Abstract
Many woody perennials, such as poplar (Populus deltoides), are not able to form flower buds during the first several years of their life cycle. They must undergo a transition from the juvenile phase to the reproductive phase to be competent to produce flower buds. After this transition, trees begin to form flower buds in the spring of each growing season. The genetic factors that control flower initiation, ending the juvenile phase, are unknown in poplar. The factors that regulate seasonal flower bud formation are also unknown. Here, we report that poplar FLOWERING LOCUS T2 (FT2), a relative of the Arabidopsis thaliana flowering-time gene FT, controls first-time and seasonal flowering in poplar. The FT2 transcript is rare during the juvenile phase of poplar. When juvenile poplar is transformed with FT2 and transcript levels are increased, flowering is induced within 1 year. During the transition between vegetative and reproductive growth in mature trees, FT2 transcripts are abundant during reproductive growth under long days. Subsequently, floral meristems emerge on flanks of the axillary inflorescence shoots. These findings suggest that FT2 is part of the flower initiation pathway in poplar and plays an additional role in regulating seasonal flower initiation that is integrated with the poplar perennial growth habit.
INTRODUCTION
Arabidopsis thaliana is an annual herbaceous plant, whereas poplar (Populus species) is a perennial tree. Although these model species are both angiosperms and dicots, closely related within the eudicots (Soltis et al., 1999; Wikstrom et al., 2001) and best characterized by a monopodial shoot system (Bradley et al., 1997; Reinhardt and Kuhlemeier, 2002; Yuceer et al., 2003), they differ in many ways. Arabidopsis completes its life cycle in 2 months, with a short juvenile developmental phase followed by the production of flowers and seeds at the reproductive developmental phase (Somerville and Koornneef, 2002). A decision to flower is made only once during its life cycle, upon receiving inductive signals that are developmental, physiological, environmental, or a combination of these. The main shoot and axillary buds continuously produce reproductive organs, and then the plant dies. Thus, Arabidopsis plants do not undergo specific phases of seasonal vegetative and floral development at the reproductive phase.
By contrast, poplar trees have life spans of 100 to 200 years and a long juvenile phase, indicating slow maturation (Braatne et al., 1996). Consequently, they begin flowering after 7 to 10 years. After flowering for the first time in their life span, annual or seasonal flowering occurs during the reproductive developmental phase. Shoots then initiate early vegetative buds (vegetative zone I), floral buds (floral zone), and late vegetative buds (vegetative zone II) in a sequential manner, suggesting that the shoot goes through repeated phase-change cycles between vegetative and reproductive growth periods (Yuceer et al., 2003). Thus, each seasonal flowering period is interrupted by a long vegetative period. These reiterating developmental cycles between vegetative and reproductive growth periods are absent in Arabidopsis (Boss et al., 2004). However, it is not known what genetic factors control first-time and seasonal flowering in poplar.
Under the influence of environmental and developmental signals, the identity of the shoot meristem in annual plants is controlled by CENTRORADIALIS (CEN) in Antirrhinum majus (Bradley et al., 1996a, 1996b), SELF-PRUNING (SP) in tomato (Solanum lycopersicum) (Pnueli et al., 1998), Heading-date3a (Hd3a) in rice (Oryza sativa) (Kojima et al., 2002), and FLOWERING LOCUS T (FT) (Kardailsky et al., 1999; Kobayashi et al., 1999) and TERMINAL FLOWER1 (TFL1) (Alvarez et al., 1992; Bradley et al., 1997) in Arabidopsis. TFL1 and FT have opposite actions in determining the identity of the shoot meristem. In the juvenile phase of Arabidopsis, the vegetative identity of the shoot meristems is promoted by TFL1. Upon receiving floral signals, CONSTANS (CO) and FT promote the transition of the shoot meristem to an inflorescence meristem that produces cauline leaves and flowers (Putterill et al., 1995; Kardailsky et al., 1999; Kobayashi et al., 1999). Then, Arabidopsis plants cease producing true leaves and end their life cycle with reproductive growth.
