induced conformational change of the LRR. Autoactive chimeras might mimic this process if the resulting interaction between the ARC and LRR domains no longer forms a perfect fit and the resulting interface lacks the structural constraint to prevent the molecule from progressing to its active conformation. The ARC2 subdomain appears to be involved in both activation and autoinhibition. Thus, we envisage a mechanism whereby elicitor recognition by the LRR alters the interface between the LRR and ARC2 domains and repositions critical residues such that the NTP binding and/or hydrolysis activity of the protein is altered, leading to signal initiation, as has been suggested previously (Moffett et al., 2002; Tameling et al., 2006). Such a model is presented in Figure 7.
Figure 7.
Perturbed LRR Model.
In this model, the avirulence determinant alters the conformation of, or perturbs, the LRR in a direct or indirect manner. This alteration allows the protein to overcome the autoinhibition conditioned by the ARC2 subdomain and activate downstream signaling (represented as a yellow star). Disruption of the LRR–ARC interaction would proceed simultaneously or subsequent to this activation, allowing the domains of the molecule to reassociate and undergo further rounds of recognition and signaling. In such a model, the combination of the GPA2 ARC2 and Rx LRR domains would lack the structural constraints required to retain the molecule in the autoinhibited state, and the protein would proceed directly to the activated form. However, some additional conformational change induced by CP recognition is required to trigger the dissociation of the Rx LRR from the GPA2 ARC for the process to reiterate.
Given that activation can occur either by recognition or by multiple amino acid substitutions, we suggest that elicitor recognition results in a perturbed LRR domain (Figure 7). This perturbed LRR does not necessarily represent a specific molecular conformation but one that has either overcome or is not subjected to structural restraints, allowing the protein to progress to an activated state. The ease of obtaining autoactivation also predicts that perturbation leading to activation could occur at many different sites within the protein, even allowing a given R protein to recognize multiple Avr elicitors via different interactions. Although this report does not directly address the initial recognition event, the mechanisms of activation we suggest are compatible with models of recognition that predict either direct interaction between the Avr and the NB-LRR proteins (Jia et al., 2000; Deslandes et al., 2003; Dodds et al., 2006) or indirect recognition events (Mackey et al., 2002, 2003; Axtell and Staskawicz, 2003; Shao et al., 2003).
METHODS
Plasmid Construction
The pBIN61-based vectors with HA, FLAG plus 6×His (FH), and six c-Myc (MYC) epitope tags, as well as mutations in the Rx P-loop (GK175AA), Kinase 2 (DD244AA), and GxP (GLPL330ALAL) motifs, Rx 1-382, Rx 1-293, Rx LRR, Rx CC-NB-ARC (previously designated CC-NBS), Bs2 LRR, Bs2 CC-NB-ARC, and Rx CC-NB-ARC(D460V) have been described (Bendahmane et al., 2002; Moffett et al., 2002). The Rx 1-452 deletion construct was generated using the same strategy as for the other deletion constructs (Bendahmane et al., 2002). All site-directed mutants or Rx/GPA2 fusions described here were generated with extension-overlap PCR (Vallejo et al., 2003) using KOD high-fidelity thermostable polymerase according to the manufacturer's instructions (Novagen). All site-directed mutants had the same start and termination sites as the wild-type constructs. The GPA2 CC-NB-ARC was generated using the same primers as the Rx CC-NB-ARC construct (Bendahmane et al., 2002), and the K3 mutation was identified as a PCR error resulting in the mutation L270P. N-terminal LRR deletions incorporated a start codon before the first LRR residue described in each deletion in Figure 6. C-terminal deletions incorporated a BamHI site after each final residue described in Figure 6. The HRT CC-NB-ARC construct includes residues 1 to 513, whereas the HRT LRR construct consists of residues 508 to 909 and was constructed using the same PCR-based strategy as for Rx deletions (Bendahmane et al., 2002). The GPA2 sequence used in the Rx-GPA2 swap experiments was amplified by PCR from a BAC containing the GPA2 genomic locus (kindly donated by A. Goverse). All constructs generated by PCR were verified by sequencing. The autoactivating Y712H mutation was generated during a screen wherein Rx was subjected to PCR random mutagenesis followed by transient agroinfiltration-mediated expression in Nicotiana benthamiana (Farnham, 2003).
