Abstract
Retroposition is widely found to play essential roles in origination of new mammalian and other animal genes. However, the scarcity of retrogenes in plants has led to the assumption that plant genomes rarely evolve new gene duplicates by retroposition, despite abundant retrotransposons in plants and a reported long terminal repeat (LTR) retrotransposon-mediated mechanism of retroposing cellular genes in maize (Zea mays). We show extensive retropositions in the rice (Oryza sativa) genome, with 1235 identified primary retrogenes. We identified 27 of these primary retrogenes within LTR retrotransposons, confirming a previously observed role of retroelements in generating plant retrogenes. Substitution analyses revealed that the vast majority are subject to negative selection, suggesting, along with expression data and evidence of age, that they are likely functional retrogenes. In addition, 42% of these retrosequences have recruited new exons from flanking regions, generating a large number of chimerical genes. We also identified young chimerical genes, suggesting that gene origination through retroposition is ongoing, with a rate an order of magnitude higher than the rate in primates. Finally, we observed that retropositions have followed an unexpected spatial pattern in which functional retrogenes avoid centromeric regions, while retropseudogenes are randomly distributed. These observations suggest that retroposition is an important mechanism that governs gene evolution in rice and other grass species.
INTRODUCTION
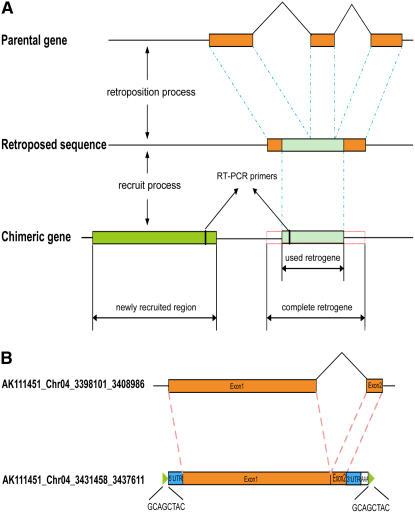
Retroposition is a cellular molecular process in which transcribed and spliced mRNAs are fortuitously reverse-transcribed and inserted into new genomic positions to form a retrogene (Figure 1A). The hallmarks of these retrosequences are usually the presence of a poly(A) tract, the loss of introns, and the presence of target site duplications that vary both in size and sequence (Figure 1B). The fate of such retrogenes is often to become nonexpressed pseudogenes because of lack of regulatory sequences (Brosius, 1991). However, if retrosequences by chance recruit certain regulatory sequences and acquire a new function by expression, then a new functional retrogene originates. The structures of such functional retrogenes are usually chimerical: they can either have a mosaic structure with the retrogene coding regions combined with novel regulatory sequences that do not exist in parental genes (Betran et al., 2002; Wang et al., 2002) or they can possess a hybrid coding region consisting of exons from unrelated genes near the insertion site in addition to new regulatory sequences (Nisole et al., 2004; Sayah et al., 2004; Zhang et al., 2004) (Figure 1A). Such chimerical structures would likely confer a function that parental genes do not have, thus often leading to adaptive evolution (Brosius, 1991; Long et al., 2003). New functional retrogenes have been reported in various organisms, especially mammals and Drosophila melanogaster (Long et al., 2003; Betran et al., 2004; Emerson et al., 2004), whereas retroposition also generated a large number of processed pseudogenes (5000 to 10,000 in human) (Venter et al., 2001; Torrents et al., 2003; Zhang et al., 2003). Spatial patterns were often found to be associated with retrogenes; for example, both human and Drosophila retrogenes escaped from X chromosomes and evolved new testis functions (Betran et al., 2002; Emerson et al., 2004). These support the hypothesis that retroposition may be an important force in driving evolution of genes and genomes (Brosius, 1991).
Figure 1.
Formation and Example of Retrogenes.
(A) General model for formation of chimerical structure of retrogenes. Orange boxes, parental and retrogene CDS regions; light-blue boxes, used retrogene regions in a new chimerical gene; green box, newly recruited region in a new chimerical gene.
(B) Example of a retrogene (AK111451_Chr04_3431458_3437611) that has the three signatures of retroposition (i.e., loss of introns, poly(A) tract, and flanking direct repeats).
Previous studies have shown dramatic differences in the numbers of retroposed sequences in plant and animal genomes, and only few plant retrogenes have been identified since the first was observed in the actin gene family of potato (Solanum tuberosum) (Drouin and Dover, 1990) and the alcohol dehydrogenase gene family in Leavenworthia (Charlesworth et al., 1998). A recent search of the Arabidopsis thaliana genome identified 69 retrosequences, of which more than one-third were found to be pseudogenes and the remaining had unknown functions (Zhang et al., 2005). The scarcity of functional retrogenes in plants was attributed to low activity of retrotransposon reverse transcription and integration functions on normal cellular mRNAs (Kumar and Bennetzen, 1999). By contrast, mammalian genomes have a high proportion of long interspersed elements (LINEs) and are estimated to contain 5000 to 11,000 retrosequences (Torrents et al., 2003; Kazazian, 2004), and 16% of these potentially may be able to contribute to the origin of new functional retrogenes (Vinckenbosch et al., 2006). These observations have led to the notion that retroposition may have played an insignificant role in evolution of plant genes and genomes, which would thus have to follow evolutionary routes different from those in animals.
However, when analyzing the indica and japonica rice (Oryza sativa) genomes, we unexpectedly observed that retroposition was involved in generating large numbers of genes, which suggested several problems for further pursuit. For example, one would wonder whether or not insertions of these retrosequences would create a high proportion of chimerical genes by recruiting unrelated gene regions near the insertion sites, as we observed previously in Drosophila (Long and Langley, 1993). If the chimerical structures are conserved across different species in the monocot lineage, does this suggest that a large number of novel gene functions were acquired during the evolution of grasses? On the other hand, we also wonder whether or not there are any rice-specific chimerical genes that would reveal an ongoing process of retrogene origination. Finally, we asked whether or not the movement of these retrogenes followed a spatial pattern defined by chromosomal positions, as previously observed in Drosophila and mammals (Betran et al., 2002; Emerson et al., 2004). Investigation of these questions will reveal the role of retroposition in genome evolution of rice and other grass species and present an important mechanism for interpreting a well-known but poorly understood massive diversification and broad adaptation of grasses to unusual diverse environments (Kellogg, 2001).
RESULTS
Abundant Retroposed Genes in the Rice Genome
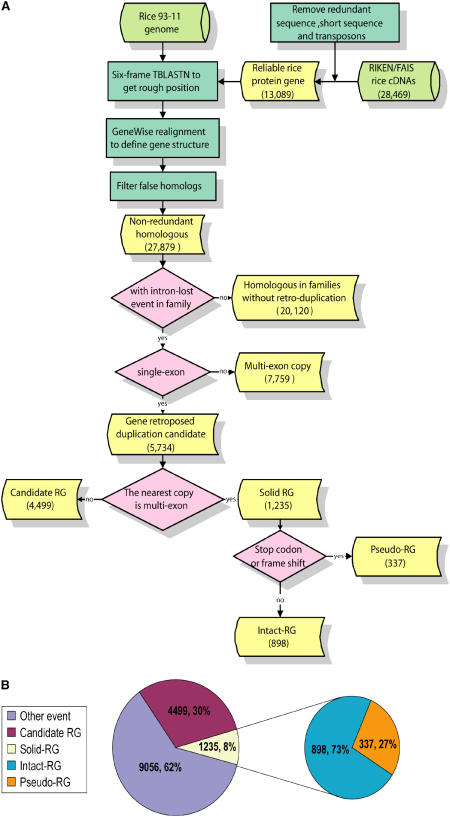
The 28,469 full-length rice cDNA sequences from RIKEN and the Foundation of Advancement of International Science (FAIS) centers (Kikuchi et al., 2003) were used for detecting retrogenes in the rice genome using a computational search scheme for retroposed genes that is depicted in the flow chart shown in Figure 2A. After removing redundancy, unlikely protein-coding genes, and the transposon-related genes, 13,089 reliable protein-coding genes were retained.
