Abstract
It is well established that polarized exocytosis is essential for fungal virulence. By contrast, the contribution of endocytosis is unknown. We made use of a temperature-sensitive mutant in the endosomal target soluble N-ethylmaleimide–sensitive factor attachment protein receptor Yup1 and demonstrate that endocytosis in Ustilago maydis is essential for the initial steps of pathogenic development, including pheromone perception and cell–cell fusion. Furthermore, spore formation and germination were drastically reduced, whereas colonization of the plant was only slightly inhibited. The function of endocytosis in the recognition of mating pheromone through the G protein–coupled pheromone receptor Pra1 was analyzed in greater detail. Biologically active Pra1–green fluorescent protein localizes to the plasma membrane and is constitutively endocytosed. Yup1ts mutants that are blocked in the fusion of endocytic transport vesicles with early endosomes are impaired in pheromone perception and conjugation hyphae formation. This is attributable to an accumulation of Pra1-carrying endocytic vesicles in the cytoplasm and the depletion of the receptor from the membrane. Consistently, strong Pra1 expression rescues the signaling defects in endocytosis mutants, but subsequent cell fusion is still impaired. Thus, we conclude that endocytosis is essential for recognition of the partner at the beginning of the pathogenic program but has additional roles in mating as well as spore formation and germination.
INTRODUCTION
Fungal pathogenicity often requires a transition from yeast-like to a hyphal growth form, which allows the pathogen to invade the host tissue (Gow et al., 2002). The dimorphic maize (Zea mays) smut fungus Ustilago maydis is amenable to molecular genetics and cell biological methods and thus became an excellent model system for fungal plant pathogenicity (Bölker, 2001; Martinez-Espinoza et al., 2002; Garcia-Pedrajas and Gold, 2004; Kahmann and Kämper, 2004). Pathogenic development is initiated by a mating reaction that involves two compatible haploid yeast-like cells, which recognize each other at the plant surface through a pheromone (mfa1/2)/pheromone receptor (Pra1/2) system (Bölker et al., 1992). Upon perception of compatible pheromone secreted by the mating partner, the endogenous mating pheromone (mfa) and the pheromone receptor genes are upregulated (Urban et al., 1996a). As a consequence, the yeast-like cells undergo a morphological switch and form conjugation hyphae that grow toward the pheromone source (Spellig et al., 1994; Snetselaar et al., 1996), fuse, and establish a dikaryon, which invades the epidermis and continues the pathogenic program inside the plant (Kahmann et al., 1995; Kämper et al., 1995). As polarized growth of hyphae has a key role in all of these steps, it is not surprising that the cytoskeleton and molecular motors, such as myosin-V, are essential for this initial phase of pathogenic development of U. maydis (Weber et al., 2003; Fuchs et al., 2005). Based on results in other fungi, it appears likely that they mediate the transport of growth supplies and lytic exoenzymes for exocytosis at the expanding hyphal tip (Geitmann and Emons, 2000). In addition, staining the endocytic pathway with the amphipathic dye FM4-64 provided indirect evidence for a role of endocytic membrane recycling in polar fungal growth (Hoffmann and Mendgen, 1998; Fischer-Parton et al., 2000; Wedlich-Söldner et al., 2000; Atkinson et al., 2002). Such endocytic recycling is well known in animal cells (Maxfield and McGraw, 2004). However, neither the actual cargo that might be recycled nor the impact of this process on fungal pathogenicity is known. Here, we elucidate the role of endocytosis and recycling from the early endocytic compartment in fungal pathogenesis. Using a temperature-sensitive endocytosis mutant that is defective in the target soluble N-ethylmaleimide-sensitive factor attachment protein receptor (t-SNARE) Yup1, which localizes to early endosomes (EEs) (Wedlich-Söldner et al., 2000), we investigated the role of EEs in the pathogenic development of U. maydis. We found that EEs are essential for the initiation of pathogenic development, cell–cell fusion, and spore formation and germination, while colonization of the plant is still possible. The initial defect in pheromone sensing is mainly attributable to a role of EEs in the recycling of endocytosed pheromone receptor back to the plasma membrane.
RESULTS
The t-SNARE Yup1 Colocalizes with the Early Endosomal Marker Rab5
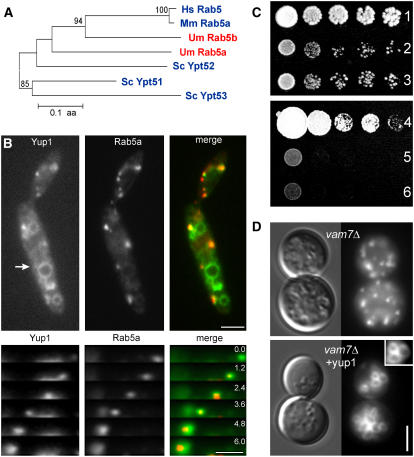
In a previous study, we identified Yup1 as an early endosomal t-SNARE protein required for functional endocytosis in U. maydis (Wedlich-Söldner et al., 2000). However, this was solely based on the use of the endocytic marker dye FM4-64 (Vida and Emr, 1995) that colocalized with Yup1-GFP (for green fluorescent protein) on EEs (Wedlich-Söldner et al., 2000). To further strengthen this conclusion, we generated a strain containing a fusion of GFP to Rab5a, a Rab5-like GTPase in U. maydis (Figure 1A) (Fuchs and Steinberg, 2005), and Yup1 fused to a double tag of red fluorescent protein (RFP2) (strain FB2GRab5aYup1R2). Both proteins colocalized and comigrated on small moving dots (94.8%, n = 94 Yup1-carrying endosomes; Figure 1B) but not on vacuoles (Figure 1B, arrow). As Rab5-GTPases are characteristic for EEs (Bucci et al., 1992), these results add strong support to our conclusion that Yup1 acts on EEs in U. maydis.
Figure 1.
Yup1 Colocalizes with the Early Endosomal RabGTPase Rab5a.
(A) Dendrogram of EE-specific Rab5GTPases from Homo sapiens (Hs), Mus musculus (Mm), Saccharomyces cerevisiae (Sc), and Ustilago maydis (Um).
(B) Colocalization and comovement (top panels) of Yup1 (red) with the early endosomal marker Rab5a (green), resulting in a light yellow color in the merged image. The arrow indicates the vacuolar localization of Yup1. Elapsed time is given in seconds. Bars = 3 μm (top) and 1 μm (bottom).
(C) Complementation of S. cerevisiae vam7Δ with Yup1. Row 1, SEY6210 + vector; row 2, vam7Δ + vector; row 3, vam7Δ + Yup1; row 4, FB2 + vector; row 5, FB2yup1ts + vector; row 6, FB2yup1ts + VAM7.
(D) FM4-64 staining of S. cerevisiae vam7Δ and vam7Δ complemented with Yup1. For details of FM 4-64 staining, see inset. Bar = 3 μm.
We noted previously that Yup1 and the vacuolar t-SNARE Vam7p from Saccharomyces cerevisiae (Wada and Anraku, 1992) share significant sequence similarity and a PX domain (Wedlich-Söldner et al., 2000). Ectopic expression of Yup1-GFP led to localization on EEs and the vacuolar tonoplast (Wedlich-Söldner et al., 2000) (Figure 1B, arrow). To visualize native levels of Yup1, we fused the endogenous yup1 to a double RFP tag (strain FB1Yup1R2), which resulted in a similar localization on EEs and the vacuole (data not shown). This argues for a nonessential function of Yup1 on vacuoles. To check whether Yup1 can substitute for Vam7p in homeotypic vacuolar fusion in S. cerevisiae, we transformed the vam7Δ strain with a CEN6 plasmid containing the Yup1 gene under the control of the ura3 promoter. The resulting mutant (for strain information, see Table 1) grew slightly better (Figure 1C, row 3) compared with vam7Δ carrying the empty vector (Figure 1C, row 2) but was still reduced in growth compared with the wild type transformed with empty vector (Figure 1C, row 1). Pulse–chase experiments using FM4-64 in vam7Δ mutants and vam7Δ cells expressing yup1 (Figure 1D, vam7Δ and vam7Δ + yup1) demonstrated that Yup1 is also able to rescue the defect in vacuole fragmentation, which is typical for vam7Δ mutants (Wada and Anraku, 1992). By contrast, expression of Vam7 did not rescue the phenotype in yup1ts mutants (Figure 1C, row 4 [wild-type control with empty vector], row 5 [yup1ts with empty vector], and row 6 [yup1ts + VAM7]), indicating that Yup1 performs additional essential roles in U. maydis that are most likely associated with endocytosis.
Table 1.