Agrobacterium-Mediated Transient Expression (Agroinfiltration)
Binary vectors were transformed into Agrobacterium tumefaciens strain C58C1 carrying the virulence plasmid pCH32. Agroinfiltration was performed as described (Bendahmane et al., 2000) at OD600 = 0.2. HR phenotypes generally presented at 24 to 48 h. The PVX:GFP resistance assay consisted of agroinfiltrating Rx variants at OD600 = 0.2 with Agrobacterium GV3101 carrying the plasmid pGr208, which expresses the PVX:GFP cDNA (Peart et al., 2002) at OD600 = 0.001. GFP fluorescence was monitored 5 d later using a handheld UV lamp.
Immunoprecipitation and Immunoblotting
Protein extracts from agroinfiltrated N. benthamiana leaves were prepared by grinding 1 g of leaf tissue in 2.5 mL of extraction buffer (25 mM Tris-HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 10% glycerol, and 5 mM DTT) in the presence of plant protease inhibitor cocktail (Sigma-Aldrich) and 2% polyvinylpolypyrrolidone. Extracts were spun at 12,000g at 4°C for 15 min, and 1 mL of extract was subsequently passed through a 5-mL Sephadex G-25 spin column (preequilibrated with extraction buffer). Immunoprecipitation was performed as follows: 400 to 800 μL of extract in a total volume of 1400 μL of extraction buffer plus 0.15% Nonidet P-40 (immunoprecipitation buffer) was precleared with 50 μL of IgG agarose (Sigma-Aldrich) at 4°C for 30 min. Extracts were spun for 2 min at 12,000g, and the supernatant was added to 25 μL of anti-HA conjugated agarose beads from either Roche (3F10) or Sigma-Aldrich (HA-7), anti-FLAG (M2; Sigma-Aldrich) agarose beads, or anti-Myc (9E10; Sigma-Aldrich) agarose beads. Extracts were incubated end over end at 4°C for 1 h and washed four times with immunoprecipitation buffer, and the pellet was resuspended in 1× SDS-PAGE loading buffer. Immunoprecipitated samples were separated by SDS-PAGE and blotted to Immun-Blot polyvinylidene difluoride membranes (Bio-Rad). Blots were preblocked with 5% skim milk powder in Tris-buffered saline plus Tween 20 and probed with 40 to 200 ng/mL antibody in Tris-buffered saline plus Tween 20. HA epitope tags were detected with 3F10 (Roche), and Myc tags were detected with either 9E10 or A-14 (Santa Cruz Biotechnology), followed by washing and incubation with an appropriate horseradish peroxidase–conjugated secondary antibody. FLAG epitope tags were detected with horseradish peroxidase–conjugated M2 antibody (Sigma-Aldrich). Proteins were visualized with ECL-Plus (Amersham).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: Rx, AJ011801; Gpa2, AJ249449; PVX-CP, AF172259; Bs2, AF202179; and HRT, AF234174.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. PVX:GFP Resistance Assay with Rx Variants.
Supplemental Figure 2. Comprehensive List of Results from Functional Assays Performed with Rx/GPA2 Chimeras Generated during the Course of These Experiments.
Supplemental Figure 3. Stable Expression of Nontransactivating Proteins.
Supplementary Material
Acknowledgments
We thank Melanie Sacco, Greg Martin, and David Baulcombe for critical review of the manuscript and the Boyce Thompson Institute greenhouse staff for plant care. We are grateful to Garry Farnham for the Rx(Y712H) mutant, to Dan Klessig for the HRT cDNA, and to Diana Taft for technical assistance. Funding for this work was provided by the National Science Foundation (Grant IOB-0343327) and The Triad Foundation. We are also grateful for financial support during preliminary stages of this work from the Biotechnology and Biological Sciences Research Council and the Sainsbury Laboratory (supported by the Gatsby Charitable Foundation).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Peter Moffett (pm99@cornell.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.106.042747.