Figure 2.
Identification of Retroposition in the Rice Genome.
(A) The flow chart of the search scheme for identification of potentially functional retroposed genes and processed pseudogenes. We mapped KOME cDNAs to the finished 93-11 genome sequences and got the transcript unit. To get reliable coding genes, we filtered genes with <300 bp CDS. For the identification of retrogenes, we did the following analysis: (1) six-frame TBLASTN searches for homologs of the KOME cDNAs in the 93-11 genome; (2) realignment of KOME proteins to genomic sequence by GeneWise to get the intron-exon structure of every homolog; (3) filtered homologs with overlapped genomic position and defined all gene duplications; (4) identified retrogenes from duplicate genes; (5) based on the existence of stop codon or frame shift, we defined retropseudogenes; and (6) other retrogenes were defined as intact retrogenes.
(B) Proportion of different categories of genes in the identified 14,790 duplication events. In total, there are 1235 primary retrogenes (RGs), each of which derived from a single retroposition event, and the other homologs are assumed to have been generated by regular DNA-based gene duplication. For retrogenes, if a premature stop codon or frame shift occurs within the CDS region, they are defined as retropseudogenes; others are called intact retrogenes.
The 27,879 homologs were identified after using a combined approach of TBLASTN and GeneWise with the 13,089 full-length cDNAs as queries against the indica rice genome (Yu et al., 2005) (see Supplemental Table 1 online). Among these 27,879 homologs, 14,790 duplicates were identified. Surprisingly, 5734 candidate retroposition duplicates were found based on the criteria commonly used (Torrents et al., 2003; Zhang et al., 2003) (see Methods); these account for 20.6% of the detected rice genes and 38% of the duplicates (Figure 2B). The retrogenes retain one or more structural hallmarks of retroposition, including loss of introns, flanking direct repeats, and poly(A) tracts (Torrents et al., 2003; Zhang et al., 2003). Given the genome size of rice, the observed number of retroposed genes in rice is comparable to that of the human genome. A number of these intronless candidate retrogenes may have been formed by duplication of existing retrogenes. To explore the retrogene evolution at higher resolution, more recently evolved primary retrogenes were identified. By applying the conservative criterion that the closest ancestral copy of the primary retrogene must be an intron-containing parental gene, we deleted the intronless genes that were probably generated from duplication of existing retrogenes. Out of the 5734 candidate retrogenes, we eventually identified 1235 primary retrogenes, each of which represents a single retroposition event (Figure 2B; see Supplemental Table 2 online). These genes were denominated primary retrogenes. Most of these have tandem duplicates (on average, 4.64 per primary retrogene), while a small proportion (2%) have no tandem duplication. In addition, we searched for these retrogenes in the Syngenta draft sequence database (http://www.tmri.org/en/Site/home.aspx) and recently published Oryza japonica genome sequences (Goncalves et al., 2000; International Rice Genome Sequencing Project, 2005) using BLAT and identified 1159 in japonica genomes (see Supplemental Table 2 online). Although the actual number of primary retrogenes most likely is higher than this sample set, for the reason that some intronless copies could have originated through retroposition of old retrogenes, we used it for further analyses of evolutionary and structural features.
The Bs1 retroelement in maize (Zea mays) provided evidence that a retrotransposon can incorporate and transmit a nuclear gene transcript within its genomic structure (Jin and Bennetzen, 1994). This posed an interesting question of what proportion of the identified retrogenes is within a retrotransposon or a free retrogene. We analyzed the genomic structures (40-kb regions surrounding a retrogene) of the 1235 retrogenes using the LTR_STRUC program (McCarthy and McDonald, 2003). We identified 84 retrogenes near or within a long terminal repeat (LTR) retrotransposon, 27 retrogenes within a retrotransposon, and 25 and 32 immediately upstream and downstream, respectively, of the LTR structures (see Supplemental Table 3 online). An unknown proportion of retrogenes within retrotransposons may escape detection because the retroelement structures can degenerate rapidly (Ma et al., 2004). Therefore, cases similar to Bs1 in maize with a cellular retrogene segment within a retrotransposon are not rare in rice, although most identified retrogenes are not within currently active LTR retrotransposons.
Several factors have been found to be associated with retroposition. Goncalves et al. (2000) found that genes with poor GC content were more easily retroposed in the human genome. However, in rice, this seems not to be the case. We found that genes with retrocopies have higher GC content than those without retrocopies, which is particularly reflected by the GC content of the third positions of codons (see Supplemental Figure 1 online). This result is more consistent with the observations that genes with high GC content have high expression levels (Arhondakis et al., 2004) and that highly expressed genes may have a higher chance of being retroposed, as shown for the retrogene families derived from the alcohol dehydrogenase gene in Drosophila (Long and Langley, 1993; Jones and Begun, 2005; Nozawa et al., 2005). The stability of transcripts and translational control were also found to impact the transposition frequency of a gene (Pavlicek et al., 2006).
High Functionality of Retroposed Genes
Out of the 1235 primary retrogenes, we only identified 337 processed pseudogenes (27%) that contain premature stop codons or frame shift mutations (Figure 2B). This is in sharp contrast with the human genome, where the vast majority of retrogenes are processed pseudogenes (Zhang et al., 2003; Emerson et al., 2004). In rice, we tested whether or not the remaining 898 (73%) retrogenes are functional by pursuing three lines of evidence.
First, these retrogenes have intact open reading frames larger than 100 codons. Many of these retrogenes are probably very old because 856 of them have synonymous values >0.69 that is equivalent to an age of 53 million years, assuming a substitution rate of 0.65 per 100 million years per synonymous sites that was estimated from the Adh loci of grasses (Gaut et al., 1996). The sequences of retropseudogenes usually accumulate deletions and will eventually disappear from the genome. Based on the estimates of the half-life of LTR retrotransposon sequences in rice, <6 million years (Ma et al., 2004), most rice retropseudogenes would be completely eliminated within 53 million years. This is in accordance with the fact that the smaller genome of rice would accumulate deletions more rapidly than mammals, by the prediction made from a comparison between mammals and Drosophila (Petrov et al., 1996). Thus, the vast majority of rice retrogenes would not be preserved, and the younger retrogenes would accumulate indels rapidly (Ma and Bennetzen, 2004), which would often lead to changes in their reading frames if they were pseudogenes without evolutionary pressure. These pseudogene-based predictions are not supported by the age of these retrogenes and the observed fact that these genes have intact reading frames.