Genotypes of Strains Used in This Study
| Strains/Plasmids | Genotype | Reference |
|---|---|---|
| FB2GRab5aYup1R2 | a2b2/pOGFPRab5a/pOyup1RFP2 | This study |
| FB1Yup1R2 | a2b2 yup1-rfp2, hygR | This study |
| SEY6210 + vec | MATα leu 2-3, 112 ura3-52 his 3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9/ pRS316 | This study |
| Δvam7 + vec | MATα leu 2-3, 112 ura3-52 his 3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 vam7∷HIS3/pRS316 | This study |
| Δvam7 + YUP1 | MATα leu 2-3, 112 ura3-52 his 3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 vam7∷HIS3/pRS316YUP1 | This study |
| FB2+Vector | a2b2/pNEBUH | This study |
| FB2Yup1ts + Vector | a2b2 yup1ts/pNEBUH | This study |
| FB2Yup1ts + Vam7 | a2b2 yup1ts/pNEBUH_OVam7 | This study |
| FB1 | a1b1 | Banuett and Herskowitz (1989) |
| FB2 | a2b2 | Banuett and Herskowitz (1989) |
| FB1Yup1ts | a1b1 yup1ts | Wedlich-Söldner et al. (2000) |
| FB2Yup1ts | a2b2 yup1ts | Wedlich-Söldner et al. (2000) |
| FB1mG | a1b1/pmfa1GFP | Spellig et al. (1996) |
| FB1Yup1tsmG | a1b1 yup1ts/pmfaGFP | This study |
| FB1Pra1G | a1b1 pra1-gfp, hygR | This study |
| FB1Pra1G Yup1R | a1b1 pra1-gfp, hygR, /pOyup1RFP2 | This study |
| FB1Yup1tsPra1G | a1b1 yup1tspra1-gfp, hygR | This study |
| FB1Yup1tsoPra1G | a1b1 yup1tspra1-gfp, hygR/pOPra1GFP | This study |
| FB1Yup1tsmRPra1G | a1b1 yup1tspra1-gfp, hygR/pmfaRFP | This study |
| FB1Yup1tsmRoPra1G | a1b1 yup1tspra1-gfp, hygR/pmfaRFP/pOPra1GFP | This study |
| FB1oPra1 | a1b1/pOPra1 | This study |
| FB2oPra2 | a2b2/pOPra2 | This study |
| FB1Yup1tsoPra1 | a1b1 yup1ts/pOPra1 | This study |
| FB2Yup1tsoPra2 | a2b2 yup1ts/pOPra2 | This study |
| FB1oPra1_G | a1b1/pOPra1/pOGFP3 | This study |
| FB2oPra2_R | a2b2/pOPra2/pORFP2 | This study |
| FB1Yup1tsoPra1_G | a1b1 yup1ts/pOPra1/pOG | This study |
| FB2Yup1tsoPra2_R | a2b2 yup1ts/pOPra2/pOR | This study |
| pRS316 | URA3, CEN6 | Sikorski and Hieter (1989) |
| pRS316Yup1 | URA3, CEN6, Pura3yup1 | This study |
| pNEBUH | hygR, Uars | Weinzierl (2001) |
| pNEBUH_OVam7 | hygR, Uars, Potefvam7 | This study |
| pOyup1RFP2 | Potef-yup1-2x mrfp, cbxR | Lenz et al. (2006) |
| pmfa1GFP | Pmfa1-egfp, cbxR | Spellig et al. (1996) |
| pmfa1RFP | Pmfa1-mrfp, bleR | This study |
| pOPra1GFP | Potef-Pra1-gfp, cbxR | This study |
| pOPra1 | Potef-Pra1, cbxR | This study |
| pOPra2 | Potef-Pra2, cbxR | This study |
| pOG | Potef-3xgfp, hygR | This study |
| pOR | Potef-2xrfp, hygR | This study |
a, b, mating-type loci; P, promotor; -, fusion; bleR, phleomycin resistance; cbxR, carboxin resistance; hygR, hygromycin resistance; /, ectopically integrated; otef, constitutive promotor; egfp, enhanced green fluorescent protein; mrfp, monomeric RFP; pra1, pheromone receptor 1; m, mfa1, mating pheromone 1; Uars, Ustilago autonomously replicating sequence; yup1ts, temperature-sensitive allele of the endosomal t-SNARE yup1.
Endocytosis Is Required for the Pathogenicity of U. maydis
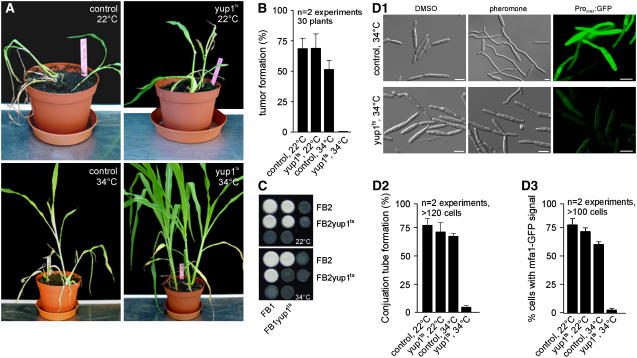
To determine the importance of endocytosis for pathogenicity, we infected 6-d-old maize plants with a mixture of control strains FB1 and FB2 as well as the temperature-sensitive endocytosis mutant strains FB1Yup1ts and FB2Yup1ts. Infected plants were incubated at both permissive (22°C) and restrictive (34°C) temperatures, and tumor formation and plant death were monitored at 14 d after infection. At 22°C, infection symptoms were found at similar rates in both wild-type and mutant infected plants (Figure 2A, control, 22°C and yup1ts, 22°C, and Figure 2B). However, at the restrictive temperature, yup1ts mutant cells failed to induce symptoms, whereas almost normal infection was observed for plants infected with control strains (Figure 2A, control, 34°C and yup1ts, 34°C, and Figure 2B). Plant infection requires the formation of dikaryotic filaments, which can be monitored on charcoal-containing agar plates. To further analyze the impaired virulence of yup1ts strains, we performed these mating assays using strains FB1Yup1ts and FB2Yup1ts as well as compatible control strains. At the permissive temperature, both compatible control and yup1ts cells fused to form a fuzzy white colony consisting of dikaryotic filaments (Figure 2C, top). By contrast, at 34°C, only the control cells formed fuzzy colonies, whereas yup1ts mutants were unable to form dikaryotic hyphae (Figure 2C, bottom), suggesting that EEs are required early in the infection process. However, a cross of a wild-type strain and a yup1ts mutant strain was dominated by the wild-type phenotype and resulted in fuzzy filaments.
Figure 2.
Characterization of Wild-Type and Conditional yup1ts Mutant Strains in Plant Infection Assays.
(A) Maize plants were infected with wild-type control strains and yup1ts mutants at 22 and 34°C.
(B) Quantification of tumor formation on infected maize plants at 22 and 34°C
(C) Cross of control strains FB1 × FB2 and yup1ts mutant strains FB1Yup1ts × FB2Yup1ts on charcoal-containing agar plates at 22 and 34°C.
(D) Filamentous growth and formation of conjugation hyphae were induced in strain FB1 and in FB1Yup1ts at 34°C. DMSO was used as a solvent control.
(D1) Pheromone perception was visualized using the mfa1 promotor–GFP fusion construct as a reporter at 34°C. Both conjugation hyphae formation and pheromone perception were quantified at 22 and 34°C. Bars = 5 μm.
(D2) Quantitative analysis of conjugation hyphae formation at 22 and 34°C.
(D3) Quantification of cells expressing pmfa-GFP after pheromone stimulation.
Pheromone Perception Is Defective in yup1ts Mutants
As a prerequisite for mating, compatible cells have to recognize the mating partner pheromone, which triggers a signal cascade and finally leads to increased expression of their own pheromone and the formation of conjugation hyphae (Urban et al., 1996b). Our mating assays on yup1ts mutants suggested that EE function is essential for the formation of dikaryotic filaments. Therefore, we tested whether the hyphal growth of conjugation tubes is also mediated by this process. We mimicked the presence of a mating partner by the addition of synthetic pheromone (Spellig et al., 1994). After ∼6 h of incubation with synthetic pheromone at 22°C, both wild-type and mutant cells had formed long conjugation filaments to similar extents (Figure 2D2). In the presence of pheromone, conjugation hyphae were also formed in wild-type strains at 34°C (Figures 2D1, control, pheromone, and 2D2, control). However, pheromone did not induce the formation of conjugation hyphae in yup1ts mutants at 34°C (Figures 2D1 and 2D2, pheromone, yup1ts). Instead, mutant cells were thickened and showed the previously described abnormal morphology (Figure 2D1) (Wedlich-Söldner et al., 2000). This raised the possibility that the absence of mating hyphae in yup1ts mutants is attributable to a morphological defect. Alternatively, we considered it possible that the impaired formation of conjugation hyphae is a consequence of defects in pheromone sensing. Thus, we made use of a strain that expressed GFP under the control of the promoter of the mating pheromone gene (mfa1; strain FB1mG) (see Table 1 for genotypic details). In this strain, the addition of external pheromone or the presence of a mating partner induces the expression of GFP, which is visible at ∼2 h after pheromone addition (Spellig et al., 1996). Wild-type and yup1ts mutant cells were able to perceive supplemented pheromone, as indicated by the cytoplasmic GFP signal at the permissive temperature (Figure 2D3). A similar situation was found in wild-type cells at 34°C (Figures 2D1, Promfa1:GFP, control, and 2D3). By contrast, mutant cells did not express GFP at 34°C (Figures 2D1, Promfa1:GFP, Yup1ts, and 2D3). Thus, it is most likely that EE function is required for pheromone perception and, consequently, for conjugation hyphae formation.