CO and FT, in the phloem companion cells of mature leaves, initiate flowering in Arabidopsis (Takada and Goto, 2003; An et al., 2004; Ayre and Turgeon, 2004). When CO was driven by a companion cell–specific promoter in Arabidopsis, early flowering was induced and the late-flowering phenotype of co mutant plants was rescued (An et al., 2004; Ayre and Turgeon, 2004). By contrast, CO driven by shoot apical meristem (SAM)–specific promoters did not induce flowering. When FT was expressed from phloem- and SAM-specific promoters, early flowering and complementation of co mutant plants were observed (An et al., 2004). Expression of FT in companion cells was essential for floral induction and was controlled by CO expression (Takada and Goto, 2003; An et al., 2004). These results suggest that CO protein activates FT in the phloem cells and that the FT protein might move out of the phloem to the SAM, where floral development is induced. Experiments showed that FT expression driven by a heat shock–inducible promoter in a single Arabidopsis leaf was sufficient to initiate flowering under short days, suggesting that FT mRNA acts as a floral signal from leaf to the SAM (Huang et al., 2005). However, movement of the FT protein from phloem to the SAM has not been demonstrated. In the SAM, FT protein formed a complex with FD protein (basic domain/leucine zipper motif transcription factor), which upregulated the floral meristem identity gene APETALA1 (AP1) to induce floral development (Abe et al., 2005; Wigge et al., 2005). All of these findings support the classical studies showing that a photoinduced systemic signal, or florigen, is synthesized in leaves and transported to the SAM, where floral development is induced (Zeevaart, 1976).
It is not known whether genes similar to those in Arabidopsis control flower initiation in long-lived woody perennial plants with a different shoot architecture. To test this idea, we isolated and cloned one of the FT homologs (FT2) from Populus deltoides. FT2 is a good choice for studying the transition from the vegetative to the reproductive stage in poplar because transcriptional activation of FT is closely associated with the induction of flowering in response to photoperiod in Arabidopsis and rice. This study examines the roll of FT2 in regulating the transition from the juvenile to the reproductive stage in poplar and investigates its potential function in seasonal flowering in mature shoots.
RESULTS
FT2 Induces Flowering in Juvenile Poplar Trees
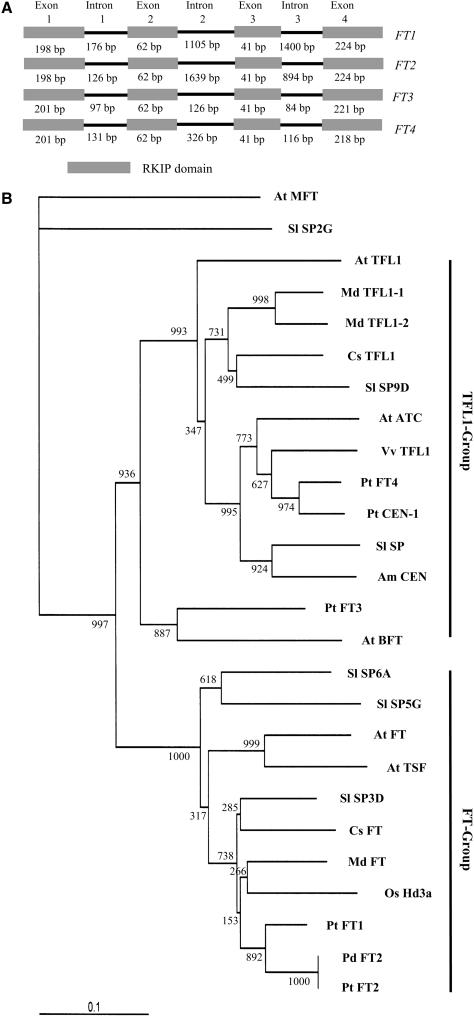
Using a PCR approach, we cloned the FT2 cDNA from P. deltoides. The recently released genome sequence of Populus trichocarpa (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html) confirmed the identity of the cDNA. P. deltoides FT2 (Pd FT2) is one of the four poplar FT/TFL1-like transcripts (Figure 1A) and is 100% identical to P. trichocarpa FT2 (Pt FT2) at the amino acid level (see Supplemental Figure 1 online). The deduced amino acid sequence of Pd FT2 shows 81 and 78% similarity to that of Hd3a in rice and FT in Arabidopsis, respectively. The sequence similarity between Pd FT2 and TFL1 in Arabidopsis is 59%. Phylogenetic analysis of deduced FT amino acid sequences from nine plants indicated that the proteins fall into two groups: the FT group, which promotes flowering; and the TFL1 group, which prolongs vegetative identity (Figure 1B). Pd FT2, Pt FT2, and Pt FT1 are clustered with Hd3a, Cs FT (citrus), and At FT, whereas Pt FT3 and Pt FT4 fall into the TFL1 group. This grouping suggests that Pd FT2 may promote flowering.