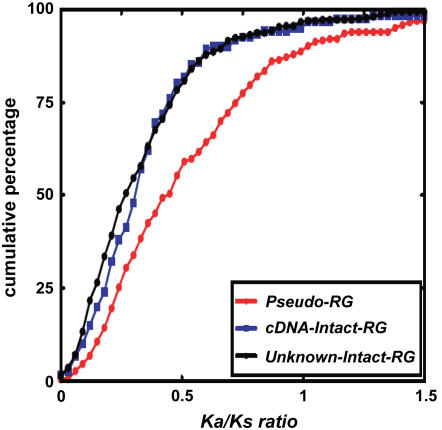
Second, we investigated functional constraints using an evolutionary methodology of comparing nonsynonymous (Ka) and synonymous (Ks) substitution rates (Ka:Ks ratios) between retrogenes and their parental copy. In general, the Ka:Ks ratio for pseudogenes, which evolved following neutrality, is expected to be 1, which is lower than unity for genes subject to functional constraint and higher than unity for genes subject to strong positive selection (Torrents et al., 2003). This method has been increasingly and efficiently used in identifying functionality of genes (Li, 1997; Nekrutenko et al., 2002; Moore and Purugganan, 2003). Figure 3 shows that intact and functional retrogenes have different Ka:Ks distributions from retropseudogenes. The Ka:Ks distribution of intact genes that still lack full-length cDNA is very similar to that of the 233 retrogenes that transcribe full-length cDNAs, but obviously different from that of retropseudogenes, which tend to have much higher Ka:Ks values, suggesting that intact retrogenes have been subject to more functional constraint. In the comparison that involves retrogenes with unknown constraints and parental copies that are known to be functional, a more conservative criterion should be Ka:Ks ≤ 0.5 to detect functionality of the retrogenes (Betran et al., 2002; Emerson et al., 2004). We observed that 81% of the intact retrogenes have Ka:Ks ratios significantly lower than 0.5 (Figure 3; see Supplemental Table 2 online). In conjunction with the 233 functional retrogenes that transcribe full-length cDNAs, application of this very conservative standard suggests that the vast majority of the intact retrogenes are subject to strong functional constraints (Figure 3; see Supplemental Table 2 online).
Figure 3.
Ka:Ks Distributions of Retropseudogenes and Intact Retrogenes with and without Full-Length cDNAs.
The Ka:Ks ratio is obtained between the retrogenes and its parental sequences. Bin size is 0.03.
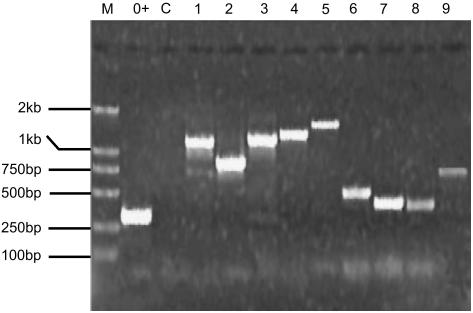
Third, consistent with the functionality of these intact retrogenes, more than half (495/898 = 55%) of them have been found to be expressed with support of either full-length cDNAs, ESTs (Boguski et al., 1993), microarray analysis of the transcriptome (Ma et al., 2005), or our RT-PCR experiments (Table 1). In Figure 4, we show examples of expression of these retrogenes based on our RT-PCR results.
Table 1.
Expression Summary of Retrogenes
| Total | Full-Length cDNA | DNA Array | EST | RT-PCR | Merged | |
|---|---|---|---|---|---|---|
| Retropseudogene | 337 | 0 | 37 | 10 | 8 | 47 |
| Functional retrogene | 898 | 233 | 281 | 205 | 57 | 495 |
Figure 4.
Examples of RT-PCR Results for Nine Chimerical Retrogenes.
M, DNA ladder (DL2000; Invitrogen); 0+, actin gene used as positive control; C, RT-PCR negative control in which the only difference from the positive lanes is that reverse transcriptase was not added in the RNA template. Lanes 1 to 9, RT-PCR–amplified fragments of the following nine chimerical retrogenes: AK072907_Chr03_25572510_25577541, AK064442_Chr06_25446009_25451022, AK070283_Chr04_33398346_33404211, AK072552_Chr07_27330570_27335745, AK073972_Chr04_3993417_3998040, AK064488_Chr04_19699106_19703477, AK059235_Chr07_11072250_11076879, AK064415_Chr07_16520248_16524877, and AK064641_Chr04_4027771_4032364. The RNAs for the amplification were the mixture of the equal amount of RNAs extracted from roots, shoots, leaves, and flowers (see Methods).
These independent lines of evidence suggest that the vast majority of the retrogenes are functional in rice. In comparison, it was recently estimated that 575 (16%) of 3590 human retroposed copies are potentially functional, and 117 of these retrocopies were shown to be bona fide retrogenes (Vinckenbosch et al., 2006). Thus, retroposition plays an unexpectedly important role in shaping the rice genome.
High Proportion of Chimerical Genes
When manually inspecting the retrogene sequences for quality by aligning retrogenes and their parental copies, we observed that many intact retrogenes had lost their start codon and/or stop codon (see Supplemental Table 4 online). One interpretation is that these retrogenes may be functionless pseudogenes, so mutations would have accumulated and deleted the start and stop codons. However, such an interpretation is inconsistent with the above evidence of functionality of these retrogenes. We therefore hypothesize that these retrogenes have recruited nearby exons and regulatory sequences and reincarnated into a new chimerical gene structure for new functions. We conducted experiments and analyses to test this prediction.
Out of the 898 intact retrogenes, 380 were predicted to have chimerical protein coding sequence (CDS) structures (see Supplemental Table 5 online) (see details in Methods). In this data set, the chimerical CDS structures of 73 (19.2%) retrogenes can be confirmed by either full-length cDNA or EST sequences in the public databases. To get expression evidence for the remaining predicted chimerical retrogenes, we used an RT-PCR assay. Primers cannot be designed for 59 genes due to the shortness of the recruited flanking sequences. On the remainder, cDNAs of 57 genes could be amplified with primers targeting the chimerical regions from any of the four tissues of rice plants, roots, shoots, leaves, or flowers (see Supplemental Table 5 online; Figure 4). By sequencing 21 RT-PCR products picked randomly, we confirmed the chimerical sequences that consist of retrosequence and recruited regions. Taken together, the expression data represent a large sample (130; ∼35%) of chimerical retrogenes, suggesting that many predicted rice retrogenes are bona fide chimerical CDS structures consisting of recruited regulatory elements, new exon-introns, and retrogene sequences.