The U. maydis Pheromone Receptor Pra1 Is Internalized via EEs
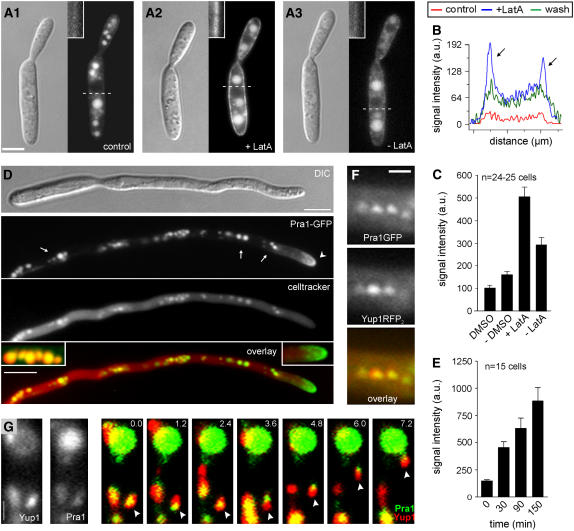
Pheromone perception in U. maydis wild-type strain FB1 requires the activity of the G-protein–coupled pheromone receptor Pra1 (Bölker et al., 1992), which is predicted to contain seven transmembrane segments and shares an overall similarity of 45% with Ste3, one of the two pheromone receptors of S. cerevisiae. Ste3 contains a signal sequence for constitutive endocytosis and ligand-induced endocytosis (Chen and Davis, 2000) that is not present in Pra1. Nevertheless, we considered it possible that impaired endocytosis of Pra1 via Yup1-tagged endosomes is responsible for the reduced pheromone sensing of the yup1ts mutant. Therefore, we fused GFP to the C terminus of the endogenous copy of Pra1 and observed its cellular distribution in strain FB1Pra1G. In nonstimulated yeast-like cells, the receptor is only weakly expressed (Urban et al., 1996a); consequently, only faint signals of the Pra1-GFP fusion protein were observed in the plasma membrane (Figure 3A1, inset), whereas most Pra1-GFP localized in the vacuoles (Figure 3A1), where the receptor is most likely degraded. However, disruption of F-actin by the inhibitor latrunculin A (LatA) enriched Pra1-GFP in the plasma membrane (Figures 3A2, inset, and 3C), and after washout of the drug, the receptor disappeared from the cell surface (Figures 3A3, inset, and 3C). The increase in Pra1-GFP content in the plasma membrane is best illustrated by line-scan analysis of the intensities of Pra1-GFP in the mother cell (Figure 3B; scanning lines are indicated in Figures 3A1 to 3A3). Whereas only faint signals were detected in DMSO-treated control cells (Figure 3B, red line), LatA treatment drastically increased the signal in the plasma membrane at the edge of the cell (Figure 3B, blue line, arrows), which decreased after removal of the drug (Figure 3B, green line). Disruption of actin is known to inhibit the endocytic removal of receptors from the cell surface (Kaksonen et al., 2003), suggesting that the increase in Pra1-GFP after the disruption of microfilaments is attributable to a defect in endocytic internalization. These results indicate that Pra1 is constitutively endocytosed in yeast-like cells of U. maydis, even when the ligand (mating pheromone) is not present.
Figure 3.
Localization of Pra1-GFP.
(A) Strain FB1PraG was treated with 10 μM of the actin inhibitor LatA (A2) or with the solvent DMSO (A1) for 45 min before washout of LatA (A3). Insets highlight the amounts of Pra1-GFP in the membrane. Bar = 3 μm.
(B) Line-scan analysis of Pra1-GFP signal intensities. Scanning lines (dashed lines) are indicated in (A). a.u., arbitrary units.
(C) Quantitative analysis of the intensity of Pra1-GFP in the tip of conjugation hyphae of strain FB1Pra1G in the presence or absence of LatA.
(D) Incubation of yeast-like cells of strain FB1PraG with synthetic pheromone for 3 h induced the formation of conjugation hyphae (DIC [differential interference contrast]). Pra1-GFP (green) localized to the tip of the hyphae (arrowhead), where it formed an apical cap (overlay, right inset). In addition, Pra1-GFP localized in subapical organelles (Pra1-GFP, arrows) that costained with CellTracker Blue (red; celltracker), indicating that they were vacuoles (overlay, left inset; colocalization results in yellow). Bar = 5 μm.
(E) Analysis of Pra1-GFP signal intensities at the tip of conjugation hyphae at different times after treatment with LatA.
(F) Colocalization of Pra1-GFP (green) with the double RFP-tagged EE markerYup1 (red) on small motile organelles (yellow in overlay). Bar = 1 μm.
(G) Time-lapse microscopy of Yup1RFP2 (red) with Pra1GFP (green), resulting in yellow in the merged images. Arrowheads indicate moving vesicles. Bar = 1 μm.
After the addition of pheromone, cells of strain FB1Pra1G formed conjugation hyphae (Figure 3D, DIC), indicating that the Pra1-GFP fusion protein was fully functional. In these conjugation hyphae, Pra1-GFP localized to the plasma membrane in a cap-like manner at the growing hyphal apex (Figure 3D, Pra1-GFP, arrowhead and right inset in the overlay image). In addition, Pra1-GFP accumulated in vacuoles that were stained with the vacuolar dye CellTracker Blue (Figure 3D, arrows and left inset in the overlay image). Similar to yeast-like cells, disruption of F-actin by 10 μM LatA for up to 150 min led to a significant increase of Pra1-GFP in the plasma membrane at the tip region (Figure 3E), indicating that internalization of Pra1 is actin-dependent in conjugation hyphae as well, whereas the delivery of new receptor does not require the actin cytoskeleton.
Endocytic Recycling of Pra1 Is Impaired in the yup1ts Mutant
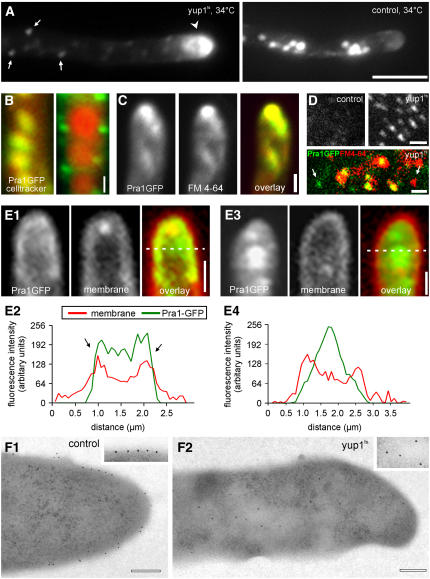
We next asked whether endocytosis of Pra1-GFP involves the Yup1-carrying EEs. This notion was supported by the observation that Pra1-GFP colocalized with Yup1RFP2 (Figure 3F) and rapidly moved on EEs (Figure 3G; strain FB1Pra1GYup1R2). To obtain further evidence for a role of EEs in Pra1-GFP processing, control cells and yup1ts mutants were stimulated with pheromone under permissive conditions for 2 h and subsequently were shifted to the restrictive temperature for an additional 2 h. Although this treatment had no effect on the Pra1-GFP distribution of control hyphae (Figure 4A, control), Pra1-GFP accumulated in yup1ts mutant hyphae in small immobile dots within the cytoplasm (Figure 4A, yup1ts, arrows), with most Pra1-GFP found in the hyphal tip (Figure 4A, yup1ts, arrowhead). In yup1ts mutants, Pra1-GFP no longer colocalized with vacuoles that were stained with CellTracker Blue (Figure 4B, cf. yup1ts and control) but accumulated in small aggregates that might represent clusters of primary endocytic vesicles. Consistently, the apical Pra1-GFP cluster colocalized with the endocytic marker dye FM 4-64 (Figure 4C), suggesting that Pra1-GFP is internalized in yup1ts mutant hyphae but accumulates in small primary endocytic vesicles within the cytoplasm. This notion is supported by the fact that whole cell extracts of strain FB1Yup1tsPra1G contained numerous small Pra1-GFP–carrying vesicles (Figure 4D), which mainly colocalized with FM4-64 (Figure 4D, bottom; colocalization results appear yellow). These vesicles were much less abundant in extracts of strain FB1PraG that was treated in similar ways (Figure 4D, control), again indicating that primary endocytic vesicles accumulate in yup1ts mutants as a result of the fusion defect at EEs. However, it is important to note that these cell extracts also contain vesicles that either carried Pra1-GFP or were only stained with FM4-64 (Figure 4D, arrows). Although the Pra1-GFP–stained vesicles could be secretory vesicles, the existence of endocytic transport vesicles that do not contain the pheromone receptor argues for additional endocytic pathways for the uptake of FM 4-64.
Figure 4.
Localization of Pra1-GFP in the yup1ts Mutant Background.
(A) Localization of Pra1-GFP in strain FB1Yup1tsPra1G and the control strain FB1Pra1G that were stimulated with pheromone for 2 h at 22°C before the shift to 34°C for 2 h. Arrows indicate primary endocytic vesicles. The arrowhead points to the accumulation of Pra1-GFP at the tip. Bar = 5 μm.
(B) Colocalization of vacuoles stained with CellTracker Blue (red) and Pra1-GFP (green) in strains FB1Pra1G (control) and FB1Yup1tsPra1G at 34°C. Colocalization results are in yellow. Bar = 1 μm.
(C) Double labeling experiments in yup1ts mutants demonstrate that Pra1-GFP (green) and endocytic membranes stained with FM 4-64 (red) colocalize in the apical cytoplasm in yup1ts cells. Bar = 1 μm.
(D) Pra1-GFP–containing vesicles obtained from the protein extracts of pheromone stimulated and shifted the control strain and FB1Yup1tsPra1G at 34°C. The merged image represents vesicles isolated from FB1Yup1tsPra1G at 34°C, which was also incubated with FM 4-64. Bars = 1 μm.
(E) Analysis of the localization of Pra1-GFP (green) with respect to the plasma membrane (red) in control cells and yup1ts mutant hyphae at 34°C. A line scan of the intensities demonstrates that FM4-64 and Pra1-GFP colocalized at the edges of the cell ([E2], arrows; scanning line indicated in [E1], overlay). In yup1ts cells, Pra1-GFP was depleted from the plasma membrane and accumulated within the cytoplasm in the hyphal tip ([E3] and [E4]). Bars = 1 μm.
