Abstract
We investigated the role of the oilbody proteins in developing and germinating Arabidopsis thaliana seeds. Seed oilbodies are simple organelles comprising a matrix of triacylglycerol surrounded by a phospholipid monolayer embedded and covered with unique proteins called oleosins. Indirect observations have suggested that oleosins maintain oilbodies as small single units preventing their coalescence during seed desiccation. To understand the role of oleosins during seed development or germination, we created lines of Arabidopsis in which a major oleosin is ablated or severely attenuated. This was achieved using RNA interference techniques and through the use of a T-DNA insertional event, which appears to interrupt the major (18 kD) seed oleosin gene of Arabidopsis and results in ablation of expression. Oleosin suppression resulted in an aberrant phenotype of embryo cells that contain unusually large oilbodies that are not normally observed in seeds. Changes in the size of oilbodies caused disruption of storage organelles, altering accumulation of lipids and proteins and causing delay in germination. The aberrant phenotypes were reversed by reintroducing a recombinant oleosin. Based on this direct evidence, we have shown that oleosins are important proteins in seed tissue for controlling oilbody structure and lipid accumulation.
INTRODUCTION
Oilseeds accumulate lipids to supply the energy requirements for the growth of the seedling after germination. Such lipids are generally stored as triacylglycerols (TAGs) in spherical compartments referred to as spherosomes (Frey-Wyssling et al., 1963), oleosomes (Murphy, 1990), or most frequently, oilbodies. These organelles arise from the endoplasmic reticulum (ER), which is responsible for the synthesis of TAG (Murphy, 1993; Murphy and Vance, 1999; Hsieh and Huang, 2004). In exalbuminous oilseeds, such as Brassica spp, oilbodies are found in cotyledons and the embryonic axis. In castor bean (Ricinus communis) and other albuminous oilseeds, oilbodies can be found in the endosperm (Anil et al., 2003). In cereals and other monocotyledonous species, oilbodies are present in the scutellum (Aalen et al., 1994). Pollen grains, the tapetum, and oleaginous fruits also store TAGs in similar organelles (Evans et al., 1992; Ross et al., 1993). During final stages of seed maturation, when the water potential decreases, oilbodies experience cytoplasmic compression and are forced into contact with each other. Surprisingly, these organelles resist coalescence and remain as small individual units (Murphy, 1990). Comparison of different species shows that oilbodies have variable diameters with a narrow range between 0.5 and 2.0 μm (Tzen et al., 1993). It has been tacitly assumed that seeds maintain oilbodies as small individual units to provide a high surface-to-volume ratio that would facilitate access by lipases during germination. In the mesocarp of oleaginous fruits, which do not undergo dehydration and are not destined to act as the plants energy supply, the diameter of oilbodies can be as large as 20 μm (Platt-Aloia and Thompson, 1981).
Chemical and ultrastructural analysis reveals that seed oilbodies are surrounded by a phospholipid monolayer where the aliphatic chains are oriented to the triglyceride lumen and the phosphate groups toward the cytoplasm (Yatsu and Jacks, 1972; Jacks et al., 1990). Chemical analysis also reveals that 1 to 4% by weight of seed oilbodies is composed of proteins (Huang, 1992; Tzen and Huang, 1992). Oleosins are the major proteins associated with oilbodies and are usually present as two or more isoforms. These isoforms are commonly classified simply as high and low molecular weight forms (Tzen et al., 1990). Oleosins are found ubiquitously in seeds and form a family with similar structural properties that include a long hydrophobic core organized around a unique 12–amino acid motif called a proline knot (Abell et al., 1997). This hydrophobic domain is flanked by hydrophilic or amphipathic N and C termini (Moloney, 1999). A high degree of similarity is located in but not restricted to the hydrophobic domain and proline knot motif, both of which are essential for the correct targeting to oilbodies (van Rooijen and Moloney, 1995b; Abell et al., 1997). Protease protection assays revealed that oleosins are anchored in oilbodies by the hydrophobic domain, exposing the hydrophilic N- and C-terminal ends to the cytoplasm (Tzen et al., 1992). The expression of oleosins is regulated by the transactivator ABI3 (Crowe et al., 2000) and is restricted to certain tissues at specific developmental stages (Keddie et al., 1994).
Several experiments with oilbodies have led to predictions on the function of oleosins. In vitro experiments, using isolated oilbodies, suggested that the oleosin coat promotes steric hindrance and electrical repulsion between these organelles, preventing them from coalescing (Tzen et al., 1992; Tzen and Huang, 1992). Correlations between oilbody size and oleosin levels suggest that accumulation of oleosins might modulate the size of oilbodies in vivo (Ross et al., 1993; Tzen et al., 1993; Ting et al., 1996), although until now this has never been experimentally tested. Species containing higher amounts of oleosin (e.g., canola [Brassica napus]) have smaller oilbodies compared with those with lower oleosin content (e.g., sesame [Sesamum indicum]) (Tzen et al., 1993). Similar results were described in the inbred maize (Zea mays) lines Illinois High Oil and Illinois Low Oil. The Illinois High Oil line has a higher TAG-to-oleosin ratio and has larger oilbodies compared with the Illinois Low Oil line, which has a lower TAG-to-oleosin ratio (Ting et al., 1996). Moreover, studies in mesocarp from avocado (Persea americana) and olive (Olea europaea) fruits revealed that oleosins were not present in fruit oilbodies (Ross et al., 1993). Typically, oleaginous fruit cells contain larger oilbodies than seeds. These comparisons led researchers to postulate that higher TAG-to-oleosin ratios result in larger oilbodies. However, these observations are circumstantial since comparisons were always made between different genotypes, different stages of development, or different tissues and organs.
We have investigated this question by directly manipulating oleosin expression levels. We present unequivocal evidence that the accumulation of oleosins modulate the size of oilbodies. Using reverse genetics methods (Bird et al., 1991), we suppressed the production of the major oleosin in Arabidopsis thaliana (OLEO1). OLEO1 suppression resulted in a dramatic phenotype in which the seed cells contained very large oilbodies. This phenotype could be reversed by overexpressing a recombinant oleosin in the OLEO1 suppressed line. We also show that beyond this clear phenotype, there are several ancillary consequences of this morphological variation on germination, mobilization of TAGs, and accumulation of lipids and proteins in the seed.
RESULTS
Arabidopsis contains four seed oleosin isoforms (Richmond et al., 1997; Jolivet et al., 2004). The 18-kD isoform, also called OLEO1 (At4g25140), accumulates at higher levels and accounts for ∼65% of oilbody-associated proteins. OLEO2 and OLEO4 (At5g40420 and At3g27660, respectively) are higher molecular mass isoforms that together represent ∼30% of oilbody proteins (Figure 1C). The other proteins are represented by the isoform OLEO3 with lower molecular mass (At5g51210) and by other oilbody-associated proteins, such as caleosin or steroleosin (Chen et al., 1999; Lin et al., 2002). These proteins can be detected only in overloaded gels visualized with Coomassie blue or silver staining.
Figure 1.
Suppression of Oleosins in Arabidopsis.
(A) Scale diagram of the constructs used to suppress OLEO1 using RNAi methods. The Antisense, Hairpin, and Loop cassettes are shown.
(B) Scale diagram of the insertion of a T-DNA element into OLEO1 and OLEO2 genes (lines KnockOLEO1 and KnockOLEO2, respectively). Each gene has two exons (thick line) and one intron (thin line). Both T-DNA insertions are located in the middle of the second exon.
(C) SDS-PAGE profile of oilbody-associated proteins in different plants. The first lane (L) contains the protein ladder (Benchmark; Invitrogen). The second lane contains the oilbody-associated proteins from wild-type (C24) plants. Lanes numbered 1 to 3 contain the oilbody-associated proteins from SupOLEO1-Loop, SupOLEO1-Hairpin, and SupOLEO1-Antisense plants, respectively. Lanes 4 and 5 contain the oilbody-associated proteins from KnockOLEO1 and KnockOLEO2 lines, respectively. The positions of the three most abundant oleosin isoforms are indicated by arrows.