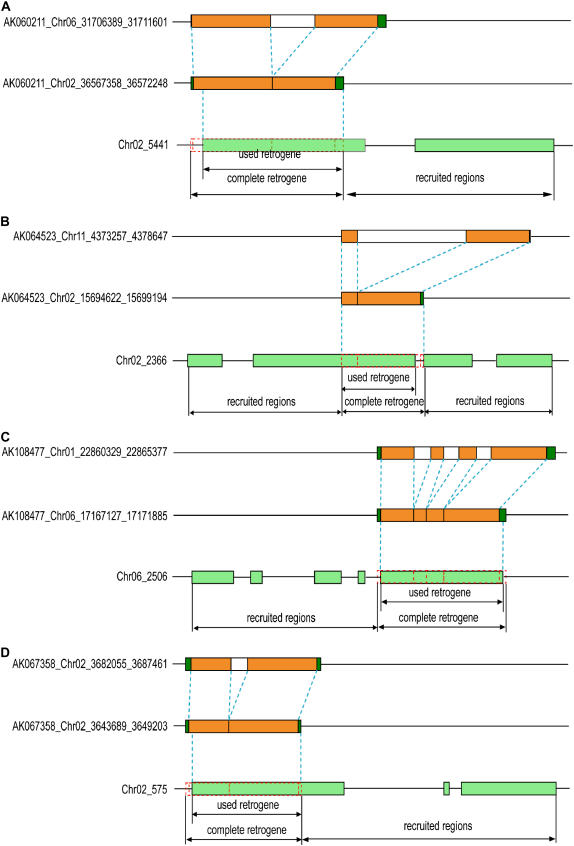
We have shown that more than one-third of the rice retrogenes identified in this study have evolved chimerical gene structures by recruiting additional coding exons after the insertion of retroposed sequences. If regulatory regions and untranslated regions (UTRs) were taken into account, we would see an even higher proportion of chimerical retrogenes in the rice genome; however, because of unavailable annotation of regulatory and UTR sequences, we were unable to include this in this study. In remarkable contrast with mammalian retrogenes that have rare recent chimerical retrogenes (Kazazian, 2004; Nisole et al., 2004; Sayah et al., 2004), rice retrogenes did not only recruit novel regulatory sequences but also new coding regions leading to a high proportion of hybrid proteins. For example, in the retrogenes AK064523_Chr02_15694622_15699194 and AK108477_Chro6_17167127_17171885, a new peptide was added to the N terminus, whereas in two other retrogenes, AK060211_Chr02_36567385_36572248 and AK067358_Chr02_3643689_3649203, a new peptide was added to the C terminus (Figure 5). There is experimental evidence that some of these rice hybrid genes might have evolved new protein functions, rendering a rich data set for future protein function studies. For example, the chimerical gene AK101897_Chr07_20914264_20919259 was shown to encode a polygalacturonase-inhibiting protein with testable enzymatic activities against Aspergillus niger polygalacturonase (Jang et al., 2003). Silencing of this gene in antisense transgenic plants was found to have an obvious phenotypic effect (e.g., increases in the numbers of floral organs, such as stamens, carpels, palea/lemmas, stigmas, and lodicules) (Jang et al., 2003).
Figure 5.
Examples of Four Chimerical Genes That Have Chimerical Protein Structures.
Orange boxes, parental and retrogene CDS regions; blue dashed boxes, used retrogene regions in new chimerical genes; green boxes, newly recruited regions in new chimerical genes; and red dashed boxes, unaligned CDS regions.
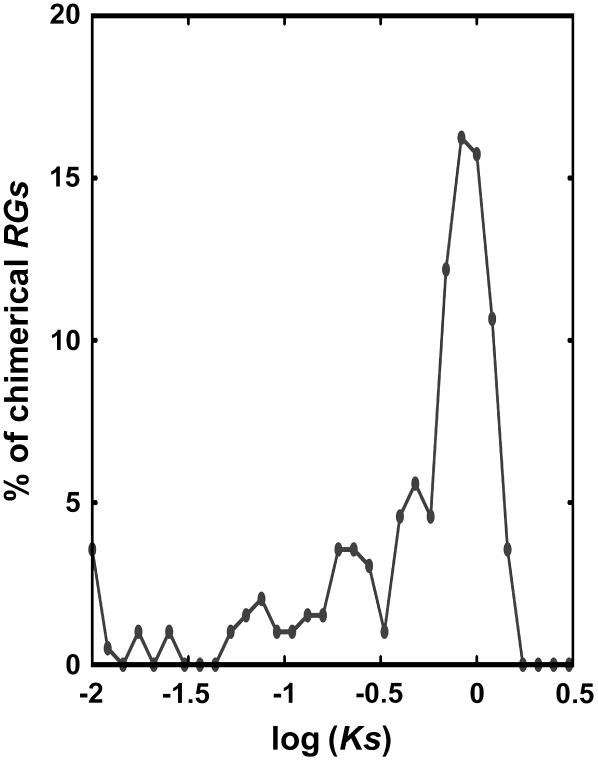
To assess if the creation of new chimerical retrogenes is a continuous process or not, we first plotted the distribution of Ks for these chimerical genes. After excluding those retrogenes with Ks above 1.5, 197 chimerical retrogenes were used to plot the distribution. Among them, seven of the chimerical genes have Ks values lower than 0.013 (Figure 6). If we assume the synonymous substitution rate of rice genes is 6.5 × 10−9 substitutions per silent site per year (Gaut et al., 1996), then approximately seven chimerical retrogenes appeared in the past 1 million years, which is 50 times the rate of 0.14 chimeric protein genes per million years in the primate lineage toward humans (Marques et al., 2005; Vinckenbosch et al., 2006), the highest rate of chimeric gene origination to our knowledge. If other nonchimerical retrogenes were taken into account, the rate would be even higher. Second, considering the great variation of Ks in different genes, we also used phylogenetic distribution of the retrogenes to help determine their ages. We searched these chimerical retrogenes in the maize and sorghum (Sorghum bicolor) genomes (Whitelaw et al., 2003; Barbazuk et al., 2005; Bedell et al., 2005). Only 97 of the 380 (25.5%) chimerical genes could be found in these grass species (see Supplemental Table 5 online). Because of the limited coverage of available maize and sorghum genome sequences, homologs for some rice chimerical retrogenes may not have been identified yet. On the other hand, it has been estimated that 96% of sorghum genes have been sequence tagged, with an average coverage of 65% across their length (Bedell et al., 2005). Therefore, not all of these nonhomolog chimerical genes can be attributed to incompleteness of genome sequences. These observations suggest that retroposition has kept the grass genome in constant flux and that many new chimerical retrogenes appeared recently after the rice lineage diverged from maize and sorghum and formed a young group of chimerical genes.
Figure 6.
Ks Distribution of Chimerical Retrogenes.
Bin size is 0.07. RGs, retrogenes.
Gene Movements and Patterns of Retropositions
The detected large numbers of functional retrogenes and functionless retropseudogenes allowed us to investigate the effects of genomic positions in determining fixation of retrogenes. A retroposition event has an easily defined direction pointing from the location of the parental gene to the landing location of the daughter retrogenes (Betran et al., 2002; Emerson et al., 2004). Because retropseudogenes evolve under neutrality, their distribution most likely represents the mutational pattern of retroposition and can therefore be compared with the distribution of functional retrogenes to infer the effects of genomic positions in determining fixation of retrogenes.
First, we investigated chromosomal distributions of retrogenes and retropseudogenes. We observed that retropseudogenes are randomly distributed among and within chromosomes, suggesting that retropseudogenes are neutral mutations as expected. However, functional retrogenes are rare in centromeric regions, showing an obvious selection of genomic positioning toward euchromatic regions (Table 2).
Table 2.