(F) Immunolocalization of Pra1-GFP in control (F1) and yup1ts mutant (F2) cells. Bars = 0.5 μm.
To gain a more detailed insight into the defects of yup1ts mutants, we investigated the spatial relation between the plasma membrane and Pra1-GFP signals in the hyphal apex. Simultaneous addition of FM4-64 and 0.5% formaldehyde to wild-type and yup1ts conjugation hyphae allowed the dye to incorporate into the plasma membrane but blocked further internalization, whereas the GFP-tagged receptor was still detectable. In control cells, Pra1-GFP colocalized with FM4-64 in the plasma membrane at the hyphal apex (Figure 4E1), which is nicely illustrated by line-scan analysis of the signal intensities of Pra1-GFP and FM-4-64 (Figure 4E2; the scanning line is indicated in Figure 4E1). By contrast, in yup1ts mutant, Pra1-GFP was depleted from the plasma membrane but accumulated in the apical cytoplasm (Figures 4E3 and 4E4). Immunogold labeling of Pra1-GFP confirmed that the receptor was located in the cell periphery in control hyphae at 34°C (Figure 4F1), whereas the majority of receptor molecules in the yup1ts mutant hyphae were found within the cytoplasm (Figure 4F2).
Recycling of the Pheromone Receptor
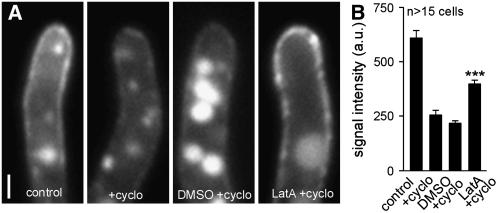
The results described above indicate that the uptake of Pra1-GFP into EEs is required to maintain enough active receptor in the plasma membrane, suggesting that the receptor is recycled to the surface for additional rounds of pheromone binding. We next attempted to gain more direct evidence for Pra1 recycling in conjugation hyphae of U. maydis. In a first set of control experiments, we confirmed that 100 μg/mL cycloheximide for 45 min fully inhibited protein biosynthesis in U. maydis (see Supplemental Figure 1 online). Next, we incubated conjugation hyphae of strain FB1Pra1G with or without cycloheximide for 45 min. The block of protein synthesis led to a drastic decrease in Pra1-GFP signal intensity in the plasma membrane (Figures 5A, control and +cyclo, and 5B), which demonstrates that the synthesis and secretion of new receptor was the major source for the exposed receptor. When this treatment was followed by an additional 120-min incubation in DMSO/cycloheximide, the amount of Pra1-GFP decreased further and intensively stained vacuoles appeared (Figures 5A, DMSO + cyclo, and 5B). By contrast, 120 min of cycloheximide treatment in combination with LatA led to a significant increase of Pra1-GFP in the plasma membrane (Figures 5A, LatA + cyclo, and 5B) (P = 0.0001). This increase, under our artificial conditions in which the synthesis of new receptor and initial endocytic uptake are blocked, indicates that a significant amount of Pra1-GFP is stored in the endocytic pathway and recycles back to the plasma membrane and EEs.
Figure 5.
Quantitative Analysis of Pra1-GFP Recycling.
(A) Pra1-GFP signals in the tip of conjugation hyphae. Cells were treated with water (control) or cycloheximide (+cyclo) for 45 min. Hyphae were incubated for an additional 120 min with LatA/cycloheximide or DMSO/cycloheximide. Bar = 1 μm.
(B) Quantitative analysis of corresponding Pra1-GFP signals in (A). *** P < 0.001. a.u., arbitrary units.
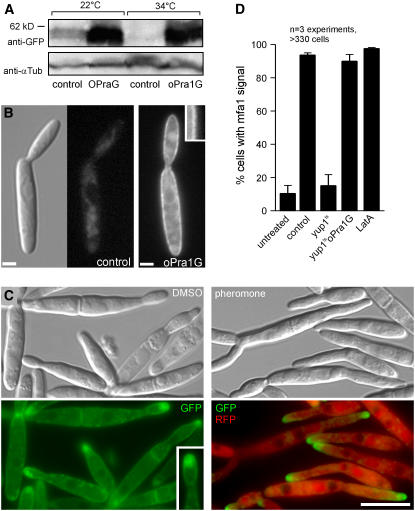
Constitutive Expression of Pra1 Restores Pheromone Perception in the yup1ts Mutant
Our results indicated that yup1ts mutants are defective in receptor recycling and that the pheromone receptor accumulates in the cytoplasm, which decreases the amount of exposed Pra1 on the cell surface. To test whether reduced amounts of Pra1-GFP are indeed responsible for the perception defects in yup1ts mutants, we increased the amount of receptor by expressing pra1-gfp under the control of the strong otef promoter (strain FB1Yup1tsoPraG). Protein gel blot analysis confirmed that receptor protein levels were increased ∼10-fold in the mutant strain at both 22 and 34°C (Figure 6A) and that most Pra1-GFP was concentrated in the plasma membrane (Figure 6B2, oPra1G, inset), whereas Pra1-GFP was not visible in the control strain FB1Yup1tsPra1G at 22°C (Figure 6B1). To monitor the ability to perceive pheromone, we next integrated the RFP under the control of the mfa1 promoter into this mutant. After 2 h at 34°C, this strain (FB1Yup1tsmRoPraG) showed a morphology defect (Figure 6C, DMSO) that was characteristic of mutants impaired in EE function (Wedlich-Söldner et al., 2000). Under these conditions, Pra1-GFP clustered in cytoplasmic accumulation near the growth region (Figure 6C, Pra1-GFP, inset), reminiscent of the cytoplasmic Pra1-GFP cluster in yup1ts conjugation hyphae (Figure 4A). In addition, Pra1-GFP was still localized to the plasma membrane, indicating that endocytosis is not able to remove the high excess of constitutively expressed Pra1-GFP from the surface (Figure 6C, Pra1-GFP, inset). Consistent with the notion that a lack of receptor is responsible for the defect in pheromone perception, treatment with synthetic pheromone now induced the expression of RFP (Figures 6C, pheromone, and 6D, yup1tsoPra1G). By contrast, but consistent with the results described above, pheromone treatment induced RFP expression only in a very minor fraction of mutant cells when Pra1-GFP was expressed under its native pra1 promoter in the yup1ts background (Figure 6D, yup1ts; strain FB1Yup1tsmRPraG). These results demonstrate that high levels of the pheromone receptor restore the defect in pheromone perception. Finally, we tested whether endocytosis of Pra1-GFP is required for pheromone perception. We disrupted F-actin in strain FB1mG with 10 μM LatA for 45 min and added synthetic pheromone/LatA to these cells. Confirming similar results in S. cerevisiae (Rohrer et al., 1993), in these cells the pheromone induced the mfa1 promoter, as indicated by GFP expression (Figure 6D, LatA), suggesting that initial steps of receptor internalization are not needed for the detection of the mating partner. Together, these results strongly support the notion that recycling via EEs is required to maintain steady state levels of the receptor at the cellular surface during the initial step of pheromone perception.
Figure 6.
Complementation of yup1ts Mutants.
(A) Protein gel blots showing levels of Pra1-GFP in cell extracts of the yup1ts mutant background at native levels and Pra1-GFP expressed under the control of the constitutive otef promotor.
(B) Localization of Pra1-GFP in yup1ts (FB1Yup1tsPraG) and with additional expression of Pra1-GFP under the control of the constitutive otef promotor (FB1Yup1tsoPra1GFP) at 22°C. High levels of pra1-gfp expression increased the amount of receptor in the plasma membrane (inset). Bars = 2 μm.
(C) Strain FB1Yup1tsmRoPraG was grown at 34°C and stimulated with synthetic pheromone or DMSO as a control. The inset highlights the increased membrane signal of Pra1-GFP. Bar = 10 μm.
(D) Quantification of the response to synthetic pheromone of control and yup1ts cells treated with synthetic pheromone or synthetic pheromone and LatA. Bars represent the percentage of cells that show mfa promoter–induced RFP expression. Error bars represent values ± sd.