To determine the biological function of oleosins, we used RNA interference (RNAi) methods to suppress the expression of OLEO1, since it encodes for the most abundant oleosin. Three different constructs were produced: an Antisense, an inverted repeat of the entire cDNA (Hairpin), and an inverted repeat containing the unique intron of OLEO1 between the repeats (Loop) (Figure 1A; Smith et al., 2000; Wesley et al., 2001). These constructs were introduced into an Agrobacterium tumefaciens binary vector under the regulation of the phaseolin promoter and terminator (Bustos et al., 1991). Several transgenic plants were selected and grown to maturity. Seeds were recovered from the first generation, and oilbody preparations were performed. The profile of oleosins in these preparations was analyzed through SDS-PAGE. Plants containing the Hairpin or Loop constructs (SupOLEO1-Hairpin and SupOLEO1-Loop) displayed lower accumulation of OLEO1 compared with plants transformed with the Antisense construct (SupOLEO1-Antisense). In Figure 1C (lanes 1 to 3), we show the oleosin profile from transgenic lines displaying the most effective suppression obtained for each construct. In many cases, we could detect ablation of OLEO1 using RNAi methods (Hamilton and Baulcombe, 1999; Meins, 2000).
To eliminate the function of oleosins, we have also used an insertional mutagenesis approach. Public collections of T-DNA insertion in Arabidopsis were screened: the Wisconsin collection (Madison, WI) and the T-DNA Express database (Alonso et al., 2003), which includes several collections. We have found insertions in two genes encoding oleosins. One insertion was found in the second exon of OLEO1 (line SM_3_29875) in the collection from the Exon Trapping Insert Consortium (Figure 1B). Another insertion was found in the second exon of OLEO2 (line SALK_072403) in the collection from the Salk Institute of Technology (San Diego, CA) (Figure 1B). These lines were requested from the Nottingham Arabidopsis Stock Centre and the ABRC, respectively, and propagated under selective media. Several plants were obtained, and oilbody-associated proteins were analyzed through SDS-PAGE. Lines exhibiting ablation of OLEO1 or OLEO2 polypeptides (Figure 1C, lanes 4 and 5, respectively) were isolated and denominated KnockOLEO1 and KnockOLEO2. These lines were grown for two more generations for further analysis.
A Decrease in OLEO1 Accumulation Results in Larger Oilbodies
To investigate the effects of oleosin accumulation on the morphology of oilbodies, we conducted microscopy analysis. Embryos from mature Arabidopsis seeds were isolated and fixed with paraformaldehyde and glutaraldehyde solution. Postfixation with osmium tetroxide was performed to preserve lipids in the specimens (Yeung, 1990).
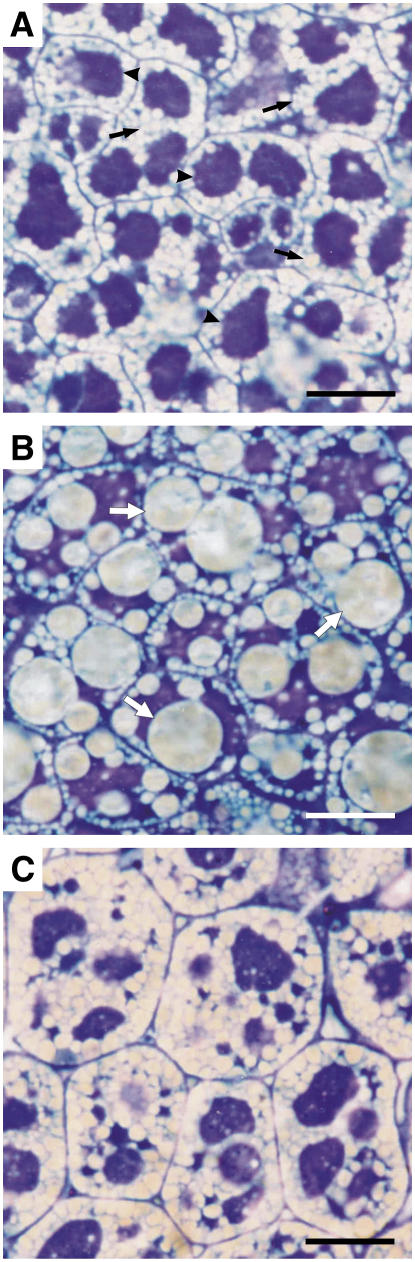
In wild-type mature embryos, oilbodies were mostly present in the periphery of the cells and between protein bodies (Figure 2A). This is the pattern usually reported for oilseeds (Mansfield and Briarty, 1991). When OLEO1 was suppressed, a very distinct phenotype was observed. In the Arabidopsis SupOLEO1-Loop line, heterogeneous oilbodies were formed (Figure 2B). Some of these lipid structures were found with similar size to those in wild-type embryos, while others were several times bigger. The organization of other subcellular compartments (mainly consisting of protein bodies) was also altered. Protein bodies that were normally found as larger structures in the center of the cells were present as very irregular units in SupOLEO1-Loop embryos. The overall cellular shape remained unaltered in this line. As the microscopy observations were performed from similar cell types, we considered this a clear indication that accumulation of oleosins determines the size of oilbodies. To ensure that this phenotype was a result of OLEO1 suppression and not an artifact, we have isolated a wild-type-like (null) Arabidopsis line segregated from the heterozygous SupOLEO1-Loop line. Seventy-six seeds from the first generation of SupOLEO1-Loop were propagated in soil. These plants were harvested, and 50 seeds from each plant were sown in media containing the herbicide marker phosphinothricin. We selected one line displaying no resistance to phosphinothricin (a null) and performed microscopy analysis. This line does not exhibit the phenotype found in the parental transgenic plant (Figure 2C), proving that the effects found in oilbody morphology are directly correlated with the decrease of OLEO1 content.
Figure 2.
Phenotype of Arabidopsis Mature Embryos.
Bright-field microscopy of osmicated embryos thin-sectioned and stained with toluidine blue. Bars = 10 μm.
(A) The wild type. Black arrows indicate oilbodies; arrowheads indicate protein bodies.
(B) SupOLEO1-Loop. White arrows indicate larger oilbodies.
(C) Plant segregated from SupOLEO1-Loop (null).
To investigate the size of oilbodies in vivo, we performed an analysis using confocal microscopy. This technique allowed the examination of whole mount of embryo sections and provided a more accurate size comparison between Arabidopsis lines. Embryos were isolated from mature seeds and stained with Nile red (Molecular Probes), a neutral lipid stain. TAGs represent the vast majority of neutral lipids in most oilseeds; hence, oilbodies are selectively stained by Nile red.
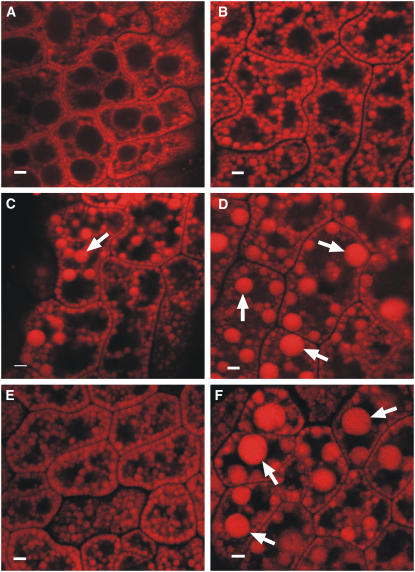
As demonstrated in bright-field microscopy, embryos from SupOLEO1-Loop (Figure 3D) held larger oilbodies compared with the wild type (Figure 3A). KnockOLEO1 embryos presented a similar phenotype (Figure 3F). In some cells, these abnormal oilbodies were as much as 5 μm in diameter. These are much larger than the average size of Arabidopsis seed oilbodies reported in the literature so far (Schumann et al., 2003; Niwa et al., 2004). SupOLEO1-Hairpin plants displayed a similar phenotype (Figure 3C). SupOLEO1-Antisense and KnockOLEO2 plants displayed oilbodies that were slightly larger than those found in the wild type (Figures 3B and 3E). In these cases, oilbodies were uniform in size compared with SupOLEO1-Hairpin, SupOLEO1-Loop, and KnockOLEO1 embryos.
Figure 3.
In Vivo Analysis of Oilbody Morphology.