Comparisons of Distributions between Retropseudogenes and Intact Retrogenes in the Centromeric and Noncentromeric Regions
| Chromosome Length | Centromere Region % | RG No. | RG Near Centromere
|
RG-Ψ No. | RG-Ψ Near Centromere
|
RG-f No. | RG-f Near Centromere
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | ||||||
| Chr1 | 47,283,185 | 10.57 | 123 | 14 | 11.38 | 33 | 8 | 24.24 | 90 | 6 | 6.67 |
| Chr2 | 38,103,930 | 13.12 | 123 | 16 | 13.01 | 50 | 10 | 20.00 | 73 | 6 | 8.22 |
| Chr3 | 41,884,883 | 11.94 | 120 | 12 | 10.00 | 29 | 3 | 10.34 | 91 | 9 | 9.89 |
| Chr4 | 34,718,618 | 14.40 | 142 | 18 | 12.68 | 36 | 8 | 22.22 | 106 | 10 | 9.43 |
| Chr5 | 31,240,961 | 16.00 | 92 | 15 | 16.30 | 19 | 4 | 21.05 | 73 | 11 | 15.07 |
| Chr6 | 32,913,967 | 15.19 | 114 | 12 | 10.53 | 21 | 4 | 19.05 | 93 | 8 | 8.60 |
| Chr7 | 27,957,088 | 17.88 | 112 | 15 | 13.39 | 30 | 3 | 10.00 | 82 | 12 | 14.63 |
| Chr8 | 30,396,518 | 16.45 | 90 | 12 | 13.33 | 26 | 4 | 15.38 | 64 | 8 | 12.50 |
| Chr9 | 21,757,032 | 22.98 | 72 | 16 | 22.22 | 22 | 10 | 45.45 | 50 | 6 | 12.00 |
| Chr10 | 22,204,031 | 22.52 | 98 | 15 | 15.31 | 25 | 7 | 28.00 | 73 | 8 | 10.96 |
| Chr11 | 23,035,369 | 21.71 | 74 | 16 | 21.62 | 28 | 9 | 32.14 | 46 | 7 | 15.22 |
| Chr12 | 23,049,917 | 21.69 | 75 | 10 | 13.33 | 18 | 3 | 16.67 | 57 | 7 | 12.28 |
| Total | 374,545,499 | 1.33 | 1235 | 171 | 13.85 | 337 | 73 | 21.66 | 898 | 98 | 10.91 |
RG-Ψ refers to retroseudogenes, and RG-f refers to intact retrogenes. RG, retrogene.
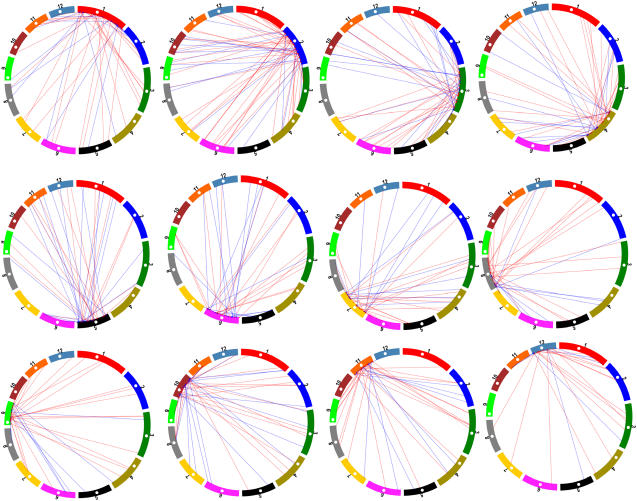
Furthermore, for simplicity, we only defined gene movement directions for the 337 retropseudogenes and the 233 functional retrogenes with full-length cDNAs. Drawing the chromosomal directions in circular movement maps shows that processed pseudogenes insert into chromosomes randomly, suggesting that the mutation events that led to these pseudogenes are random (Figure 7). Out of a total of 278 interchromosomal retropositions leading to retropseudogenes, we observed 51 insertions in centromeres or nearby regions. However, in the 168 interchromosomal retropositions generating functional retrogenes, we only observed 11 events that inserted functional genes in the nearby region of centromeres and none into the centromeric regions. These two distributions are significantly different (Fisher's exact test P = 0.00037), suggesting that the functional retrogenes tend to avoid centromeric regions under selection (Figure 7). In other words, retrosequences outside centromeric regions have a higher chance to be fixed as functional genes. These analyses reveal that plant functional retrogenes show significant spatial bias in the genome. However, this plant positional effect differs from the sex/autosomal preference effects in mammalian and Drosophila retrogenes (Betran et al., 2002; Emerson et al., 2004).
Figure 7.
Positional Effect of Retropositions between Chromosomes.
The white points in the chromosomes represent centromeres. Retropositions from other chromosomes into each chromosome are shown by colored lines, which suggest the directions from parental copies to the inserted sites of retrogenes in a particular chromosome. Blue lines suggest intact retrogenes, and red lines suggest retropseudogenes. Blue lines are rarely directed to centromeric regions, while red line are randomly directed (P = 0.00037).
DISCUSSION
This study reveals a large number of retrogenes in rice. One likely mechanism involving plant cellular gene retroposition was observed to be retrotransposon-mediated. In maize, the Bs1 retrotransposon (Jin and Bennetzen, 1989) was found to have recruited a portion of a cellular gene encoding the plasma membrane proton ATPase (Mha1) within the retroelement structure (Jin and Bennetzen, 1994). This acquired gene segment lost all introns that exist in its paralogous parental copy, which is a hallmark of retroposition. This case clearly reveals the existence of a mechanism for cellular gene retroposition in plants and thus suggests the importance of distinguishing between retrogenes within LTR retrotransposons and those that are free retrogenes. Such a comparison will provide insight into mechanisms that are involved in origination of retrogenes. Our in silico search of the rice genome using LTR_STRUC identified a number of retrogenes within a retrotransposon (see Results), describing generality of the Bs1-like mechanism in the generation of retrogenes. However, the vast majority of identified retrogenes appear to be outside of retrotransposons. In addition, we found that most of these retrogenes have tandem duplicates (on average, 4.64 per primary retrogene), and a small proportion of retrogenes (2%) have no tandem duplication, suggesting an ongoing retroposition in these retrogene families.
Furthermore, we find that this abundance of retrogenes is not seen in rice genomes only. The maize and sorghum genomes also encode many retrogenes, suggesting that retroposition shapes the genomes of grass species in general. On the other hand, the observed abundant retrogenes in the rice genome stand in sharp contrast with the lack of retrogenes in Arabidopsis and other dicotyledonous plants (Arabidopsis Genome Initiative, 2000). This is reminiscent of the previously observed correlation between LTR retrotransposons and genome size, suggesting that retrotransposons are a major factor in determining the size of plant genomes (Kumar and Bennetzen, 1999). In mammalian genomes, the generation of retrogenes is considered closely related to the activities of non-LTR retrotransposons (Goodier et al., 2000; Ostertag and Kazazian, 2001; Kazazian, 2004). The human LINE-1 elements were shown to be able to generate retropseudogenes (processed pseudogenes) in trans as demonstrated by Esnault et al. (2000), Pickeral et al. (2000), and Wei et al. (2001). Moran et al. (1999) showed that the LINE-1 elements (L1) could mobilize sequences from their 3′ flanking regions to new genomic locations, thus potentially resulting in new chimerical genes. In plants, there is evidence that the L1-analogous element Cin4 in maize likely provides reverse transcriptase for retroposition of cellular genes (Schwarz-Sommer et al., 1987).