References
- Adams, S.E., Jones, R.A.C., and Coutts, R.H.A. (1986). Expression of potato virus X resistance gene Rx in potato leaf protoplasts. J. Gen. Virol. 67 2341–2345. [Google Scholar]
- Albrecht, M., and Takken, F.L. (2006). Update on the domain architectures of NLRs and R proteins. Biochem. Biophys. Res. Commun. 339 459–462. [DOI] [PubMed] [Google Scholar]
- Axtell, M.J., and Staskawicz, B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112 369–377. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A., Farnham, G., Moffett, P., and Baulcombe, D.C. (2002). Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 32 195–204. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A., Kanyuka, K., and Baulcombe, D.C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane, A., Kohn, B.A., Dedi, C., and Baulcombe, D.C. (1995). The coat protein of potato virus X is a strain-specific elicitor of Rx1-mediated virus resistance in potato. Plant J. 8 933–941. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A., Querci, M., Kanyuka, K., and Baulcombe, D.C. (2000). Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: Application to the Rx2 locus in potato. Plant J. 21 73–81. [DOI] [PubMed] [Google Scholar]
- Cannon, S.B., Zhu, H., Baumgarten, A.M., Spangler, R., May, G., Cook, D.R., and Young, N.D. (2002). Diversity, distribution, and ancient taxonomic relationships within the TIR and non-TIR NBS-LRR resistance gene subfamilies. J. Mol. Evol. 54 548–562. [DOI] [PubMed] [Google Scholar]
- Cooley, M.B., Pathirana, S., Wu, H.J., Kachroo, P., and Klessig, D.F. (2000). Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell 12 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L., Dietrich, R.A., and Richberg, M.H. (1996). Death don't have no mercy: Cell death programs in plant-microbe interactions. Plant Cell 8 1793–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente van Bentem, S., Vossen, J.H., de Vries, K.J., van Wees, S., Tameling, W.I., Dekker, H.L., de Koster, C.G., Haring, M.A., Takken, F.L., and Cornelissen, B.J. (2005). Heat shock protein 90 and its co-chaperone protein phosphatase 5 interact with distinct regions of the tomato I-2 disease resistance protein. Plant J. 43 284–298. [DOI] [PubMed] [Google Scholar]
- Deslandes, L., Olivier, J., Peeters, N., Feng, D.X., Khounlotham, M., Boucher, C., Somssich, I., Genin, S., and Marco, Y. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA 100 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., Catanzariti, A.M., Teh, T., Wang, C.I., Ayliffe, M.A., Kobe, B., and Ellis, J.G. (2006). Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 103 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnham, G. (2003). A Structure-Function Analysis of the Plant Disease Resistance Protein Rx. PhD dissertation (Norwich, UK: University of East Anglia).
- Flor, H.H. (1971). Current status of gene-for-gene concept. Annu. Rev. Phytopathol. 9 275–296. [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D. (1996). Resistance gene-dependent plant defense responses. Plant Cell 8 1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howles, P., Lawrence, G., Finnegan, J., McFadden, H., Ayliffe, M., Dodds, P., and Ellis, J. (2005). Autoactive alleles of the flax L6 rust resistance gene induce non-race-specific rust resistance associated with the hypersensitive response. Mol. Plant Microbe Interact. 18 570–582. [DOI] [PubMed] [Google Scholar]
- Hwang, C.F., Bhakta, A.V., Truesdell, G.M., Pudlo, W.M., and Williamson, V.M. (2000). Evidence for a role of the N terminus and leucine-rich repeat region of the Mi gene product in regulation of localized cell death. Plant Cell 12 1319–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, C.F., and Williamson, V.M. (2003). Leucine-rich repeat-mediated intramolecular interactions in nematode recognition and cell death signaling by the tomato resistance protein Mi. Plant J. 34 585–593. [DOI] [PubMed] [Google Scholar]
- Jia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, B.C., Yeam, I., and Jahn, M.M. (2005). Genetics of plant virus resistance. Annu. Rev. Phytopathol. 43 581–621. [DOI] [PubMed] [Google Scholar]
- Keen, N.T. (1990). Gene-for-gene complementarity in plant-pathogen interactions. Annu. Rev. Genet. 24 447–463. [DOI] [PubMed] [Google Scholar]
- Leipe, D.D., Koonin, E.V., and Aravind, L. (2004). STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: Multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol. 343 1–28. [DOI] [PubMed] [Google Scholar]
- Leister, R.T., Dahlbeck, D., Day, B., Li, Y., Chesnokova, O., and Staskawicz, B.J. (2005). Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell 17 1268–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Clarke, J.D., Zhang, Y., and Dong, X. (2001). Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol. Plant Microbe Interact. 14 1131–1139. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Belkhadir, Y., Alonso, J.M., Ecker, J.R., and Dangl, J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112 379–389. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Holt, B.F., Wiig, A., and Dangl, J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108 743–754. [DOI] [PubMed] [Google Scholar]