Confocal sections of living mature Arabidopsis embryos stained with Nile red. Large oilbodies in (C), (D), and (F) are shown by arrows. Bars = 2 μm.
(A) Wild-type plant.
(B) SupOLEO1-Antisense plant.
(C) SupOLEO1-Hairpin plant.
(D) SupOLEO1-Loop plant.
(E) KnockOLEO2 plant.
(F) KnockOLEO1 plant.
Reversion of the OLEO1-Suppressed Phenotype
To explore the possibility of restoring the oilbody phenotype by increasing the amount of oleosins, we introduced a gene coding for a recombinant oleosin in the SupOLEO1-Loop line. To avoid cross-suppression through RNAi, we selected an oleosin from maize (OLE16 accession number U13701) to restore the function because it is phylogenetically distant from OLEO1 (Lee et al., 1994; Huang, 1996). Furthermore, OLE16 is already known to target correctly to oilbodies of dicotyledonous plants (Lee et al., 1991). SupOLEO1-Loop was manually crossed with MaizeOle, an Arabidopsis line expressing OLE16 under control of a linin seed-specific promoter (this line was generously provided by Chao Jiang, SemBioSys Genetics) (Figure 4B). The linin promoter is derived from a gene encoding a major seed storage protein, 2S albumin from Linum usitatissimum. This promoter was previously shown to be a strong, seed-specific promoter with maximal rates of transcription during the cotyledonary stage of seed development in wild-type flax but also when regulating a GUS transgene in transgenic Arabidopsis (Chaudhary et al., 2004). OLE16 has a distinct molecular mass (15.8 kD) compared with all of the Arabidopsis oleosins (Lee et al., 1991). We used this property to analyze the progeny of the crossed lines. The lines showing presence of OLE16 and suppression of OLEO1 were selected and propagated for two more generations. A homozygous line was obtained, and the seeds were analyzed using confocal microscopy (Figure 4C). Oilbodies in this line did not display the phenotype found in SupOLEO1-Loop. Such oilbodies were still somewhat larger than the wild-type ones but uniform in size, like those found in the SupOLEO1-Antisense line, suggesting at least a partial reversal of the phenotype.
Figure 4.
Introduction of a Recombinant Oleosin to Rescue the Oleosin-Deficient Phenotype.
SDS-PAGE profiles of oilbody-associated proteins are shown in left panels and confocal sections of mature embryos in right panels.
(A) SupOLEO1-Loop plant. OLEO1 polypeptide is indicated by the black arrow. White arrows show large oilbodies. Bar = 5 μm.
(B) MaizeOle line. OLEO1 and OLE16 are indicated by the black arrows. Bar = 8 μm.
(C) Progeny from crossing between SupOLEO1-Loop and MaizeOle plants. OLEO1 and OLE16 are indicated by the black arrows. Bar = 5 μm.
Oleosin Deficiency Retards Germination
Microscopy analysis clearly showed that suppression of oleosins results in a perturbation of oilbody biogenesis. This also affected the organization of protein storage vacuoles. TAGs and storage proteins are the main compounds used by Arabidopsis after germination. Therefore, we investigated the effects of this aberrant subcellular morphology during germination and seedling growth. We conducted germination tests in different conditions of carbohydrate availability and light exposure and noted a substantial delay in germination of SupOLEO1-Loop compared with wild-type seeds (Figure 5). The most prominent difference was found during the second and third days for seeds germinated on moistened filter paper (Figure 5A). However, this difference was masked for seeds germinated under light exposure with sucrose supplement, suggesting that the delay in germination was related to available carbon sources (Figure 5D). The effect on germination could also be reverted by submitting seeds to a prior cold treatment (stratification) (Figure 5B). A similar delay in germination was observed with the KnockOLEO1 line (see Supplemental Figure 1 online). After germination, no difference in seedling development was observed for oleosin-suppressed seedlings (see Supplemental Figure 2 online).
Figure 5.
Comparison of Germination Frequency between Wild-Type and SupOLEO1-Loop Plants in Various Conditions.
Arabidopsis seeds were germinated in different conditions of light and sucrose availability.
(A) Wet filter paper; light.
(B) Wet filter paper; stratified seeds; light.
(C) Half-strength Murashige and Skoog (MS) medium + sucrose; light.
(D) Half-strength MS medium + sucrose; light.
(E) Half-strength MS medium − sucrose; dark.
(F) Half-strength MS medium + sucrose; dark.
Oleosin Depletion Affects the Accumulation of Seed Reserves
To investigate whether alterations in oilbody biogenesis would affect the accumulation of TAG and other seed reserves, we performed biochemical analyses of seed composition. Comparison of SupOLEO1-Loop and KnockOLEO1 lines with their respective wild-type backgrounds (C24 and Col-0, respectively) shows a significant decrease in the amount of lipids in oleosin-suppressed seeds (Table 1). This reduction was also accompanied by an increase in accumulation of total protein, in an almost compensatory manner, such that the total weight of protein and oil combined remained constant within the seed (Table 1). Analysis of sucrose and starch content revealed no significant difference in the accumulation of these carbohydrates.
Table 1.
Lipid, Protein, and Carbohydrate Composition of Arabidopsis Seeds
| Lipid (%) | Protein (%) | Starch (%) | Sucrose (%) | |
|---|---|---|---|---|
| Wild type (C24) | 40.3 ± 1.4 | 25.1 ± 1.7 | 0.5 ± 0.3 | 3.2 ± 0.4 |
| SupOLEO1-Loop | 32.9 ± 2.0 | 33.9 ± 1.6 | 0.8 ± 0.4 | 2.8 ± 0.2 |
| Wild-type (Col-0) | 36.1 ± 1.6 | 35.9 ± 2.4 | 0.7 ± 0.1 | 2.9 ± 0.3 |
| KnockOLEO1 | 30.3 ± 0.9 | 39.9 ± 1.3 | 0.8 ± 0.3 | 2.9 ± 0.1 |
| KnockOLEO2 | 34.1 ± 1.5 | 35.8 ± 2.8 | 0.8 ± 0.4 | 2.2 ± 0.3 |
Mean values are given in percentage of seed fresh weight with standard deviations. For lipid, starch, and sucrose analysis, n = 5, and for protein analysis, n = 8.
Oleosin Depletion Causes Minor Changes in Fatty Acid Composition of TAGs
Although there are quantitative differences in the accumulation of storage reserves in seeds showing suppression of oleosin accumulation, it was not clear whether this would also qualitatively affect the fatty acid profiles of TAGs in these seeds. Recent work by Lu et al. (2006) suggested that the overexpression of oleosin in transgenic Arabidopsis could change the overall fatty acid composition of transgenic Arabidopsis seed modified metabolically to make ricinoleic acid. In that case, it was shown that overexpression of a castor bean oleosin in Arabidopsis resulted in a 20% increase in ricinoleic acid in TAGs. We therefore measured fatty acid composition of TAGs in oleosin-suppressed lines of Arabidopsis. The results are shown in Figures 6A and 6B. When compared with wild-type Arabidopsis of the same background, the suppressed lines (KnockOLEO1 and KnockOLEO2; Figure 6A) or SupOLEO1-loop (Figure 6B) did not show any dramatic differences in overall fatty acid profiles in TAG, with the exception of a statistically significant preference for the accumulation of C20:1 (eicosenoic acid) at the expense of C18:1 (oleic acid). While this was a relatively small effect, it was consistent between the suppressed line (SupOLEO1-loop) and the insertional mutant (KnockOLEO1) even though these are both in different genetic backgrounds. Thus, consistent with the report of Lu et al. (2006), oleosins may also exert some influence on qualitative and quantitative aspects of TAG accumulation in seeds.
Figure 6.
Fatty Acid Profiles in Wild-Type, Nulls, and Oleosin-Suppressed Seeds of Arabidopsis.
(A) Comparison of major fatty acids (FA) in TAG in wild-type Columbia and oleosin T-DNA insertional knockouts of OLEO1 and OLEO2 in a Columbia background.
(B) Comparison of C24 and segregating nulls from the oleosin-suppressed line SupOLEO1-loop.