We found that the functional retrogenes and nonfunctional retrosequences showed different spatial distributions, in which the former tends to avoid centromeric regions and the latter has no such biased distribution. This phenomenon may be discussed in the light of a recent observation that the LTR retrotransposons in the centromeric region exhibit a low rate of unequal homologous recombination outside of the centromeric core (Ma et al., 2004; Bennetzen, 2005). Thus, the nonfunctional sequences can stay in the centromeric region longer, whereas a functional retrogene would accumulate deleterious mutations and eventually become a nonfunctional processed pseudogene because of reduced ability to discard deleterious mutations due to lack of recombination (e.g., Charlesworth et al., 2005). A further possibility is that the landing of retrogenes in centromeric regions may lead to epigenetically suppressed expression (e.g., Karpen and Allshire, 1997; May et al., 2005), consequently resulting in gene degeneration.
Chimerical gene structures have been reported in various organisms and are considered to be an important component of protein diversity (reviewed in Patthy, 1999; Long et al., 2003). Recent case studies of several young chimerical genes in Drosophila (e.g., jingwei, sphinx, and Adh-Twain) and primates (e.g., TRIM5, PMCHL, and TRE2) have cast light onto the molecular evolutionary process that generated chimerical genes. The genomic search of chimerical genes in this study demonstrates that their formation is driven by the merging of retrogenes and existing unrelated gene regions.
Our results suggest that many novel functions may have originated in rice and grass genomes through retroposition at a remarkably rapid evolutionary pace that is an order of magnitude faster than the primate lineage toward humans. The recently identified abundant transposons Pack-MULE in rice (Jiang et al., 2004) and Helitrons in maize (Morgante et al., 2005) that contain gene fragments also support the possibility of rapid gene recombination in grass species. These findings provide insights into the genetic basis for the broad adaptation of grasses to a diverse range of environments and the evolution of a great range of biological diversity (Kellogg, 2001).
METHODS
Defining the Rice Retrogenes
The procedure for finding rice retrogenes is described below, and the flow chart for this is shown as Figure 2A. The initial 28,469 full-length cDNA sequences were downloaded from RIKEN and FAIS centers (Kikuchi et al., 2003). We aligned these cDNAs to the Beijing Oryza sativa indica genome using BLAT (Kent, 2002), and the smaller cDNA is considered to be redundant if there are two alignments that overlap by at least 100 bp. Consequently, we generated a database that contains 16,524 nonredundant genes. To eliminate unlikely protein coding genes, cDNAs with small open reading frames <100 amino acids were discarded (Okazaki et al., 2002; Wang et al., 2004). Of the remaining 15,631 genes, we removed genes with >10% transposon-like sequences using RepeatMasker with the RepBase database (Jurka, 1998). Eventually, 13,089 reliable protein coding genes were obtained for the following analyses.
Using the combined procedures of TBLASTN (Altschul et al., 1997) and the GeneWise program (Birney et al., 2004), we searched the 13,089 cDNAs against the rice indica genome. TBLASTN with an E-value threshold at 10−8 was used for the rough alignment. Homologous hits of the exons were chained together by a dynamic programming algorithm. GeneWise with default settings and a filtration score at 35 was then used for defining the intron-exon boundaries of every homologous chain. After that, only homologous genes with a minimum length of 70% of the query protein sequence were kept for the assurance of the high-quality alignments. In some cases, false hits happened to the shared domains of different genes. To exclude this, we picked up the best homolog with the highest identity if any two homologs overlapped by >60% of the length. Finally, we got 27,879 homologs (14,790 duplication events), and the details are shown in Supplemental Table 1 online. Out of these 27,879 homologs, the genes that have more than two homologs in the genome will be used for the retrogene searching.
The following criteria were set to define the candidate retrogenes (Birney et al., 2004): (1) minimum 70% coverage of the protein coding regions (CDS) of the gene; (2) homologs have to be intronless; and (3) the families have to have both multi-exon and single-exon copies, which can suggest occurred intron loss events.
Ultimately, this produced a total of 5734 retrogene candidates satisfying all the above criteria. To divide these into primary retrogenes and retrogenes evolving from the duplication of another retrogene, we examined the closest copy of the retrogenes for introns. This produced 1235 primary retrogenes. We did not take into account that some real retrogenes originated through retroposed intronless genes, and these were neglected by this treatment. Out of these 1235 retrogenes, 337 retrogenes were defined as retropseudogenes with the occurrence of either premature stop codons or frame-shift mutations. The remaining retrogenes (898) are defined as intact retrogenes, 233 of them have the full-length cDNA support (Figure 2B).
Ks and Ka substitution rates and Ka:Ks ratios were calculated between the retrogenes and their intron-containing parental copies using the maximum likelihood–based program in the PAML package following the Nei-Gojobori method (Nei and Gojobori, 1986; Yang, 1997).
We also searched the positions of retrogenes relative to the structure of LTR retrotransposons using the LTR_STRUC program (McCarthy and McDonald, 2003) to quantify how many of the identified retrogenes are within the structure of LTR retrotransposons, as shown in Bs1 (Jin and Bennetzen, 1994), and how many of them are outside the retroelements. A technical consideration is the size of window for the genomic search. McCarthy et al. (2002) conducted a genomic search for LTR retrotransposons in the rice genome and found that copia-like elements are usually 5 to 6 kb in length, and gypsy-like elements are typically 10 to 13 kb long. We thus chose 40 kb as a window (i.e., 20 kb upstream of a retrogene and 20 kb downstream of a retrogene) to search LTR structures.
Identification of Chimerical Retrogenes
We noticed that many rice retrogenes lost either original start or stop, or both start and stop codons (see Supplemental Table 2 online). This suggests that many retrogenes have formed new chimerical gene structures with flanking sequences after they retroposed to their new loci. For the 233 intact retrogenes with full-length cDNA support, 44 chimerical retrogenes were identified by the criterion that the retrogene shares no more than 80% sequence with its parental intron-containing copy. For the intact retrogenes without the full-length cDNA support, we used the current rice annotation (20) to represent the retrogene. If the flanking sequence(s) that the retrogene recruits is larger than 50 bp of the CDS, we considered it as a chimerical retrogene. Out of 665 retrogenes, 336 met the criterion to be chimerical, and 29 of them have available ESTs, which confirmed their chimerical structures.
Adding up these two types of intact retrogenes, we got 380 chimerical retrogenes. For the 307 unconfirmed chimerical retrogenes, RT-PCR primers were designed for further RNA expression experimental tests. The primers for 175 genes met the standards of good experimental conditions, the primers for 73 genes were designed with compromise of stringency, and no primer could be obtained for the rest of the 59 genes due to the shortness of the recruited flanking sequences.
To examine the homologous chimerical structures for these 380 retrogenes, the same procedure of combining TBLASTN and GeneWise was used to search against the sorghum and maize genomes (the extensive maize gene enrichment sequence database at The Institute for Genomic Research) (Whitelaw et al., 2003; Barbazuk et al., 2005; Bedell et al., 2005). The homologous chimerical structures were identified in the sorghum/maize genome when they met the following criteria: (1) the best homolog was larger than 50% of the chimerical retrogene; (2) the homolog had new recruited flanking sequences; and (3) the homolog had the same gene structure as the chimerical retrogene.