Figure 1.
Poplar FT/TFL1-Like Genes and Their Phylogenetic Analysis.
(A) Scheme of the poplar FT/TFL1-like genes located in the recently sequenced poplar genome. Shown are the positions and sizes in base pairs of exons and introns. RKIP denotes Raf Kinase Inhibitor Protein, also known as Phosphatidylethanolamine Binding Protein.
(B) Phylogenetic relationship of FT/TFL1-like proteins from poplar (Pt/Pd), tomato (Sl), apple (Md), citrus (Cs), grape vine (Vv), rice (Os), Arabidopsis (At), and Antirrhinum (Am). Arabidopsis MFT was used as an outgroup. Bootstrap numbers are shown at nodes.
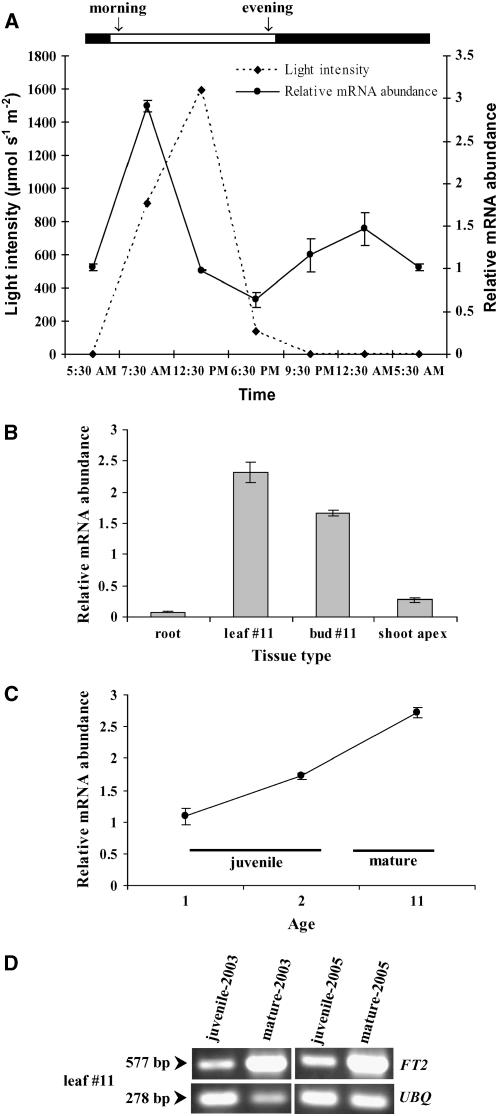
We first determined whether the abundance of FT2 mRNA fluctuated throughout the day and in which tissues in P. deltoides the FT2 transcripts were most abundant. FT2 mRNA was most abundant in leaf 11 during sunrise and declined to predawn levels by midday, when the light fluence rate reached 1600 μmol·m−2·s−1 (Figure 2A). Thus, we collected tissues 2.5 h after sunrise throughout the project period. In mid May, FT2 mRNA was most abundant in leaf 11 and its axil bud, which was destined to be floral (Figure 2B). However, its expression was detected at background levels in roots and at the shoot apex. These data suggest that FT2 expression may be upregulated in the leaves and axillary buds.
Figure 2.
FT2 Transcript Accumulation in P. deltoides Tissues.
(A) Over a 24-h period, FT2 transcript was more abundant in the morning, when the rate of light intensity increased in leaf 11 (n = 3). Leaves were sampled on May 16, and the rate of light intensity (μmol·m−2·s−1) at each leaf sampling time is given. Error bars show sd.
(B) FT2 transcripts were abundant in leaves and buds (number 11) that were destined to be floral. Tissues were sampled on May 16 (n = 3). Error bars show sd.
(C) Abundance of FT2 transcripts increased in leaf 11 from the juvenile to the mature developmental stage. Leaves were sampled on May 16 (n = 3). Error bars show sd.
(D) Abundance of FT2 transcripts was also measured in leaf 11 sampled from different trees at the juvenile (1 year old) and mature (11 years old) developmental stages in 2003 and 2005. The poplar UBQ transcript was also amplified to verify that similar amounts of cDNA were used in the RT-PCR. Numbers at left represent the size of the amplified cDNA fragments in base pairs. The RT-PCR data are representative of at least three experiments.