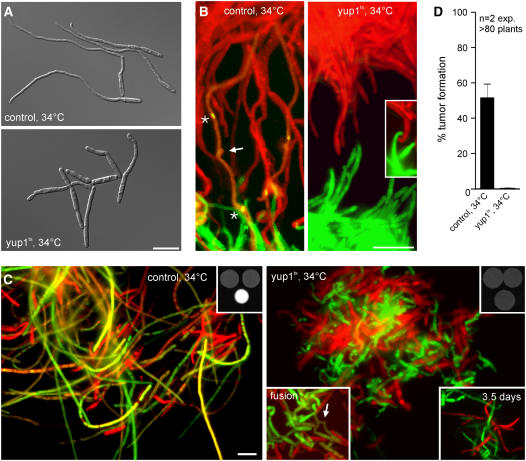
Endocytosis Is Essential for Cell–Cell Fusion
Restored pheromone perception in FB1yup1tsoPra1 (Figure 7A, yup1ts) led to the formation of short and irregular conjugation hyphae in the presence of synthetic pheromone (Figure 7A, cf. with control [strain FB1oPra1]). We next asked whether these conjugation hyphae are able to orient themselves in a gradient of pheromone and mediate cell–cell fusion. Therefore, we spotted compatible strains that expressed cytoplasmic GFP or RFP across from each other on water agar slides at 34°C. In these confrontation assays, control cells (strains FB1oPra1_G and FB2oPra2_R) formed long filaments that grew toward the partner and fused, which resulted in a faint yellow color in the merged image (Figure 7B, control; the arrow indicates the site of fusion, and asterisks mark nuclei). Cells of the compatible yup1ts mutants (FB1Yup1tsoPra1_G and FB2Yup1tsoPra2_R) grew toward the mating partner (Figure 7B, yup1ts, inset) but were unable to bridge large distances (see Supplemental Figure 2 online). However, under these conditions, no cell–cell fusion was detected in the yup1ts mutants, suggesting that endocytosis might be required during this step. In agreement with this, a mixture of the compatible yup1ts mutants did not form a fuzzy white colony, and only rarely were yellow dikaryotic cells found at 34°C (Figure 7C, yup1ts; the top right inset shows colony appearance, fusion). This was in striking contrast with the result of control experiments (Figure 7C, control). Cell–cell fusion did not increase in yup1ts cells even after 3.5 d at 34°C (Figure 7C, yup1ts, bottom right inset), indicating that endocytosis is essential for cell–cell fusion during early pathogenic development. Consequently, in plant infection assays, no symptoms were seen after infection with compatible yup1ts cells that constitutively express the pheromone receptor Pra1 or Pra2 at 34°C (Figure 7D). Together, these results demonstrate that endocytosis is essential for mating in U. maydis.
Figure 7.
Mating and Fusion Ability of the yup1ts Mutant Constitutively Expressing Pra1.
(A) Conjugation hyphae formation of wild-type and yup1ts cells expressing otef Pra1 with synthetic pheromone to analyze the ability to form conjugation hyphae.
(B) Confrontation assay of mating partners expressing otef Pra1 as well as cytoplasmic GFP or RFP to identify their mating types. Asterisks in the control panel indicate the nuclei in the haploid conjugation hyphae. Their fusion site to form the dikaryotic filament is marked by the arrow. Fused hyphae express both GFP and RFP, resulting in yellow. yup1ts cells did not fuse (inset).
(C) Assay for the fusion of GFP- or RFP-labeled mating partners on charcoal plates after incubation overnight. Fused hyphae express both RFP and GFP, resulting in yellow. The top insets show the overall colony morphology observed after overnight incubation at 34°C. The bottom left inset in the yup1ts panel shows a rare fusion event of mutant cells. The bottom right inset depicts hyphae that have been incubated for 3.5 d. Bars = 10 μm.
(D) Quantification of tumor formation after infection of maize plants with control and yup1ts mutant strains both expressing additional Pra1/Pra2 under the control of the constitutive otef promotor. Error bars represent values ± sd.
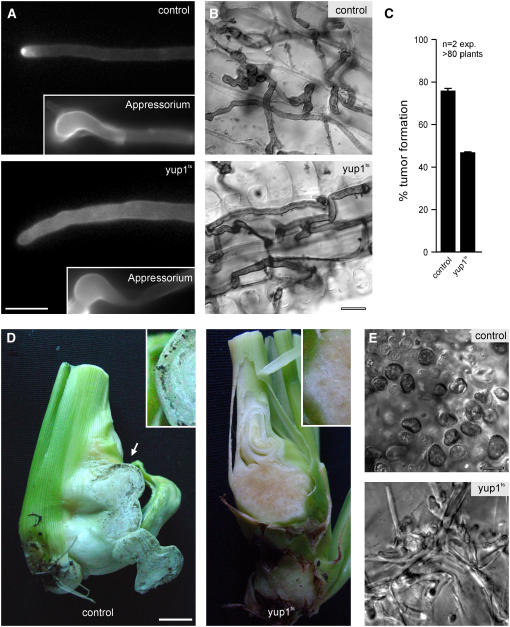
Plant Colonization Is Only Slightly Affected in yup1ts Cells
Next, we analyzed the importance of endocytosis at later stages of pathogenic development. We inoculated plants with compatible wild-type strains (FB1 and FB2) and yup1ts mutants (FB1Yup1ts and FB2Yup1ts) at the permissive temperature and allowed them to complete the mating reaction before shifting them to higher temperature (see Methods). Under these conditions, yup1ts mutants formed thicker and more irregularly shaped hyphae (Figure 8A, cell wall stained with calcofluor white; cf. yup1ts with control), which were nevertheless able to form appressoria (Figure 8A, insets) and entered the plant at the restrictive temperature (Figure 8B). Further time course experiments covering the first 7 d did not reveal any significant differences between control hyphae and yup1ts mutant hyphae inside the plant tissue (Figure 8B), although yup1ts cells appeared slightly thicker and were sometimes slower in colonization of the host. Consequently, yup1ts infected plants that were incubated for 3 d at 22°C before the shift to the restrictive temperature showed tumor formation (Figure 8C). Interestingly, tumors in the yup1ts mutant infected plants were observed only at the stem and not in higher parts of the plant (data not shown). However, these tumors did not contain any teliospores even at 19 d after infection (Figures 8D and 8E), although fungal material was detected in the tumors of plants infected with yup1ts mutant strains (Figure 8E; cf. with control). In summary, these findings indicate that endocytosis is less important for growth within the plant but is essential for the formation of teliospores.
Figure 8.
Analysis of yup1ts Mutants in Planta.
(A) Control and yup1ts hyphae stained with calcofluor after incubation at 22°C for 14 h and subsequent shift to 34°C. Appressoria that penetrate the plant surface are formed by control and yup1ts hyphae (insets). Bar = 5 μm.
(B) Images of Chlorazole Black E–stained control and yup1ts hyphae after 1 d of incubation at 22°C and subsequent shift to 34°C for 1 to 2 d. Bar = 10 μm.
(C) Quantitative analysis of tumor formation in plants infected with control strains and yup1ts strains after incubation at 22°C for 3 d and subsequent shift to 34°C for a total of 14 d after infection. Error bars represent values ± sd.
(D) Whole tumors of infected plants were harvested 3 weeks after infection. Wild-type tumors contain black teliospores (arrow) that form at the edge of the tumor (control, inset). By contrast, yup1ts tumors are devoid of teliospores in the center as well as at the edge of the tumor (yup1ts). Bar = 1 cm.
(E) Higher magnification of tumor tissue confirmed the presence of teliospores in wild-type tumors (control), whereas teliospores were absent in yup1ts tumors and only hyphal fragments were visible (yup1ts). Bar = 10 μm.
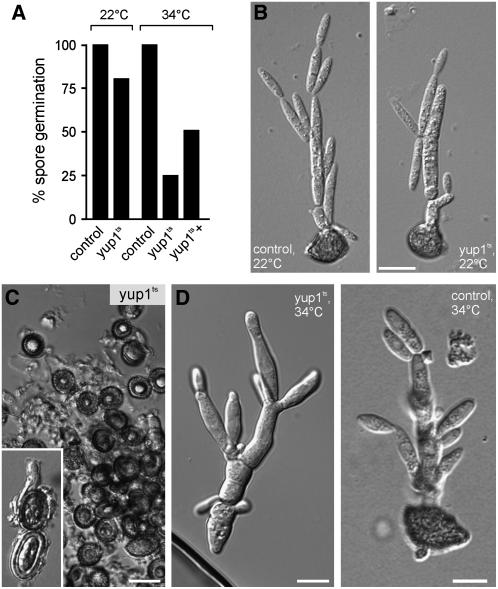
Teliospore Germination Is Mediated by Yup1
Finally, we investigated the ability of yup1ts mutants to germinate from teliospores generated at 22°C. Both wild-type and yup1ts teliospores germinated after ∼24 h at 22°C on CM-glucose–containing agar layers (Figures 9A, 22°C, and 9B). However, germination rates of yup1ts teliospores were greatly reduced at 34°C (Figures 9A, 34°C [cf. yup1ts and control], and 9C) and only slowly increased after prolonged incubation for 3 d (Figure 9A, 34°C, yup1ts+). In those cases in which yup1ts teliospores managed to germinate, the promycelium showed morphological alterations compared with the wild type (Figure 9D; note different magnifications in yup1ts and control). Thus, Yup1-mediated endocytosis participates in teliospore germination, and the functional impairment of endocytosis leads to reduced germination rates.
Figure 9.
Teliospore Germination.
(A) Quantification of teliospore germination at the permissive temperature (22°C) and the restrictive temperature (34°C) for control and yup1ts spores. At 34°C, germination of yup1ts teliospores is greatly impaired and only slightly increases after incubation (yup1ts+) for 3 d. Germination of control teliospores is set to 100%.
(B) Images of control and yup1ts teliospores germinated at 22°C on CM-glucose–containing agar slides. Bar = 10 μm.
(C) and (D) Images of control and yup1ts teliospores germinated at 34°C. Bars = 5 μm.
(C) Yup1ts spores after incubation for 1 d at 34°C (inset) and 3 d.
(D) When yup1ts teliospores managed to germinate, they again showed morphological alterations (left). Control teliospores germinated at 34°C after 1 d of incubation (right).