RT-PCR Confirmation of Chimerical Retrogenes
Total RNA was extracted from four tissue samples of the 93-11 strain of O. sativa indica (roots, shoots, leaves, and flowers) with the RNeasy plant mini kit (Qiagen) according to the manufacturer's instructions. Equal amounts of the four RNA samples were mixed together before the synthesis of the first-strand cDNA. Six micrograms of mixed total RNA was treated with DNase I (Invitrogen) to remove contamination from genomic DNA. First-strand cDNA was synthesized by SuperScript II reverse transcriptase (Invitrogen) at 42°C for 1 h with oligo(dT)18 as primer. Expression of chimerical retrogenes was checked by RT-PCR using the cDNA as template. The rice actin gene (GenBank accession no. X16280) was used as internal control for the quality of cDNA using the primers 5′-TCCATCTTGGCATCTCTCAG-3′ (sense) and 5′-GTACCCGCATCAGGCATCTG-3′ (antisense).
Gene Structures of the 380 Chimerical Retrogenes
Chimerical retrogene structures can be accessed at the following site: http://retrogene.genomics.org.cn/380_chimeric_strcture.zip. The first line is for the retrogene and the second line the annotated gene (or that predicted by the gene annotation tool FgeneSH). The green lines suggest ESTs. In the first line, the block regions show gene locations, including the UTR regions, red for aligned CDS regions, white for unaligned CDS regions, and yellow for UTR regions. For annotated genes, only the CDS is shown; thus, the very beginning and end of the blue blocks are the start and stop codons.
Accession Numbers
Sequence from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AK064523, AK10847, AK060211, AK067358, AK101897, AK111451,AK072907, AK064442, AK070283, AK072552, AK073972, AK064488, AK059235, AK064415, and AK064641.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Results of Homolog Searches in the indica Rice Genome Using the 13,089 cDNAs as Query Sequences.
Supplemental Table 2. The 1235 Primary Retrogenes Identified in This Study.
Supplemental Table 3. The 84 Retrogenes Near or within LTR Retrotransposons.
Supplemental Table 4. Summary of Loss of Start and Stop Codons in the Retrosequences.
Supplemental Table 5. List of 380 Chimerical Retrogenes.
Supplemental Figure 1. GC Content Comparisons between Genes with and without Retrogenes.
Supplementary Material
Acknowledgments
This work was sponsored by the CAS-Max-Planck Society Fellowship, a Chinese Academy of Sciences key project grant (KSCX2-SW-121), a National Science Foundation of China (NSFC) award (30325016) to distinguished young scientists, and a NSFC key grant (30430400) to W.W. Funding was from the Chinese Academy of Sciences (GJHZ0518), the Ministry of Science and Technology of China (CNG1-04-15-7A), the National Natural Science Foundation of China (90208019, 90403130, and 30221004), the China National Grid, the Danish Platform for Integrative Biology, Ole Rømer grants from the Danish Natural Science Research Council to J.W., a National Science Foundation CAREER award (MCB0238168), a National Institutes of Health R01 grant (R01GM065429-01A1), and from the Packard Fellowship for Science and Engineering from the David and Lucile Packard Foundation to M.L. We thank two anonymous reviewers for their generous and insightful suggestions. We also appreciate the valuable discussions with Elizabeth Kellogg (University of Missouri, St. Louis, MO) and Ming-Che Shih (University of Iowa, Iowa City, IA).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Wen Wang (wwang@mail.kiz.ac.cn), Manyuan Long (mlong@uchicago.edu), and Jun Wang (wangj@genomics.org.cn).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.106.041905.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Arhondakis, S., Auletta, F., Torelli, G., and D'Onofrio, G. (2004). Base composition and expression level of human genes. Gene 325 165–169. [DOI] [PubMed] [Google Scholar]
- Barbazuk, W.B., Bedell, J.A., and Rabinowicz, P.D. (2005). Reduced representation sequencing: A success in maize and a promise for other plant genomes. Bioessays 27 839–848. [DOI] [PubMed] [Google Scholar]
- Bedell, J.A., et al. (2005). Sorghum genome sequencing by methylation filtration. PLoS Biol. 3 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J.L. (2005). Transposable elements, gene creation and genome rearrangement in flowering plants. Curr. Opin. Genet. Dev. 15 621–627. [DOI] [PubMed] [Google Scholar]
- Betran, E., Emerson, J.J., Kaessmann, H., and Long, M. (2004). Sex chromosomes and male functions: Where do new genes go? Cell Cycle 3 873–875. [PubMed] [Google Scholar]
- Betran, E., Thornton, K., and Long, M. (2002). Retroposed new genes out of the X in Drosophila. Genome Res. 12 1854–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E., Clamp, M., and Durbin, R. (2004). GeneWise and Genomewise. Genome Res. 14 988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski, M.S., Lowe, T.M., and Tolstoshev, C.M. (1993). dbEST–Database for “expressed sequence tags”. Nat. Genet. 4 332–333. [DOI] [PubMed] [Google Scholar]
- Brosius, J. (1991). Retroposons—Seeds of evolution. Science 251 753. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., Charlesworth, B., and Marais, G. (2005). Steps in the evolution of heteromorphic sex chromosomes. Heredity 95 118–128. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., Liu, F.L., and Zhang, L. (1998). The evolution of the alcohol dehydrogenase gene family by loss of introns in plants of the genus Leavenworthia (Brassicaceae). Mol. Biol. Evol. 15 552–559. [DOI] [PubMed] [Google Scholar]
- Drouin, G., and Dover, G.A. (1990). Independent gene evolution in the potato actin gene family demonstrated by phylogenetic procedures for resolving gene conversions and the phylogeny of angiosperm actin genes. J. Mol. Evol. 31 132–150. [DOI] [PubMed] [Google Scholar]
- Emerson, J.J., Kaessmann, H., Betran, E., and Long, M. (2004). Extensive gene traffic on the mammalian X chromosome. Science 303 537–540. [DOI] [PubMed] [Google Scholar]
- Esnault, C., Maestre, J., and Heidmann, T. (2000). Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24 363–367. [DOI] [PubMed] [Google Scholar]
- Gaut, B.S., Morton, B.R., McCaig, B.C., and Clegg, M.T. (1996). Substitution rate comparisons between grasses and palms: Synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. USA 93 10274–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves, I., Duret, L., and Mouchiroud, D. (2000). Nature and structure of human genes that generate retropseudogenes. Genome Res. 10 672–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier, J.L., Ostertag, E.M., and Kazazian, H.H., Jr. (2000). Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum. Mol. Genet. 9 653–657. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436 793–800. [DOI] [PubMed] [Google Scholar]
- Jang, S., Lee, B., Kim, C., Kim, S.-J., Yim, J., Han, J.-J., Lee, S., Kim, S.-R., and An, G. (2003). The OsFOR1 gene encodes a polygalacturonase-inhibiting protein (PGIP) that regulates floral organ number in rice. Plant Mol. Biol. 53 357–369. [DOI] [PubMed] [Google Scholar]
- Jiang, N., Bao, Z., Zhang, X., Eddy, S.R., and Wessler, S.R. (2004). Pack-MULE transposable elements mediate gene evolution in plants. Nature 431 569–573. [DOI] [PubMed] [Google Scholar]
- Jin, Y.K., and Bennetzen, J.L. (1994). Integration and nonrandom mutation of a plasma membrane proton ATPase gene fragment within the Bs1 retroelement of maize. Plant Cell 6 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y.K., and Bennetzen, J.L. (1989). Structure and coding properties of Bs1, a maize retrovirus-like transposon. Proc. Natl. Acad. Sci. USA 86 6235–6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, C.D., and Begun, D.J. (2005). Parallel evolution of chimeric fusion genes. Proc. Natl. Acad. Sci. USA 102 11373–11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka, J. (1998). Repeats in genomic DNA: Mining and meaning. Curr. Opin. Struct. Biol. 8 333–337. [DOI] [PubMed] [Google Scholar]
- Karpen, G.H., and Allshire, R.C. (1997). The case for epigenetic effects on centromere identity and function. Trends Genet. 13 489–496. [DOI] [PubMed] [Google Scholar]
- Kazazian, H.H.J. (2004). Mobile elements: Drivers of genome evolution. Science 303 1626–1632. [DOI] [PubMed] [Google Scholar]
- Kellogg, E.A. (2001). Evolutionary history of the grasses. Plant Physiol. 125 1198–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J. (2002). BLAT—The BLAST-like alignment tool. Genome Res. 12 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, S., et al. (2003). Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301 376–379. [DOI] [PubMed] [Google Scholar]
- Kumar, A., and Bennetzen, J.L. (1999). Plant retrotransposons. Annu. Rev. Genet. 33 479–532. [DOI] [PubMed] [Google Scholar]
- Li, W.H. (1997). Molecular Evolution. (Sunderland, MA: Sinauer Associates).