Fate of Oilbodies during Embryo Development and Seedling Growth
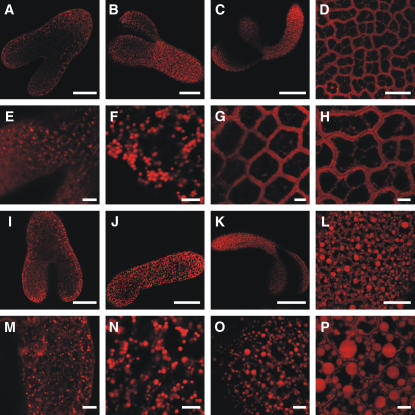
To determine the ontogeny of the abnormal oilbodies in oleosin-deficient lines, we analyzed Arabidopsis embryos at different maturity phases using confocal microscopy. The development of Arabidopsis embryos is well studied and can be divided into distinct morphological stages (Bowman, 1994). Synthesis of TAG starts in the late-heart stage and continues through the torpedo, walking-stick, and bent cotyledon stages until the seed desiccates (Mansfield and Briarty, 1991). Our analysis showed that oilbodies in late-heart and torpedo stages were morphologically similar between the wild-type (Figures 7E and 7F) and KnockOLEO1 lines (Figures 7M and 7N). Differences in the size of oilbodies started to occur in the walking-stick stage (Figures 7G and 7O), when oilbodies should be normally present in the boundaries of the wild-type cells. In mature desiccated seeds, oilbodies have reached their maximum size in the KnockOLEO1 line (Figure 7P), while in wild-type cells, they remain as single individual units (Figure 7H). Similar results could be observed using transmission electron microscopy (TEM). Embryos from wild-type and KnockOLEO1 lines held oilbodies with similar sizes during earlier stages of maturation (Figures 9A and 9B). More mature KnockOLEO1 embryos presented larger oilbodies in comparison with the wild type (Figures 9E and 9F). TEM has also revealed the presence of elongated oilbodies in some cells (Figure 9D) and the putative association with the ER (Figure 9C) as it has been previously reported (Frey-Wyssling et al., 1963). These results indicate that in both wild-type and oleosin-deficient lines, oilbodies are similar during earlier stages of embryo development. As the embryos mature, the large oilbodies are generated in the oleosin deficient cells.
Figure 7.
Fate of Oilbodies during Embryo Development.
Arabidopsis embryos were collected at different stages of development and stained with Nile red. Bars = 40 μm in (A) and (I), 60 μm in (B) and (J), 150 μm in (C) and (K), 20 μm in (D) and (L), and 5 μm in (E) to (H) and (M) to (P).
(A) to (D) Confocal sections of wild-type Arabidopsis embryos at late-heart stage, torpedo stage, walking-stick stage, and mature seed, respectively.
(E) to (H) Same as (A) to (D) in higher magnification.
(I) to (L) Confocal sections of KnockOLEO1 Arabidopsis embryos at late-heart stage, torpedo stage, walking-stick stage, and mature seed, respectively.
(M) to (P) Same as (I) to (L) in higher magnification.
Figure 9.
Transmission Electron Micrographs of Developing Arabidopsis Embryos.
(A) and (C) Wild-type embryos in the early stages of maturation. Black arrow indicates the ER.
(B) KnockOLEO1 embryo in the early stages of maturation.
(D) and (F) KnockOLEO1 embryo in the middle stages of maturation. Open arrow indicates an elongated oilbody.
(E) Wild-type embryo in the middle stages of maturation.
Bars = 1 μm in (A), (B), and (D) to (F) and 0.1 μm in (C).
Oilbodies are usually consumed during the first days after germination. Our experiments showed that 2 d after germination oilbodies are still present as small units (Figures 8A and 8B). However, from 3 to 8 d after germination, oilbodies were scarcely found as very tiny particles in wild-type seedlings (Figures 8C to 8H). In the KnockOLEO1 line, oilbodies assumed a different behavior. During the first day after germination, small and large organelles were found (Figure 8I). From 2 to 8 d after germination, only the large oilbodies could be detected in some cells (Figures 8J to 8P). These results suggest a slower mobilization of oilbodies in the KnockOLEO1 line. We have not analyzed older seedlings than 8 d after germination, but apparently these organelles could remain intact for even longer periods. Similar results were found with the SupOLEO1 line (see Supplemental Figure 3 online).
Figure 8.
Fate of Oilbodies during Seedling Growth.
Arabidopsis seeds were germinated in half-strength MS medium without sucrose supplement. Seedlings with different ages were stained with Nile red and analyzed by confocal microscopy. Nile red was detected in the red channel and autofluorescence of chlorophyll was detected in the blue channel. Bars = 10 μm.
(A) to (H) Confocal sections of wild-type Arabidopsis seedlings at 1 to 8 d after germination, respectively.
(I) to (P) Confocal sections of KnockOLEO1 Arabidopsis seedlings at 1 to 8 d after germination, respectively.
DISCUSSION
Oilseeds store lipids in oilbodies, which are relatively simple organelles comprising a matrix of TAG coated by a phospholipid monolayer embedded by oleosins. It has been established that these organelles are synthesized in the ER as nascent oilbodies, and they subsequently bud off from ER forming mature organelles (reviewed in Murphy and Vance, 1999; Hsieh and Huang, 2004). Oleosins appear to play an important role in oilseeds. This is evidenced not only by their presence in seeds from most species but also by the remarkable conservation of different isoforms in diverse species (Tzen et al., 1990). This suggests that during evolution, plants maintained oleosins in TAG storing tissues because they provided a survival advantage. There are no reports of natural mutants displaying substantial decrease of oleosin accumulation. In fact, the maize lines FR2 and CM555 are depleted in one polypeptide isoform. However, this is compensated for by higher accumulation of another isoform maintaining the overall amount of oleosins (Lee et al., 1995). Such circumstantial evidence has pointed to an important role for oleosins in seeds.
It has been speculated that oleosins stabilize oilbodies preventing coalescence of the lipid particles during seed dehydration (Cummins et al., 1993). Some authors have also suggested that oleosins prevent oilbodies from coalescing during seed imbibition and germination (Leprince et al., 1998). Reductions in oleosin content would result in decrease of surface-to-volume ratio on oilbodies due to production of larger oilbodies (Tzen et al., 1993). The maintenance of high surface-to-volume ratios could facilitate access by lipases and thus accelerate TAG mobilization. However, until now the consequences of loss of oleosin function to lipid storage, mobilization, and other cellular processes have not been directly determined. Using reverse genetics approaches, we have shown clear evidence that the accumulation of oleosins modulate the size of oilbodies. Furthermore, we have shown the consequences of a decrease in oleosin accumulation to seed germination and TAG accumulation. These consequences are all consistent with the idea that oleosins facilitate access to TAG during germination either by the simple change in surface-to-volume ratio or by a more active mechanism such as forming lipase docking sites as previously suggested (Huang, 1992).
In our first approach to study the function of oleosins, we suppressed the most abundant oleosin, OLEO1, using RNAi methods. Three different constructs were used: Antisense, Hairpin, and Loop (Figure 1A). To ensure that the interfering RNA molecules would be abundantly produced, we used the phaseolin promoter that is very active during Arabidopsis embryo development coincident with the period in which the OLEO1 gene is active. Approximately half of transgenic plants presented some degree of OLEO1 suppression, but no complete ablation event was isolated by this procedure (see Supplemental Figure 4 online). Different degrees of OLEO1 accumulation were found according to the cassette used (Figure 1C). Generally, the cassettes designed to produce RNA hairpin structures (Hairpin and Loop) were more effective than the Antisense cassette, although they could not provide ablation of the polypeptide. To completely eliminate the function of OLEO1, we have also used an insertional mutagenesis approach. T-DNA insertions were found in the genes encoding OLEO1 and OLEO2 polypeptides (Figure 1B). In KnockOLEO1 and the other OLEO1-suppressed lines, accumulation of OLEO2 and OLEO4 isoforms was similar in both transgenic and wild-type seeds. There is no apparent increase in accumulation of these peptides as it occurs in maize when one isoform is missing (Lee et al., 1995). It is unlikely, based on divergent sequence, that the Antisense, Hairpin, and Loop constructs interfere with the transcript of the other isoforms.