- McHale, L., Tan, X., Koehl, P., and Michelmore, R.W. (2006). Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 7 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Chin, D.B., Shen, K.A., Sivaramakrishnan, S., Lavelle, D.O., Zhang, Z., and Michelmore, R.W. (1998). The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell 10 1817–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Dickerman, A.W., Michelmore, R.W., Sivaramakrishnan, S., Sobral, B.W., and Young, N.D. (1999). Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 20 317–332. [DOI] [PubMed] [Google Scholar]
- Moffett, P., Farnham, G., Peart, J., and Baulcombe, D.C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21 4511–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragon-Palomino, M., Meyers, B.C., Michelmore, R.W., and Gaut, B.S. (2002). Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res. 12 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel, L., Moores, T.L., van der Biezen, E.A., Parniske, M., Daniels, M.J., Parker, J.E., and Jones, J.D. (1999). Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11 2099–2112. [PMC free article] [PubMed] [Google Scholar]
- Noutoshi, Y., Ito, T., Seki, M., Nakashita, H., Yoshida, S., Marco, Y., Shirasu, K., and Shinozaki, K. (2005). A single amino acid insertion in the WRKY domain of the Arabidopsis TIR-NBS-LRR-WRKY-type disease resistance protein SLH1 (sensitive to low humidity 1) causes activation of defense responses and hypersensitive cell death. Plant J. 43 873–888. [DOI] [PubMed] [Google Scholar]
- Peart, J.R., Cook, G., Feys, B.J., Parker, J.E., and Baulcombe, D.C. (2002). An EDS1 orthologue is required for N-mediated resistance against tobacco mosaic virus. Plant J. 29 569–579. [DOI] [PubMed] [Google Scholar]
- Rairdan, G., and Moffett, P. (2006). Brothers in arms? Common and contrasting themes in pathogen perception by plant NBARC-LRR and animal CLR proteins. Microbes Infect., in press. [DOI] [PubMed]
- Riedl, S.J., Li, W., Chao, Y., Schwarzenbacher, R., and Shi, Y. (2005). Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature 434 926–933. [DOI] [PubMed] [Google Scholar]
- Shao, F., Golstein, C., Ade, J., Stoutemyer, M., Dixon, J.E., and Innes, R.W. (2003). Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301 1230–1233. [DOI] [PubMed] [Google Scholar]
- Shen, Q.H., Zhou, F., Bieri, S., Haizel, T., Shirasu, K., and Schulze-Lefert, P. (2003). Recognition specificity and RAR1/SGT1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. Plant Cell 15 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirano, Y., Kachroo, P., Shah, J., and Klessig, D.F. (2002). A gain-of-function mutation in an Arabidopsis Toll Interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Q., Collins, N.C., Ayliffe, M., Smith, S.M., Drake, J., Pryor, T., and Hulbert, S.H. (2001). Recombination between paralogues at the Rp1 rust resistance locus in maize. Genetics 158 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai, T.H., Dahlbeck, D., Clark, E.T., Gajiwala, P., Pasion, R., Whalen, M.C., Stall, R.E., and Staskawicz, B.J. (1999). Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl. Acad. Sci. USA 96 14153–14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, H., Miller, J., Nozaki, Y., Takeda, M., Shah, J., Hase, S., Ikegami, M., Ehara, Y., and Dinesh-Kumar, S.P. (2002). RCY1, an Arabidopsis thaliana RPP8/HRT family resistance gene, conferring resistance to cucumber mosaic virus requires salicylic acid, ethylene and a novel signal transduction mechanism. Plant J. 32 655–667. [DOI] [PubMed] [Google Scholar]
- Tameling, W.I., Elzinga, S.D., Darmin, P.S., Vossen, J.H., Takken, F.L., Haring, M.A., and Cornelissen, B.J. (2002). The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell 14 2929–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling, W.I., Vossen, J.H., Albrecht, M., Lengauer, T., Berden, J.A., Haring, M.A., Cornelissen, B.J., and Takken, F.L. (2006). Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 140 1233–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe, T., et al. (2004). Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J. 23 1587–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting, J.P., and Davis, B.K. (2005). CATERPILLER: A novel gene family important in immunity, cell death, and diseases. Annu. Rev. Immunol. 23 387–414. [DOI] [PubMed] [Google Scholar]
- Vallejo, A.N., Pogulis, R.J., and Pease, L.R. (2003). Mutagenesis and synthesis of novel recombinant genes using PCR. In PCR Primer: A Laboratory Manual, C.W. Dieffenbach and G.S. Dveksler, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 467–474.
- van der Biezen, E.A., and Jones, J.D. (1998). The NB-ARC domain: A novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 8 R226–R227. [DOI] [PubMed] [Google Scholar]
- van der Vossen, E.A., van der Voort, J.N., Kanyuka, K., Bendahmane, A., Sandbrink, H., Baulcombe, D.C., Bakker, J., Stiekema, W.J., and Klein-Lankhorst, R.M. (2000). Homologues of a single resistance-gene cluster in potato confer resistance to distinct pathogens: A virus and a nematode. Plant J. 23 567–576. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.