DISCUSSION
Many fungal pathogens invade the host tissue by exocytosis of enzymes and wall material at the expanding tip (Gow et al., 2002). Thus, it is not surprising that secretion is essential for the pathogenic development of fungi. However, a role of endocytosis in hyphal growth and development, and in particular in fungal pathogenicity, is still a matter of debate (Read and Kalkman, 2003). In this study, we used the plant pathogen U. maydis to gain insights into the importance of endocytosis in plant infection. We previously showed that Yup1, a t-SNARE, localized on rapidly moving organelles that colocalize with the endocytic marker dye FM4-64, indicating that Yup1 functions on EEs (Wedlich-Söldner et al., 2000). Here, we provide further evidence for this conclusion by colocalizing Yup1-RFP2 with a fusion protein of GFP and Rab5a. Rab5 proteins are characteristic markers for EEs (Bucci et al., 1992; Pfeffer, 2001). Therefore, it is very likely that Yup1 functions as a t-SNARE for the fusion of incoming primary endocytic vesicles with EEs. EEs cluster at growth sites, suggesting that they participate in membrane recycling processes during fungal growth (Wedlich-Söldner et al., 2000). The t-SNARE domain of Yup1 shows highest similarity to Vam7p, and both proteins contain a PX domain (Wedlich-Söldner et al., 2000). However, Vam7p is a t-SNARE that functions in homeotypic vacuole fusion (Wada and Anraku, 1992; Sato et al., 1998), whereas Yup1 functions on EEs (Wedlich-Söldner et al., 2000). Interestingly, Yup1-GFP also localized to vacuoles, although the yup1ts mutant did not show any defects in vacuolar fusion (Wedlich-Söldner et al., 2000), and Yup1 is able to substitute for the deletion of VAM7. This finding indicates that both proteins share common functions. However, it appears that Yup1 participates in additional processes, which are essential and most likely linked to the endocytic pathway, whereas Vam7p has a role in vacuole organization.
The data presented here demonstrate that Yup1-carrying EEs are crucial for the initial steps of pathogenicity as well as for spore formation and germination, because of the position of these organelles at the intersection between the degradation pathway, which leads to the vacuole, and the recycling pathway back to the cell surface (summarized in Maxfield and McGraw, 2004). Consequently, yup1ts mutants are completely impaired in both endocytic pathways. However, the initial uptake of material at the plasma membrane is still functional in yup1ts mutants, which leads to an accumulation of primary endocytic vesicles in the cytoplasm (Wedlich-Söldner et al., 2000). Indeed, we found that the pheromone receptor Pra1, which is constitutively endocytosed, accumulates in small vesicular structures in yup1ts mutants. A similar phenotype was reported in rcy1Δ cells of S. cerevisiae, in which an accumulation of the pheromone receptor Ste2 was found in an uncharacterized endocytic compartment (Wiederkehr et al., 2000).
This accumulation and the lack of endocytic recycling results in a depletion of the receptor from the cell surface and, consequently, in a defect in pheromone perception. This notion is supported by the observation that high levels of additional Pra1 rescue the pheromone perception defect of yup1ts mutants. Thus, recycling of the receptor ensures a certain level of receptor at the plasma membrane, which is crucial for cell–cell recognition during early pathogenic development. In the yeast S. cerevisiae, the closest homolog of Pra1, Ste3, is also constitutively endocytosed and recycled back to the plasma membrane once pheromone ligand has bound (Chen and Davis, 2000). This process most likely allows the cell to reuse the receptor, but it was also suggested to polarize the receptor at the growth region, which guarantees high efficiency chemotropic growth of the cell within the pheromone gradient of the partner (Chen and Davis, 2000). Indeed, we found that Pra1 concentrates at the tip of conjugation hyphae, and this apical cap is lost after the block of endocytic internalization by the disruption of F-actin with LatA (see Supplemental Figure 3 online). Therefore, receptor recycling not only saves the cell energy but also might help to concentrate the receptor at certain regions in the plasma membrane to recognize the pheromone gradient provided by the mating partner.
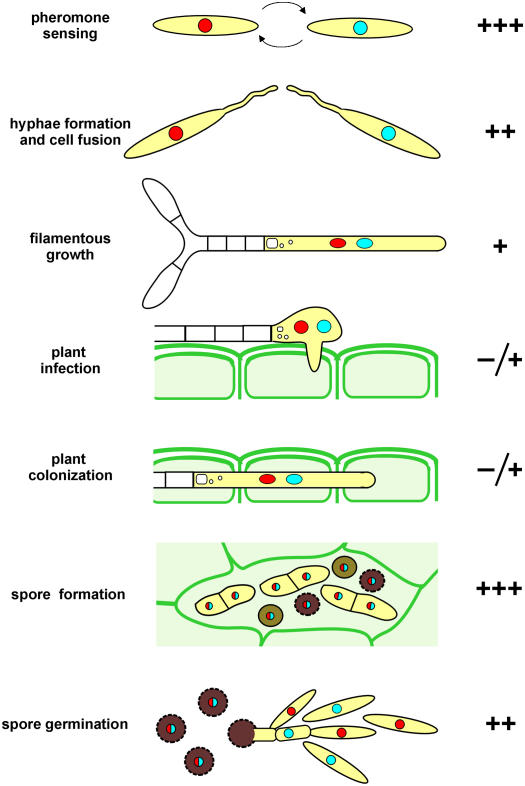
Our experiments argue that endocytosis is also essential for cell–cell fusion during mating, for spore formation, and is important for spore germination (Figure 10). By contrast, in yup1ts mutants, plant invasion and colonization were only slightly impaired, although we found that yup1ts hyphae showed morphological defects that were described previously for dikaryotic hyphae on agar plates (Wedlich-Söldner et al., 2000). At present, we can only speculate about the endocytosed cargos during these stages of pathogenic development in U. maydis. Several components required for cell fusion have been identified in the yeast S. cerevisiae (summarized in White and Rose, 2001); however, whether these compounds cycle between endosomes and the plasma membrane remains to be elucidated. The yeast α-factor transporter Ste6 undergoes endocytic recycling (Kelm et al., 2004; Schmitz et al., 2005) and has additional roles in cell–cell fusion (Elia and Marsh, 1996). Moreover, it was demonstrated that chitin synthases, which participate in wall formation, undergo endocytic recycling in S. cerevisiae (Ziman et al., 1996), and the same might hold true for U. maydis (U. Fuchs and G. Steinberg, unpublished data). Finally, experiments using FM4-64 suggest that endocytosis occurs in hydrated conidia of Magnaporthe grisea (Atkinson et al., 2002), which confirms our observation that spore germination requires endocytic processes. Again, no endocytic cargo is known, which emphasizes the need to further investigate this essential process in fungi.
Figure 10.
Importance of yup1-Mediated Endocytosis during the Life Cycle and Pathogenic Development of U. maydis.
Pathogenic development of U. maydis is initiated by pheromone sensing of two compatible mating partner cells. During this initial step, which is the basis for initiation of the pathogenic program, endocytosis is essential (+++). However, if pheromone perception is functional, the subsequent formation of conjugation tubes is possible even in the absence of endocytosis, whereas endocytosis is crucial for cell fusion (++). Growth of dikaryotic hyphae relies only to a small extent on endocytosis. Similarly, penetration of the plant and growth inside the plant are possible in the absence of endocytosis (+), whereas endocytosis becomes necessary again for the formation (+++) and germination (++) of teliospores.
METHODS
Strains and Plasmids
Standard protocols were followed for plasmid construction and DNA isolation (Sambrook et al., 1989). Complementation of the Saccharomyces cerevisiae vamΔ strain was done using the CEN6 plasmid pBR316 (Sikorski and Hieter, 1989) containing the Yup1 gene under the control of the URA3 promotor according to standard protocols (Guthrie and Fink, 1991). Complementation of the yup1ts mutant was tested using a self-replicating plasmid containing the Vam7 gene under the control of the otef promotor (Spellig et al., 1996). Ustilago maydis transformation was performed as described previously (Schulz et al., 1990). All U. maydis strains are listed in Table 1. Throughout the study, FB1 and FB2 were used as control strains (Banuett and Herskowitz, 1989), and studies in the yup1ts mutant background were done in the temperature-sensitive yup1ts mutants FB1Yup1ts and FB2Yup1ts (Wedlich-Söldner et al., 2000). We used the strain FB1mfaG (Spellig et al., 1996), which has the promotor of the pheromone gene mfa1 fused to GFP, to visualize pheromone perception in the wild-type background. We ectopically integrated pmfaG to visualize pheromone perception in the yup1ts mutant background. The pheromone receptor Pra1 was visualized by C-terminal fusion of Pra1 with eGFP using the construct pPra1GFP, which was integrated into the endogenous locus in the wild-type background (FB1Pra1G) and the yup1ts mutant background (FB1yup1tsPra1G). For supplementation of FB1Yup1tsPra1G with additional pheromone receptor molecules, a Pra1-GFP fusion construct under the control of the constitutive otef promotor (Spellig et al., 1996) (potefPra1GFP) was ectopically integrated in the strain, and the ability to perceive pheromone was assayed by ectopic integration of the mfa1 promotor fused to a single RFP (Promfa1RFP) in the same strain (FB1Yup1tsmfaRPra1GoPra1G). Integration of all constructs into the endogenous loci was confirmed by DNA gel blotting (data not shown).