Our analysis provides evidence that the accumulation of oleosins modulates the size of oilbodies. First, embryos containing the Loop cassette or embryos holding an insertion in the OLEO1 gene displayed a distinct phenotype with production of large oilbodies (Figures 2B and 3F, respectively). Second, this phenotype could be reversed by removing the gene from the plant genotype by genetic segregation (Figure 2C). Third, the phenotype could be partially reversed by introduction of a foreign oleosin (Figure 4). Fourth, there was correlation between the amount of total oleosins and the size of the oilbodies (Figure 3). Plants transformed with the Antisense cassette or KnockOLEO2 plants did not have as large oilbodies as did plants transformed with Loop and Hairpin cassettes or KnockOLEO1 plants. It should also be noticed that if oleosins regulate the size (radius) of oilbodies, the radius is proportional to the square root of the surface area. Thus, it would take a fourfold increase in the surface to record a doubling of oilbody radius. This might be difficult to detect if only a small perturbation of oleosin content is involved.
Besides the consequences in oilbody morphology, the reduction in OLEO1 content produced other effects in Arabidopsis seeds. We observed retardation in germination of OLEO1-suppressed seeds compared with the wild type. This effect could be reversed by germinating seeds in the light in the presence of sucrose, indicating a relationship to the carbon availability. However, a prior cold treatment (stratification) also reversed the phenotype, indicating that the effect in germination could be more indirect. It is possible that stratification could lead to a hormone-related elevation in lipase activity, which overrides the retardation in germination. The deficiency in OLEO1 also caused a reduction in total lipid content in the seed. This reduction did not affect the levels of carbohydrates, such as sucrose and starch, but was instead accompanied by a compensatory increase in the amount of total proteins (Table 1). An inverse relationship between oil and protein accumulation in seeds has been reported for other plant species (Cober and Voldeng, 2000; Chung et al., 2003). In this case, it is likely that a reduction in lipid accumulation is caused by interference in the biogenesis of oilbodies. This effect could be caused by a reduction in the synthesis of TAG or by an increased turnover of oilbodies during seed maturation. For instance, it has been previously described that turnover of TAG during embryo development is a common process during development of oilseeds (Chia et al., 2005). Therefore, it is possible that turnover of TAG is enhanced in the oleosin-unprotected oilbodies.
In a qualitative sense, the suppression of oleosin had a small but statistically significant effect on fatty acid preferences in TAG. The suppression of the major oleosin (OLEO1) correlated with an increase in C20:1 (eicosenoic acid) at the expense of C18:1 (oleic acid) in TAG. Although this effect was small, it is consistent with the findings of Lu et al. (2006), who showed changes in TAG composition as a function of oleosin overexpression in Arabidopsis.
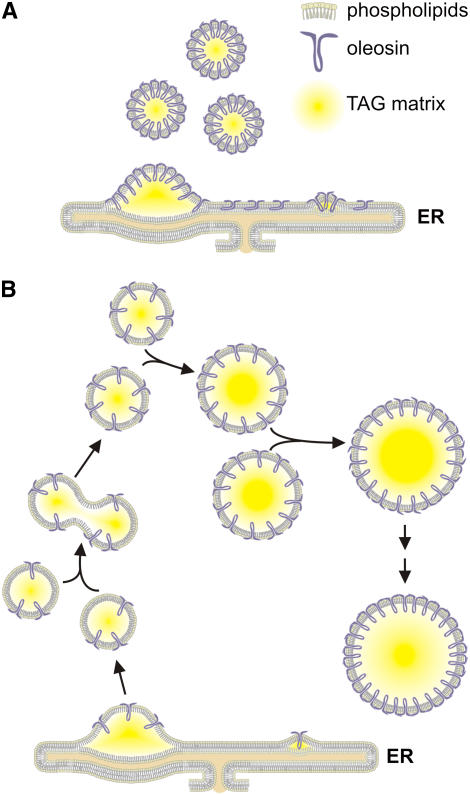
Analysis of developing Arabidopsis embryos revealed that wild-type and KnockOLEO1 lines both produced small oilbodies with comparable sizes during early developmental stages (Figures 7E, 7F, 7M, 7N, 9A, and 9B). In the walking-stick stage, however, the OLEO1-deficient cells presented much larger oilbodies (Figure 7O). In this stage, synthesis of vacuolated protein bodies is normally intense, reducing the cytoplasmic volume of the cell (Mansfield and Briarty, 1991). This would result in compression of oilbodies leading to coalescence of OLEO1-deficient lipid particles. In mature desiccated seeds, oilbodies are even bigger in the KnockOLEO1 line (Figure 7P), indicating that desiccation might play a role in oilbody coalescence. For instance, transmission electron micrographs revealed elongated lipid particles in developing KnockOLEO1 embryos, suggesting the presence of coalescing oilbodies (Figure 9D). It is interesting to note that OLEO1-deficient plants accumulated larger oilbodies (with a diameter of ∼6 μm), although smaller oilbodies were still found in mature seeds. The smaller oilbodies could have sufficient amounts of oleosin to form wild-type-like oilbodies, whereas the very large organelles would be generated by a number of coalescence events due to insufficient amounts of oleosin or the oilbody surface. SupOLEO1-Antisense plants presented larger oilbodies than the wild type but smaller than the large ones described before. In this case, the organelles were more homogeneous in size. We conclude that slight decreases in OLEO1 content result in larger and homogeneous oilbodies (SupOLEO1-Antisense plants). Further decreases produce oilbodies with diverse sizes (SupOLEO1-Hairpin, SupOLEO1-Loop, and KnockOLEO1 plants). During seedling growth in the KnockOLEO1 line, the larger oilbodies seem to increase in size during the first days of germination (Figures 8I and 8J). This is the period when oleosins start to be degraded (Sadeghipour and Bhatla, 2002), and it is reasonable to assume that coalescence would occur between the large oilbodies as previously suggested (Leprince et al., 1998). These observations suggest that the availability of oleosin polypeptide does not directly determine the size of a nascent oilbody but rather allows oilbodies of a given size to be released into the cytoplasm. Oilbodies depleted of oleosins probably coalesce with other oilbodies until they reach a critical surface density, which may correspond to a complete monolayer of oleosins. Once this situation occurs, coalescence is no longer favored, and the large oilbody stabilizes (Figure 10). According to this assumption, the amount of oleosins in the surface would increase after each fusion, and to maintain the spherical shape, the excess of phospholipids should be eliminated. We have found recently that oilbodies from SupOLEO1-Loop and KnockOLEO1 lines contain lower levels of phospholipids compared with the wild type (see Supplemental Figure 5 online). This explanation is consistent with our microscopy observations of the distribution of oilbody sizes in the oleosin-suppressed transgenics. The introduction of a foreign oleosin in the suppressed line showed that it is possible to replace modified oleosins in an oilbody's surface. This could certainly be useful in many studies concerning oleosin function and molecular interactions. Replacing natural oleosins by recombinant modified proteins could give new insights on targeting mechanisms, TAG sequestering to oilbodies, and in vivo features of the oilbody surface.
Figure 10.
Model of Oilbody Biogenesis in Oleosin-Suppressed Lines.
The three major components of oilbodies are shown as phospholipids, oleosins (purple), and the TAG matrix (yellow).
(A) Biogenesis of an oilbody in a wild-type cell. The oleosin-saturated environment in ER results in production of lipid bodies completely covered by oleosins.
(B) Biogenesis of an oilbody in an oleosin-suppressed cell. Oilbodies that do not contain enough oleosins to coat their entire surface coalesce, forming larger lipid bodies. These particles might fuse until the surface is completely covered of oleosins.