- Long, M., Betran, E., Thornton, K., and Wang, W. (2003). The origin of new genes: Glimpses from the young and old. Nat. Rev. Genet. 4 865–875. [DOI] [PubMed] [Google Scholar]
- Long, M., and Langley, C.H. (1993). Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science 260 91–95. [DOI] [PubMed] [Google Scholar]
- Ma, J., and Bennetzen, J.L. (2004). Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 101 12404–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., and Bennetzen, J.L. (2006). Recombination, rearrangement, reshuffling, and divergence in a centromeric region of rice. Proc. Natl. Acad. Sci. USA 103 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., Devos, K.M., and Bennetzen, J.L. (2004). Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 14 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., et al. (2005). A microarray analysis of the rice transcriptome and its comparison to Arabidopsis. Genome Res. 15 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques, A.C., Dupanloup, I., Vinckenbosch, N., Reymond, A., and Kaessmann, H. (2005). Emergence of young human genes after a burst of retroposition in primates. PLoS Biol. 3 e357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, B.P., Lippman, Z.B., Fang, Y., Spector, D.L., and Martienssen, R.A. (2005). Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet. 1 e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, E.M., Liu, J., Lizhi, G., and McDonald, J.F. (2002). Long terminal repeat retrotransposons of Oryza sativa. Genome Biol. 3 RESEARCH0053. [DOI] [PMC free article] [PubMed]
- McCarthy, E.M., and McDonald, J.F. (2003). LTR_STRUC: A novel search and identification program for LTR retrotransposons. Bioinformatics 19 362–367. [DOI] [PubMed] [Google Scholar]
- Moore, R.C., and Purugganan, M.D. (2003). The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 100 15682–15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, J.V., DeBerardinis, R.J., and Kazazian, H.H., Jr. (1999). Exon shuffling by L1 retrotransposition. Science 283 1530–1534. [DOI] [PubMed] [Google Scholar]
- Morgante, M., Brunner, S., Pea, G., Fengler, K., Zuccolo, A., and Rafalski, A. (2005). Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat. Genet. 37 997–1002. [DOI] [PubMed] [Google Scholar]
- Nei, M., and Gojobori, T. (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3 418–426. [DOI] [PubMed] [Google Scholar]
- Nekrutenko, A., Makova, K.D., and Li, W.H. (2002). The K(A)/K(S) ratio test for assessing the protein-coding potential of genomic regions: An empirical and simulation study. Genome Res. 12 198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisole, S., Lynch, C., Stoye, J.P., and Yap, M.W. (2004). A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. USA 101 13324–13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa, M., Aotsuka, T., and Tamura, K. (2005). A novel chimeric gene, siren, with retroposed promoter sequence in the Drosophila bipectinata complex. Genetics 171 1719–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki, Y., et al. (2002). Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420 563–573. [DOI] [PubMed] [Google Scholar]
- Ostertag, E.M., and Kazazian, H.H., Jr. (2001). Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35 501–538. [DOI] [PubMed] [Google Scholar]
- Patthy, L. (1999). Genome evolution and the evolution of exon-shuffling—A review. Gene 238 103–114. [DOI] [PubMed] [Google Scholar]
- Pavlicek, A., Gentles, A.J., Paces, J., Paces, V., and Jurka, J. (2006). Retroposition of processed pseudogenes: The impact of RNA stability and translational control. Trends Genet. 22 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov, D.A., Lozovskaya, E.R., and Hartl, D.L. (1996). High intrinsic rate of DNA loss in Drosophila. Nature 384 346–349. [DOI] [PubMed] [Google Scholar]
- Pickeral, O.K., Makalowski, W., Boguski, M.S., and Boeke, J.D. (2000). Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 10 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayah, D.M., Sokolskaja, E., Berthoux, L., and Luban, J. (2004). Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430 569–573. [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer, Z., Leclercq, L., Gobel, E., and Saedler, H. (1987). Cin4, an insert altering the structure of the A1 gene in Zea mays, exhibits properties of nonviral retrotransposons. EMBO J. 6 3873–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrents, D., Suyama, M., Zdobnov, E., and Bork, P. (2003). A genome-wide survey of human pseudogenes. Genome Res. 13 2559–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter, J.C., et al. (2001). The sequence of the human genome. Science 291 1304–1351. [DOI] [PubMed] [Google Scholar]
- Vinckenbosch, N., Dupanloup, I., and Kaessmann, H. (2006). Evolutionary fate of retroposed gene copies in the human genome. Proc. Natl. Acad. Sci. USA 103 3220–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Zhang, J., Zheng, H., Li, J., Liu, D., Li, H., Samudrala, R., Yu, J., and Wong, G.K.-S. (2004). Mouse transcriptome: Neutral evolution of ‘non-coding’ complementary DNAs. Nature 431 757. [PubMed] [Google Scholar]
- Wang, W., Brunet, F.G., Nevo, E., and Long, M. (2002). Origin of sphinx, a young chimeric RNA gene in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 99 4448–4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, W., Gilbert, N., Ooi, S.L., Lawler, J.F., Ostertag, E.M., Kazazian, H.H., Boeke, J.D., and Moran, J.V. (2001). Human L1 retrotransposition: Cis preference versus trans complementation. Mol. Cell. Biol. 21 1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw, C.A., et al. (2003). Enrichment of gene-coding sequences in maize by genome filtration. Science 302 2118–2120. [DOI] [PubMed] [Google Scholar]
- Yang, Z. (1997). PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13 555–556. [DOI] [PubMed] [Google Scholar]
- Yu, J., Wang, J., Lin, W., Li, S., Li, H., Zhou, J., Ni, P., Dong, W., Hu, S., Zeng, C., Zhang, J., Zhang, Y., et al. (2005). The genomes of Oryza sativa: A history of duplications. PLoS Biol. 3 e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Dean, A.M., Brunet, F., and Long, M. (2004). Evolving protein functional diversity in new genes of Drosophila. Proc. Natl. Acad. Sci. USA 101 16246–16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Wu, Y., Liu, Y., and Han, B. (2005). Computational identification of 69 retroposons in Arabidopsis. Plant Physiol. 138 935–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Harrison, P.M., Liu, Y., and Gerstein, M. (2003). Millions of years of evolution preserved: A comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 13 2541–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.