Growth Conditions and Formation of Conjugation Hyphae
S. cerevisiae strains were grown in yeast extract/peptone/dextrose medium or in yeast synthetic dropout medium without URA (Guthrie and Fink, 1991) for selection. U. maydis strains were grown overnight at 28°C in liquid complete medium (Holliday, 1974) complemented with 1% glucose to OD600 = 0.5 to 0.6. Temperature-sensitive strains (for details, see Table 1) were incubated in the same medium at 22°C overnight. For inhibition of Yup1 function, temperature-sensitive yup1ts strains were shifted from the permissive temperature (22°C) to the restrictive temperature (34°C) and incubated for 2 h before starting experimental setups. The formation of conjugation hyphae was induced by the addition of 0.5 μL of pheromone a2 of U. maydis (2.5 μg/μL stock in DMSO, final concentration 2.5 × 10−3 μg/μL) (Szabo et al., 2002) to 500 μL of cell suspension in a 2-mL reaction tube and incubation for 6 h at 22°C and 200 rpm. To assay the ability of yup1ts mutant strains to form conjugation hyphae and to express PPro:GFP, pheromone stimulation was induced at 34°C after preincubation of control strains and yup1ts strains for 2 h or longer (Figure 7A, 21 h) at 34°C. Conjugation tube formation of FB1Pra1G and FB1Yup1tsPra1G was allowed for 2 h at the permissive temperature (as described above), and strains were further incubated for 2 h at 34°C.
Plant Infection Assays, Mating on Charcoal, and Confrontation Assays
Pathogenic development of wild-type and yup1ts mutant strains was assayed by plant infections of the maize (Zea mays) variety Early Golden Bantam (Olds Seeds). Strains FB1, FB2, FB1Yup1ts, and FB2Yup1ts were concentrated in water to OD600 ∼ 2, and compatible control and mutant strains were equally mixed before infection. Cell suspensions were injected at the basal stem of 6-d-old maize seedlings with a syringe. Infected plants were incubated at 22 or 34°C with 16 h of light in a phytochamber for 14 d. All plant infection assays after the initial infection assays (Figure 2) were done at 31°C, because the plants tolerated the heat stress better at that temperature than at 34°C. The growth phenotype of yup1ts mutant strains was the same at 31 and 34°C (data not shown). For quantification of tumor formation, plants with one or more tumors were counted, regardless of tumor size. For time course experiments, wild-type and yup1ts infected plants were incubated for 1 to 7 d at 22°C followed by a subsequent shift to 31°C and incubation for an additional 1 or 2 d.
To assay the formation of teliospores, plants were incubated for 19 d and thin sections of tissue were observed with the microscope. For charcoal mating assays, strains were crossed on charcoal-containing potato dextrose plates (Holliday, 1974) and incubated at 22 or 34°C. To assay the growth of strains toward the mating partner, they were spotted across from each other on 2% water agar slides and incubated overnight before direct observation with the light microscope.
Calcofluor and Chlorazole Black E Staining
Staining of infected plant samples with calcofluor followed the protocol delineated previously (Weber et al., 2003). Chlorazole Black E staining was done as described previously (Brachmann et al., 2003).
Spore Germination
Spores were generated through the incubation of infected plants at 22°C for ∼3 weeks. Tumors containing spores were dried at 22°C and minced using a mortar and pestle. The tumor material was then incubated in tetracycline solution (5 mg/mL stock; GERBU) for 30 min before washing in water and incubation in 1.5% copper(II) sulfate solution (Carl Roth). Samples were washed three times in water and were plated on 2% CM-glucose–containing agar slides and/or on plates with 20 μL of each: tetracycline, chloramphenicol (34 mg/mL stock; Carl Roth), and ampicillin (10 mg/mL stock; Carl Roth) in 25 mL agar. Slides containing the spore samples were incubated in a moist chamber at 22 or 34°C.
Colocalization Experiments, Membrane Staining, and Vacuole Staining
Colocalization of Yup1RFP2 and GFPRab5a was done after fixation with 1% formaldehyde (16% stock; Polysciences) in strain FB1Pra1GYup1RFP2 or after cooling of the same strain to 10°C and observation with a precooled objective to be able to visualize the otherwise rapidly moving molecules. Plasma membranes were stained by the addition of 1 μM of the endocytic marker dye FM4-64 (16 mM stock; Molecular Probes, Invitrogen) and the addition of 0.5% formaldehyde after 20 s, thereby inhibiting further endocytosis of the dye. Endocytosis and accumulation of FM4-64 in strain FB1Yup1tsPra1G was observed after incubation for 2 h at 34°C. Vacuolar staining was done using CellTracker Blue CMCA (1 mM stock; Invitrogen) at a final concentration of 100 μM and incubation for 15 min.
Inhibitor Studies
Inhibitor studies with LatA were done as described previously (Fuchs et al., 2005), and cells were incubated for 45 to 150 min. For inhibition of protein biosynthesis, cycloheximide (5 mg/mL stock in water; Sigma-Aldrich) was added to a final concentration of 100 μg/mL, and cells were incubated for 45 min with gentle shaking at 22°C before addition and further incubation with DMSO or LatA.
Light Microscopy, Image Processing, and Quantitative Data Analysis
For in vivo observations, cells from logarithmically growing cultures were placed on a thin 1% agarose layer and immediately observed using a Zeiss Axioplan II microscope. Epifluorescence was observed using filter sets for fluorescein isothiocyanate (BP500/20, FT515, BP535/30) and DsRed (HQ565/30). All microscopic observations were done using a CoolSNAP-HQ charge-coupled device camera (Photometrics) controlled by the imaging software MetaMorph (Universal Imaging). All measurements and image processing, including adjustment of brightness, contrast, and γ-values and two-dimensional deconvolution, were performed with MetaMorph and Photoshop (Adobe Systems). Statistical analysis by two-tailed t test at α < 0.05 was performed using Prism (GraphPad). All values are given as means ± sd unless stated otherwise.
Vesicle Extraction and Protein Gel Blot Analysis
Strains FB1Pra1G and FB1Yup1tsPra1G were grown overnight at 22°C. Cells (50 mL) were stimulated with synthetic pheromone as described above in 200-mL plastic conical centrifuge tubes for 4 h at 22°C or shifted to 34°C after 2 h of stimulation at 22°C. Cultures were then quickly harvested by centrifugation at 4°C for 10 min at 3000 rpm. Cells were resuspended in 10 mL of extraction buffer (100 mM PIPES, pH 6.9, 2 mM MgCl2, 1 mM EDTA, and 1 mM EGTA), centrifuged again for 10 min at 3000 rpm, washed in 2 mL of extraction buffer supplied with complete protease inhibitor (1 tablet/10 mL buffer; Roche Applied Sciences), centrifuged again, and resuspended in 1 pellet volume of extraction buffer with complete mini. Protein extracts were obtained by disruption of the frozen cell suspensions in a mixer mill (MM200; Retsch) and centrifuged for 10 min at 16,000 rpm for vesicle observation and 8,000 rpm, 4°C in a Biofuge Stratos centrifuge (Kendro). Subsequently, protein concentrations were determined using NanoDrop analysis (Nanodrop). For analysis of Pra1 protein levels, samples were supplied with SDS and Triton X-100 to a final concentration of 20% each and incubated on ice for 40 min to extract Pra1 from membranes. Protein concentrations of the supernatants were determined using a Coomassie gel. Supernatants were supplied with 6× Laemmli buffer and incubated at 65°C for 10 min before SDS gel electrophoresis. Proteins were separated on a 10% polyacrylamide gel and transferred onto a nitrocellulose membrane for 60 min at 400 mA in a wet blot chamber. The anti-GFP antibody (Roche Applied Sciences) was used at 1:5000 to detect GFP-tagged Pra1 fusion proteins according to standard procedures.
Electron Microscopy Studies
After high-pressure freezing and cryosubstitution as described by Straube et al. (2006), pheromone-stimulated cells were embedded in K11M (Polysciences Europe). Ultrathin sections were immunolabeled with a monoclonal anti-GFP antibody (clones 7.1 and 13.1; Roche Diagnostics) and a secondary antibody conjugated with 10-nm gold (G 7777; Sigma-Aldrich). The sections were poststained with uranyl acetate and lead citrate in an EM-Stain apparatus (Leica) and subsequently observed with an EM 900 transmission electron microscope (Zeiss SMT).
Bioinformatic Analysis
Protein sequences were downloaded from public databases (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi) and aligned by ClustalX (Thompson et al., 1997). t-SNARE prediction was done using http://us.expasy.org/tools/scanprosite/. Transmembrane domain prediction was done using http://www.cbs.dtu.dk/services/TMHMM-2.0/. Phylogenetic dendrograms were constructed by the minimum evolution method (MEGA version 2.1) (Kumar et al., 2001) with a nearest-neighbor-joining tree as starting point and 1000 bootstrap replicates.
Accession Numbers
Sequence data from this article can be found in the GenBank data library under accession numbers XP_758632 (Um Rab5b), P36017 (Sc Ypt51), NP_012939 (Sc Ypt52), P36019 (Sc Ypt53), NP_080163 (Mm Rab5a), P20339 (Hs Rab5), EAK83421.1 (Pra1), P31303 (Pra2), AAF62178.1 (Yup1), AAA99765.1 (Mfa1). The sequence information of UmRab5a was obtained from the public MIPS Ustilago maydis database (http://mips.gsf.de/genre/proj/ustilago/) under accession number um10615.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Cycloheximide Treatment.
Supplemental Figure 2. Confrontation Assay with yup1ts Mutants.
Supplemental Figure 3. Effects of LatA Treatment in Pra1-GFP–Expressing Conjugation Hyphae.
Supplementary Material
Acknowledgments
We thank Hedwich Teunissen for providing the Pra1-GFP plasmid and Philip Müller for the Pra1 and Pra2 templates for construction of the otefPra1 and otefPra2 constructs. We thank Michael Bölker for fruitful discussions and supportive ideas. William T. Wickner is acknowledged for sending the yeast vam7Δ strain. Stefan Brückner is thanked for assisting the yeast complementation experiments. U.F. was supported by the International Max Planck Research School.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Gero Steinberg (gero.steinberg@mpi-marburg.mpg.de).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.039388.