METHODS
Plasmid Constructs and Arabidopsis thaliana Transformation
The OLEO1 cDNA was amplified by PCR from a clone previously isolated in our lab using the forward primer NTD (5′-TATTAAGCTTCCATGGCCGATACTGCTAGAGG-3′; the NcoI site is underlined) annealed at the beginning of the coding region and the reverse primer CTR (5′-AGCCATACTAGTAGTGTGTTGACCACCACGAG-3′; the SpeI site is underlined) annealed at the end of the coding region. The PCR product was cloned in the vector pSBS2090 using the SwaI restriction site delineating the phaseolin promoter and terminator (Slightom et al., 1983). The orientation and number of copies inserted in the vector were confirmed by digestion with different restriction enzymes. One vector containing a single copy in antisense orientation and another vector containing inverted repeats were isolated and called pAntisense and pHairpin, respectively. The unique intron from OLEO1 was amplified by PCR using the forward primer IntronD (5′-TTTTACTAGTGATTTACAATTAAGCACACATTTATC-3′; the SpeI site is underlined) and the reverse primer IntronR (5′-CTGTACTAGTTCTCCCGTTGCGTACCTATTCAC-3′; the SpeI site is underlined). The PCR product was cloned in the vector pHairpin digested with SpeI between the inverted repeats of OLEO1 cDNA. The orientation of the insertion was examined by PCR using the forward primer Phapr (5′-CCTGCATATGCGTGTCATCCATGC-3′) and the reverse primer IntronR. The plasmid containing the intron in the sense orientation was designated as pLoop. These three cassettes were cloned in the binary vector pSBS3000 between the BamHI and KpnI restriction sites. This vector was derived from the Agrobacterium tumefaciens binary plasmid pPZP (Hajdukiewicz et al., 1994). The gentamycin resistance gene of pPZP was replaced by the phosphinothricin acetyl transferase coding region (Wohlleben et al., 1988) regulated by a parsley (Petroselinum crispum) ubiquitin promoter and terminator sequences. The binary vectors were introduced into Agrobacterium strain EHA101 (Hood et al., 1986). Recombinant Agrobacterium lines were used to transform Arabidopsis plants (ecotype C24) through the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on half-strength MS agar plates (Murashige and Skoog, 1962) containing phosphinothricin (25 mg/L).
Microscopy Analysis
For light microscopy, mature embryos were isolated from dry seeds according to the method described by Perry and Wang (2003). These embryos were fixed immediately in 2.5% glutaraldehyde and 1.6% paraformaldehyde in a 0.1 M phosphate buffer, pH 6.8, for 4 h. After rinsing several times in the same buffer, the embryos were postfixed with a 2% osmium tetroxide solution for an additional 4 h. After dehydration using the acetone series, the embryos were infiltrated and subsequently embedded in the Ladd LX-112 epoxy resin. Semithin sections were obtained using a Sorvall MT-1 ultramicrotome. The sections were stained using periodic acid Schiff's reaction and counterstained with an alkaline toluidine blue O solution (Yeung, 1990).
Primary fixation for TEM was similar to that for light microscopy. The developing seeds with the seed coat partially removed were fixed at room temperature for 1 h, transferred to ice for another 3 h of fixation, washed several times in 0.05 M phosphate buffer, and postfixed in 2% osmium tetroxide in the same buffer for 4 h. The seeds were dehydrated through a graded acetone series and embedded in Ladd LX-112 epoxy resin. Ultrathin (∼90 nm) sections were cut with a diamond knife on a Leica UC6 ultramicrotome and stained serially with 2% uranyl acetate and 1% lead citrate. Sections were examined in a Jeol 1200EX transmission electron microscope operated at 80 kV and photographed using a Deben AMT 1.3k digital camera.
Dark-field microscopy analysis was used to analyze embryos and seedlings from Arabidopsis. The material was infiltrated with an aqueous solution of Nile red (Molecular Probes) to visualize neutral lipids (Greenspan et al., 1985). Images were acquired using confocal laser scanning microscopy based on the Zeiss LSM 510 system equipped with argon ion and HeNe lasers as excitation sources using a ×60 objective.
Oilbody Isolation and Analysis
Oilbodies were isolated by a flotation centrifugation method previously described (van Rooijen and Moloney, 1995a), with minor modifications. Arabidopsis seeds (50 mg) were ground in 5 mL of oilbody extraction buffer (0.4 M sucrose, 0.5 M NaCl, and 50 mM Tris-HCl, pH 8.0) and centrifuged. Oilbodies were washed once in high-stringency buffer (8 M urea and 100 mM Na2CO3, pH 10) and twice in water. Oilbody-associated proteins were analyzed by SDS-PAGE using standard protocols (Sambrook et al., 1989) and stained with Coomassie Brilliant Blue R 250.
Lipid, Protein, and Carbohydrate Analysis
The lipid, protein, and carbohydrate composition of seeds was performed with five or eight biological replicates and expressed in seed mass basis. The seeds were obtained from a randomized growth trial where 50 plants of each line were cultivated in a growth chamber with homogeneous humidity and light. Wild-type and transgenic lines were grown side by side at the same time, and the plants were equally irrigated and fertilized. Lipids were extracted according to the method described by Bligh and Dyer (1959). Fifty milligrams of Arabidopsis seeds were homogenized in liquid nitrogen and incubated at 70°C for 10 min with 5 mL of isopropanol. The isopropanol was evaporated under nitrogen, and lipids were extracted with three extractions of chloroform, methanol, and water biphasic solutions (methanol:CHCl3:H2O). The first extraction was performed with 5.8 mL of methanol:CHCl3:H2O (2:2:1.8 [v/v]), and the second and third extractions were performed with 2.0 mL of methanol:CHCl3:H2O (1:2:0.8 [v/v]). The lipid fractions were collected and the solvents were completely evaporated under a nitrogen environment. Total lipids were quantified by gravimetry after drying the samples in a desiccator for 24 h. This analysis was repeated using five biological replications.
For protein extraction, 50 mg of Arabidopsis seeds were homogenized in 1.5 mL of protein extraction buffer (2% SDS, 5 mM EDTA, and 50 mM Tris-HCl, pH 6.8). The homogenates were placed in boiling water for 5 min and centrifuged at full speed (15,000g) for 10 min. The upper phase was removed, and the debris was washed twice with 0.5 mL of extraction buffer. The fractions were pooled, and the amount of protein was measured using the BCA protein assay reagent (Pierce) using three technical replications. The total protein content was calculated using eight biological replications.
The analysis of carbohydrates was performed as described by Focks and Benning (1998) with some modifications. Five milligrams of seeds were homogenized in 80% (v/v) ethanol and incubated at 70°C for 90 min. The homogenate was centrifuged at full speed for 5 min, and the supernatant was transferred to a new test tube. The pellet was washed three times with 0.5 mL of 80% ethanol, and the solvent of the combined supernatants was evaporated at room temperature under vacuum. The residue, which represents the soluble carbohydrate fraction, was dissolved in 0.1 mL of water and used for sucrose quantification. The insoluble fraction from the ethanol extraction was suspended in 0.2 mL of 0.2 M KOH and incubated 95°C for 1 h. The solution was neutralized with 35 μL of 1 M acetic acid and centrifuged for 5 min at full speed. The supernatant was used for starch quantification. Sucrose and starch were determined in five biological replications using kits from Sigma-Aldrich.
Lipid Extraction and Fatty Acid Methyl Ester Preparation
Approximately 25 mg of seeds of each sample (n = four independent replicates) were placed in a hexane-washed, hand-held, ground-glass homogenizer and boiled in 1 mL of isopropanol (80°C) for 10 min. The seed was then cooled on ice for 5 min. Thereafter, 1 mL of hexane and 2 mL of 3:2 hexane:isopropanol (HIP) was added and the seed homogenized until completely pulverized. An additional 2 mL of 3:2 HIP was added and grinding continued. The slurry was transferred to a screw capped glass tube, and 2 mL of 3.3% (w/v) Na2SO4 was added, capped, and shaken for 2 min. The tubes were spun at 555g for 2 min, and the upper organic phase transferred to a new hexane-washed screw-capped tube. The aqueous phase was re-extracted with 4 mL of 7:2 HIP, capped, and shaken for 2 min. The tubes were spun again at 555g for 2 min, and the upper organic phase added with the first extracted organic phase. The combined organic phases were evaporated to dryness in a heating block (37°C) under a gentle N2 stream. To determine fatty acid methyl esters (FAMEs), 1.2 mL of HCl-methanol (1.5 M HCl in methanol made fresh) was added to the dried lipid and incubated at 100°C for 1 h. Then, 1 mL of double distilled water was added to quench the transesterification reaction. The FAMEs were then extracted with 2 mL of hexane. The samples were centrifuged as above and the upper organic phase containing the FAMEs transferred to a clean hexane-washed test tube. The aqueous phase was re-extracted with an addition 2 mL of hexane and centrifuged, and the resulting upper phase transferred and combined with the previously collected organic phase. The combined organic phases containing the FAMEs were then dried down completely in a heating block with N2 stream. Finally, the FAMEs were solubilized in 1 mL of hexane and transferred to gas chromatography vials and capped.
FAME Analysis by Gas–Liquid Chromatography
FAMEs were analyzed on an Agilent Technologies 6890N gas chromatograph equipped with an autosampler. FAMEs were separated and detected by flame ionization detection on a narrow-bore DB-23 column (60 m × 0.25-mm i.d. × 0.25-μm film) under constant pressure (51.75 psi) under the following oven regime: 180°C for 5 min, 180 to 240°C at 2°C/min, and hold at 240°C for 15 min. The splitless injector was set to 250°C, and the flame ionization detector was set to 300°C with nitrogen makeup gas at 25 mL/min. Integration events were detected and identified between 5 and 30 min and compared against a NuChek 502 gas–liquid chromatography standard.
Germination Assays
Seeds from wild-type Arabidopsis (ecotype C24) and the OLEO1-suppressed line (SupOLEO1-Loop) were harvested from plants cultivated in the same growth conditions at the same time. Five hundred seeds were sown in five batches of 100 seeds per batch in 14-cm Petri dishes containing half-strength MS medium with or without 3% sucrose supplement. Seeds germinated on moistened paper were sown on two layers of Whatman 595 filter paper soaked with 5 mL of distilled-deionized water. The Petri dishes were incubated at 22°C in an 8-h-dark/16-h-light cycle for light conditions and 24 h of continuous darkness for dark conditions. The germination rate of each batch was scored by visualization of radical emergence every 24 h.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers U13701 (OLE16), At4g25140 (OLEO1), At5g40420 (OLEO2), At5g51210 (OLEO3), and At3g27660 (OLEO4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Comparison of Germination Frequency between Wild-Type (Columbia) and KnockOLEO1 Plants.
Supplemental Figure 2. Comparison of Seedling Growth of Wild-Type and SupOLEO1-Loop Arabidopsis.
Supplemental Figure 3. Confocal Section of Arabidopsis SupOLEO1 Seedlings 8 d after Germination.
Supplemental Figure 4. Suppression of OLEO1 in Arabidopsis Using Different Constructs.
Supplemental Figure 5. Thin Layer Chromatography of Oilbody Lipids.
Supplementary Material
Acknowledgments
We thank Sue Bunnewell for the assistance with TEM, Chao Jiang for the provision of OLE16 Arabidopsis plants, and SemBioSys Genetics for technical assistance. We also thank the Salk Institute of Technology for releasing the knockout line SALK_072403 (KnockOLEO2) and the Nottingham Arabidopsis Stock Centre for releasing the line SM_3_29875. We acknowledge the financial support of the Natural Sciences and Engineering Research Council (Research Partnerships Program) for the award of an Industrial Research Chair to M.M.M. and for Discovery Grant 3490.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Maurice M. Moloney (mmmolone@ucalgary.ca).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.106.041269.
References
- Aalen, R.B., Opsahlferstad, H.G., Linnestad, C., and Olsen, O.A. (1994). Transcripts encoding an oleosin and a dormancy-related protein are present in both the aleurone layer and the embryo of developing barley (Hordeum vulgare L.) seeds. Plant J. 5 385–396. [DOI] [PubMed] [Google Scholar]
- Abell, B.M., Holbrook, L.A., Abenes, M., Murphy, D.J., Hills, M.J., and Moloney, M.M. (1997). Role of the proline knot motif in oleosin endoplasmic reticulum topology and oil body targeting. Plant Cell 9 1481–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Anil, V.S., Harmon, A.C., and Rao, K.S. (2003). Temporal association of Ca(2+)-dependent protein kinase with oilbodies during seed development in Santalum album L.: Its biochemical characterization and significance. Plant Cell Physiol. 44 367–376. [DOI] [PubMed] [Google Scholar]
- Bligh, E.G., and Dyer, W.J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37 911–917. [DOI] [PubMed] [Google Scholar]
- Bird, C.R., Ray, J.A., Fletcher, J.D., Boniwell, J.M., Bird, A.S., Teulieres, C., Blain, I., Bramley, P.M., and Schuch, W. (1991). Using antisense RNA to study gene function - Inhibition of carotenoid biosynthesis in transgenic tomatoes. Biotechnology 9 635–639. [Google Scholar]
- Bowman, J. (1994). Embryogenesis. In Arabidopsis: An Atlas of Morphology and Development, J. Bowman, ed (New York: Springer-Verlag), pp. 351–401.
- Bustos, M.M., Begum, D., Kalkan, F.A., Battraw, M.J., and Hall, T.C. (1991). Positive and negative cis-acting DNA domains are required for spatial and temporal regulation of gene expression by a seed storage protein promoter. EMBO J. 10 1469–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary, S., van Rooijen, G.J.H., Moloney, M.M., and Singh, S. (2004). Legumin-like storage protein promoter isolated from Flax and methods of expressing proteins in plant seeds using the promoter. U.S. Patent 6777591, issued Aug. 17, 2004.
- Chen, J.C.F., Tsai, C.C.Y., and Tzen, J.T.C. (1999). Cloning and secondary structure analysis of caleosin, a unique calcium-binding protein in oilbodies of plant seeds. Plant Cell Physiol. 40 1079–1086. [DOI] [PubMed] [Google Scholar]
- Chia, T.Y.P., Pike, M.J., and Rawsthorne, S. (2005). Storage oil breakdown during embryo development of Brassica napus (L.). J. Exp. Bot. 56 1285–1296. [DOI] [PubMed] [Google Scholar]
- Chung, J., Babka, H.L., Graef, G.L., Staswick, P.E., Lee, D.J., Cregan, P.B., Shoemaker, R.C., and Specht, J.E. (2003). The seed protein, oil, and yield QTL on soybean linkage group I. Crop Sci. 43 1053–1067. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cober, E.R., and Voldeng, H.D. (2000). Developing high-protein, high-yield soybean populations and lines. Crop Sci. 40 39–42. [Google Scholar]
- Crowe, A.J., Abenes, M., Plant, A., and Moloney, M.M. (2000). The seed-specific transactivator, ABI3, induces oleosin gene expression. Plant Sci. 151 171–181. [DOI] [PubMed] [Google Scholar]
- Cummins, I., Hills, M.J., Ross, J.H., Hobbs, D.H., Watson, M.D., and Murphy, D.J. (1993). Differential, temporal and spatial expression of genes involved in storage oil and oleosin accumulation in developing rapeseed embryos: Implications for the role of oleosins and the mechanisms of oil-body formation. Plant Mol. Biol. 23 1015–1027. [DOI] [PubMed] [Google Scholar]
- Evans, D.E., Taylor, P.E., Singh, M.B., and Knox, R.B. (1992). The interrelationship between the accumulation of lipids, protein and the level of acyl carrier protein during the development of Brassica napus pollen. Planta 186 343–354. [DOI] [PubMed] [Google Scholar]
- Focks, N., and Benning, C. (1998). wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 118 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey-Wyssling, A., Grieshaber, E., and Muhlethaler, K. (1963). Origin of spherosomes in plant cells. J. Ultrastruct. Res. 8 506–516. [Google Scholar]
- Greenspan, P., Mayer, E.P., and Fowler, S.D. (1985). Nile red - A selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 100 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25 989–994. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J., and Baulcombe, D.C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286 950–952. [DOI] [PubMed] [Google Scholar]
- Hood, E.E., Helmer, G.L., Fraley, R.T., and Chilton, M.D. (1986). The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of Ptibo542 outside of transfer DNA. J. Bacteriol. 168 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, K., and Huang, A.H.C. (2004). Endoplasmic reticulum, oleosins, and oils in seeds and tapetum cells. Plant Physiol. 136 3427–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, A.H.C. (1992). Oilbodies and oleosins in seeds. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43 177–200. [Google Scholar]