References
- Atkinson, H.A., Daniels, A., and Read, N.D. (2002). Live-cell imaging of endocytosis during conidial germination in the rice blast fungus, Magnaporthe grisea. Fungal Genet. Biol. 37 233–244. [DOI] [PubMed] [Google Scholar]
- Banuett, F., and Herskowitz, I. (1989). Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. USA 86 5878–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölker, M. (2001). Ustilago maydis—A valuable model system for the study of fungal dimorphism and virulence. Microbiology 147 1395–1401. [DOI] [PubMed] [Google Scholar]
- Bölker, M., Urban, M., and Kahmann, R. (1992). The a mating type locus of U. maydis specifies cell signaling components. Cell 68 441–450. [DOI] [PubMed] [Google Scholar]
- Brachmann, A., Schirawski, J., Müller, P., and Kahmann, R. (2003). An unusual MAP kinase is required for efficient penetration of the plant surface by Ustilago maydis. EMBO J. 22 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci, C., Parton, R.G., Mather, I.H., Stunnenberg, H., Simons, K., Hoflack, B., and Zerial, M. (1992). The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell 70 715–728. [DOI] [PubMed] [Google Scholar]
- Chen, L., and Davis, N.G. (2000). Recycling of the yeast a-factor receptor. J. Cell Biol. 151 731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia, L., and Marsh, L. (1996). Role of the ABC transporter Ste6 in cell fusion during yeast conjugation. J. Cell Biol. 135 741–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer-Parton, S., Parton, R.M., Hickey, P.C., Dijksterhuis, J., Atkinson, H.A., and Read, N.D. (2000). Confocal microscopy of FM4-64 as a tool for analysing endocytosis and vesicle trafficking in living fungal hyphae. J. Microsc. 198 246–259. [DOI] [PubMed] [Google Scholar]
- Fuchs, U., Manns, I., and Steinberg, G. (2005). Microtubules are dispensable for the initial pathogenic development but required for long-distance hyphal growth in the corn smut fungus Ustilago maydis. Mol. Biol. Cell 16 2746–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, U., and Steinberg, G. (2005). Endocytosis in the plant-pathogenic fungus Ustilago maydis. Protoplasma 226 75–80. [DOI] [PubMed] [Google Scholar]
- Garcia-Pedrajas, M.D., and Gold, S.E. (2004). Kernel knowledge: Smut of corn. Adv. Appl. Microbiol. 56 263–290. [DOI] [PubMed] [Google Scholar]
- Geitmann, A., and Emons, A.M. (2000). The cytoskeleton in plant and fungal cell tip growth. J. Microsc. 198 218–245. [DOI] [PubMed] [Google Scholar]
- Gow, N.A., Brown, A.J., and Odds, F.C. (2002). Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5 366–371. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., and Fink, G.R. (1991). Guide to Yeast Genetics and Molecular Biology. (San Diego, CA: Academic Press).
- Hoffmann, J., and Mendgen, K. (1998). Endocytosis and membrane turnover in the germ tube of Uromyces fabae. Fungal Genet. Biol. 24 77–85. [DOI] [PubMed] [Google Scholar]
- Holliday, R. (1974). Ustilago maydis. In The Handbook of Genetics, R.C. King, ed (New York: Plenum Press), pp. 575–595.
- Kahmann, R., and Kämper, J. (2004). Ustilago maydis: How its biology relates to pathogenic development. New Phytol. 164 31–42. [DOI] [PubMed] [Google Scholar]
- Kahmann, R., Romeis, T., Bölker, M., and Kämper, J. (1995). Control of mating and development in Ustilago maydis. Curr. Opin. Genet. Dev. 5 559–564. [DOI] [PubMed] [Google Scholar]
- Kaksonen, M., Sun, Y., and Drubin, D.G. (2003). A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115 475–487. [DOI] [PubMed] [Google Scholar]
- Kämper, J., Reichmann, M., Romeis, T., Bölker, M., and Kahmann, R. (1995). Multiallelic recognition: Nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell 81 73–83. [DOI] [PubMed] [Google Scholar]
- Kelm, K.B., Huyer, G., Huang, J.C., and Michaelis, S. (2004). The internalization of yeast Ste6p follows an ordered series of events involving phosphorylation, ubiquitination, recognition and endocytosis. Traffic 5 165–180. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. (2001). MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17 1244–1245. [DOI] [PubMed] [Google Scholar]
- Lenz, J.H., Schuchardt, I., Straube, A., and Steinberg, G. (2006). A dynein loading zone for retrograde endosome motility at microtubule plus-end. EMBO J. 25 2275–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Espinoza, A.D., Garcia-Pedrajas, M.D., and Gold, S.E. (2002). The Ustilaginales as plant pests and model systems. Fungal Genet. Biol. 35 1–20. [DOI] [PubMed] [Google Scholar]
- Maxfield, F.R., and McGraw, T.E. (2004). Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5 121–132. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S.R. (2001). Rab GTPases: Specifying and deciphering organelle identity and function. Trends Cell Biol. 11 487–491. [DOI] [PubMed] [Google Scholar]
- Read, N.D., and Kalkman, E.R. (2003). Does endocytosis occur in fungal hyphae? Fungal Genet. Biol. 39 199–203. [DOI] [PubMed] [Google Scholar]
- Rohrer, J., Benedetti, H., Zanolari, B., and Riezman, H. (1993). Identification of a novel sequence mediating regulated endocytosis of the G protein-coupled alpha-pheromone receptor in yeast. Mol. Biol. Cell 4 511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sato, T.K., Darsow, T., and Emr, S.D. (1998). Vam7p, a SNAP-25-like molecule, and Vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol. Cell. Biol. 18 5308–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz, C., Kinner, A., and Kolling, R. (2005). The deubiquitinating enzyme Ubp1 affects sorting of the ATP-binding cassette-transporter Ste6 in the endocytic pathway. Mol. Biol. Cell 16 1319–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, B., Banuett, F., Dahl, M., Schlesinger, R., Schafer, W., Martin, T., Herskowitz, I., and Kahmann, R. (1990). The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60 295–306. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snetselaar, K.M., Bölker, M., and Kahmann, R. (1996). Ustilago maydis mating hyphae orient their growth toward pheromone sources. Fungal Genet. Biol. 20 299–312. [DOI] [PubMed] [Google Scholar]
- Spellig, T., Bölker, M., Lottspeich, F., Frank, R.W., and Kahmann, R. (1994). Pheromones trigger filamentous growth in Ustilago maydis. EMBO J. 13 1620–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellig, T., Bottin, A., and Kahmann, R. (1996). Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol. Gen. Genet. 252 503–509. [DOI] [PubMed] [Google Scholar]
- Straube, A., Hause, G., Fink, G., and Steinberg, G. (2006). Conventional kinesin mediates microtubule-microtubule interactions in vivo. Mol. Biol. Cell 17 907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo, Z., Tonnis, M., Kessler, H., and Feldbrugge, M. (2002). Structure-function analysis of lipopeptide pheromones from the plant pathogen Ustilago maydis. Mol. Genet. Genomics 268 362–370. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban, M., Kahmann, R., and Bölker, M. (1996. a). Identification of the pheromone response element in Ustilago maydis. Mol. Gen. Genet. 251 31–37. [DOI] [PubMed] [Google Scholar]
- Urban, M., Kahmann, R., and Bölker, M. (1996. b). The biallelic a mating type locus of Ustilago maydis: Remnants of an additional pheromone gene indicate evolution from a multiallelic ancestor. Mol. Gen. Genet. 250 414–420. [DOI] [PubMed] [Google Scholar]
- Vida, T.A., and Emr, S.D. (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, Y., and Anraku, Y. (1992). Genes for directing vacuolar morphogenesis in Saccharomyces cerevisiae. II. VAM7, a gene for regulating morphogenic assembly of the vacuoles. J. Biol. Chem. 267 18671–18675. [PubMed] [Google Scholar]
- Weber, I., Gruber, C., and Steinberg, G. (2003). A class-V myosin required for mating, hyphal growth, and pathogenicity in the dimorphic plant pathogen Ustilago maydis. Plant Cell 15 2826–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner, R., Bölker, M., Kahmann, R., and Steinberg, G. (2000). A putative endosomal t-SNARE links exo- and endocytosis in the phytopathogenic fungus Ustilago maydis. EMBO J. 19 1974–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinzierl, G. (2001). Isolierung und Charakterisierung der B-Vermittelten Regulationskaskade in Ustilago maydis. PhD dissertation. (Marburg, Germany: Philipps University).
- White, J.M., and Rose, M.D. (2001). Yeast mating: Getting close to membrane merger. Curr. Biol. 11 R16–R20. [DOI] [PubMed] [Google Scholar]
- Wiederkehr, A., Avaro, S., Prescianotto-Baschong, C., Haguenauer-Tsapis, R., and Riezman, H. (2000). The F-box protein Rcy1p is involved in endocytic membrane traffic and recycling out of an early endosome in Saccharomyces cerevisiae. J. Cell Biol. 149 397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman, M., Chuang, J.S., and Schekman, R.W. (1996). Chs1p and Chs3p, two proteins involved in chitin synthesis, populate a compartment of the Saccharomyces cerevisiae endocytic pathway. Mol. Biol. Cell 7 1909–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.