- Huang, A.H.C. (1996). Oleosins and oilbodies in seeds and other organs. Plant Physiol. 110 1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks, T.J., Hensarling, T.P., Neucere, J.N., Yatsu, L.Y., and Barker, R.H. (1990). Isolation and physicochemical characterization of the half-unit membranes of oilseed lipid bodies. J. Am. Oil Chem. Soc. 67 353–361. [Google Scholar]
- Jolivet, P., Roux, E., d'Andrea, S., Davanture, M., Negroni, L., Zivy, M., and Chardot, T. (2004). Protein composition of oilbodies in Arabidopsis thaliana ecotype WS. Plant Physiol. Biochem. 42 501–509. [DOI] [PubMed] [Google Scholar]
- Keddie, J.S., Tsiantis, M., Piffanelli, P., Cella, R., Hatzopoulos, P., and Murphy, D.J. (1994). A seed-specific Brassica napus oleosin promoter interacts with a G-box-specific protein and may be bidirectional. Plant Mol. Biol. 24 327–340. [DOI] [PubMed] [Google Scholar]
- Lee, K., Bih, F.Y., Learn, G.H., Ting, J.T.L., Sellers, C., and Huang, A.H.C. (1994). Oleosins in the gametophytes of pinus and Brassica and their phylogenetic relationship with those in the sporophytes of various species. Planta 193 461–469. [DOI] [PubMed] [Google Scholar]
- Lee, K., Ratnayake, C., and Huang, A.H.C. (1995). Genetic dissection of the coexpression of genes encoding the 2 isoforms of oleosins in the oilbodies of maize kernel. Plant J. 7 603–611. [DOI] [PubMed] [Google Scholar]
- Lee, W.S., Tzen, J.T.C., Kridl, J.C., Radke, S.E., and Huang, A.H.C. (1991). Maize oleosin is correctly targeted to seed oil bodies in Brassica napus transformed with the maize oleosin gene. Proc. Natl. Acad. Sci. USA 88 6181–6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprince, O., vanAelst, A.C., Pritchard, H.W., and Murphy, D.J. (1998). Oleosins prevent oil-body coalescence during seed imbibition as suggested by a low-temperature scanning electron microscope study of desiccation-tolerant and -sensitive oilseeds. Planta 204 109–119. [Google Scholar]
- Lin, L.J., Tai, S.S.K., Peng, C.C., and Tzen, J.T.C. (2002). Steroleosin, a sterol-binding dehydrogenase in seed oilbodies. Plant Physiol. 128 1200–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C., Fulda, M., Wallis, J.G., and Browse, J. (2006). A high throughput screen for genes from castor that boost hydroxyl fatty acid accumulation in seed oils of transgenic Arabidopsis. Plant J. 45 847–856. [DOI] [PubMed] [Google Scholar]
- Mansfield, S.G., and Briarty, L.G. (1991). Cotyledon cell development in Arabidopsis thaliana during reserve deposition. Can. J. Bot. 70 151–164. [Google Scholar]
- Meins, F. (2000). RNA degradation and models for post-transcriptional gene silencing. Plant Mol. Biol. 43 261–273. [DOI] [PubMed] [Google Scholar]
- Moloney, M. (1999). Seed oleosins. In Seed Proteins, P.R. Shrewy and R. Casey, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 781–806.
- Murashige, T., and Skoog, F.A. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15 473–497. [Google Scholar]
- Murphy, D. (1993). Structure, function and biogenesis of storage lipid bodies and oleosins in plants. Prog. Lipid Res. 32 247–280. [DOI] [PubMed] [Google Scholar]
- Murphy, D.J. (1990). Storage lipid bodies in plants and other organisms. Prog. Lipid Res. 29 299–324. [PubMed] [Google Scholar]
- Murphy, D.J., and Vance, J. (1999). Mechanisms of lipid-body formation. Trends Biochem. Sci. 24 109–115. [DOI] [PubMed] [Google Scholar]
- Niwa, Y., Kato, T., Tabata, S., Seki, M., Kobayashi, M., Shinozaki, K., and Moriyasu, Y. (2004). Disposal of chloroplasts with abnormal function into the vacuole in Arabidopsis thaliana cotyledon cells. Protoplasma 223 229–232. [DOI] [PubMed] [Google Scholar]
- Perry, S.E., and Wang, H. (2003). Rapid isolation of Arabidopsis thaliana developing embryos. Biotechniques 35 278–281. [DOI] [PubMed] [Google Scholar]
- Platt-Aloia, K.A., and Thompson, W.W. (1981). Ultrastructure of mesocarp of mature avocado fruit and changes associated with ripening. Ann. Bot. (Lond.) 48 451–465. [Google Scholar]
- Richmond, S.W., vanRooijen, G.J.H., Holbrook, L.A., Abenes, L., and Moloney, M.M. (1997). Regulation of expression of oleosin isoform genes in Arabidopsis thaliana. Plant Physiol. 114 256–261. [Google Scholar]
- Ross, J.H.E., Sanchez, J., Millan, F., and Murphy, D.J. (1993). Differential presence of oleosins in oleogenic seed and mesocarp tissues in olive (Olea europaea) and avocado (Persea americana). Plant Sci. 93 203–210. [Google Scholar]
- Sadeghipour, H.R., and Bhatla, S.C. (2002). Differential sensitivity of oleosins to proteolysis during oilbody mobilization in sunflower seedlings. Plant Cell Physiol. 43 1117–1126. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schumann, U., Wanner, G., Veenhuis, M., Schmid, M., and Gietl, C. (2003). AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. USA 100 9626–9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slightom, J.L., Sun, S.M., and Hall, T.C. (1983). Complete nucleotide sequence of a french bean storage protein gene - Phaseolin. Proc. Natl. Acad. Sci. USA 80 1897–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, N.A., Singh, S.P., Wang, M.B., Stoutjesdijk, P.A., Green, A.G., and Waterhouse, P.M. (2000). Gene expression - Total silencing by intron-spliced hairpin RNAs. Nature 407 319–320. [DOI] [PubMed] [Google Scholar]
- Ting, J.T.L., Lee, K.Y., Ratnayake, C., Platt, K.A., Balsamo, R.A., and Huang, A.H.C. (1996). Oleosin genes in maize kernels having diverse oil contents are constitutively expressed independent of oil contents - Size and shape of intracellular oilbodies are determined by the oleosins oils ratio. Planta 199 158–165. [DOI] [PubMed] [Google Scholar]
- Tzen, J.T., and Huang, A.H. (1992). Surface structure and properties of plant seed oilbodies. J. Cell Biol. 117 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzen, J.T., Lie, G.C., and Huang, A.H. (1992). Characterization of the charged components and their topology on the surface of plant seed oilbodies. J. Biol. Chem. 267 15626–15634. [PubMed] [Google Scholar]
- Tzen, J.T.C., Cao, Y.Z., Laurent, P., Ratnayake, C., and Huang, A.H.C. (1993). Lipids, proteins, and structure of seed oilbodies from diverse species. Plant Physiol. 101 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzen, J.T.C., Lai, Y.K., Chan, K.L., and Huang, A.H.C. (1990). Oleosin isoforms of high and low molecular weights are present in the oilbodies of diverse seed species. Plant Physiol. 94 1282–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooijen, G.J., and Moloney, M.M. (1995. a). Plant seed oil-bodies as carriers for foreign proteins. Biotechnology (N. Y.) 13 72–77. [DOI] [PubMed] [Google Scholar]
- van Rooijen, G.J., and Moloney, M.M. (1995. b). Structural requirements of oleosin domains for subcellular targeting to the oilbody. Plant Physiol. 109 1353–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley, S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27 581–590. [DOI] [PubMed] [Google Scholar]
- Wohlleben, W., Arnold, W., Broer, I., Hillemann, D., Strauch, E., and Puhler, A. (1988). Nucleotide sequence of the phosphinothricin n-acetyltransferase gene from Streptomyces viridochromogenes-Tu494 and its expression in Nicotiana tabacum. Gene 70 25–37. [DOI] [PubMed] [Google Scholar]
- Yatsu, L.Y., and Jacks, T.J. (1972). Spherosome membranes: Half unit-membranes. Plant Physiol. 49 937–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung, E.C. (1990). A simple procedure to visualize osmicated storage lipids in semithin epoxy sections of plant tissues. Stain Technol. 65 45–47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.