We then examined the abundance of FT2 transcripts in leaf 11 from juvenile and adult trees (genetically unrelated) at 1, 2, and 11 years of age and found that their abundance increased from the juvenile to the reproductive developmental phase (Figure 2C). This experiment was expanded to leaf samples collected from different trees in 2003 and 2005 (Figure 2D). Although trees sampled in 2005 were genetically unrelated, trees sampled in 2003 came from the same clone (genetically identical). We found that the abundance of FT2 transcript was low in juvenile trees (1 year old) but at least threefold higher in mature trees (11 years old), regardless of genetic relatedness. Thus, it is possible that the low abundance of FT2 transcript during the juvenile period accounts for the lack of flowering in juvenile trees. Then, we speculated that an increase in the production of FT2 transcript should induce premature flowering in juvenile poplar trees.
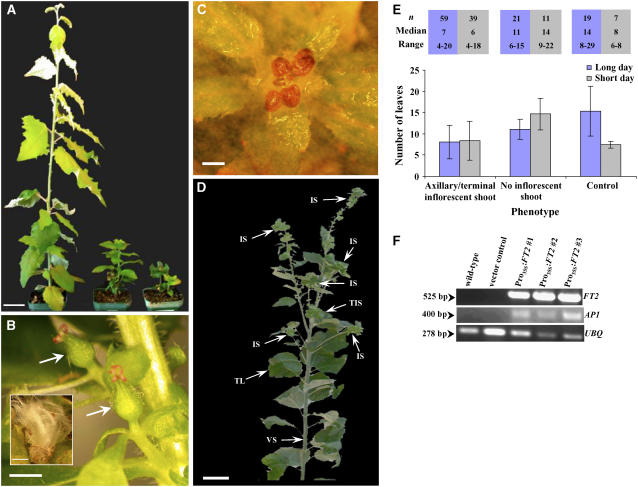
A binary vector for the overexpression of FT2 was constructed by fusing the cauliflower mosaic virus (CaMV) 35S promoter to the protein-coding region of FT2 cDNA (Pro35S:FT2). Agrobacterium tumefaciens carrying this binary vector was used to transform the juvenile female poplar clone 717-1B4 (P. alba × P. tremula). Fifty-five transformed lines were independently regenerated and planted in soil. Forty-five lines (82%) produced flowers 7 months after transformation of tissues (Figures 3A and 3B), whereas nine lines had not formed flowers after 18 months. One line produced flowers 9 months after transformation of tissues. We observed the formation of axillary flowers (in leaf axils) (Figure 3B) and terminal inflorescence shoots with terminal flowers (Figures 3C and 3D). Seeds were not found in mature capsules (Figure 3B, inset). Because poplar forms unisexual flowers and a female poplar clone was transformed, the absence of seeds was probably attributable to lack of pollen. The vector control (no promoter with β-glucuronidase) and wild-type plants had not produced flowers 18 months after the start of the experiment. Control and Pro35S:FT2 poplar trees planted in soil were grown under long and short days for 76 d. Axillary/terminal inflorescence shoots were observed on Pro35S:FT2 poplar trees under both daylength conditions (Figure 3E). Control trees did not form inflorescence shoots. When the first inflorescence was visible, Pro35S:FT2 poplar trees that formed axillary/terminal inflorescence shoots had similar numbers of leaves under both daylength conditions (Figure 3E). Formation of the terminal inflorescence shoot prevented Pro35S:FT2 poplar trees from producing more leaves.
Figure 3.
An Increase in the Production of FT2 via the Constitutive Promoter 35S (Pro35S:FT2) Induced Premature Flowering in Juvenile Poplar Trees (717-1B4; P. alba × P. tremula).
(A) Plants were photographed 9 months after transformation with Pro35S:FT2. The tallest plant is the control. The other two plants with lateral and terminal flowers are independent lines transformed with Pro35S:FT2. Bar = 2 cm.
(B) Closeup photograph of lateral flowers. Arrows point to carpellate flowers. The inset shows an open mature seed capsule. Bars = 1 mm.
(C) Pro35S:FT2 plants formed terminal flowers (arrow). Bar = 1 mm.
(D) Another independent Pro35S:FT2 poplar line shown at 13 months after transformation. IS, inflorescence shoot; TIS, terminal inflorescence shoot; TL, true leaf; VS, vegetative shoot. Bar = 2 cm.
(E) After rooting and planting in soil, Pro35S:FT2 poplar plants were grown under long (16 h of light/8 h of dark) and short (8 h of light/16 h of dark) days for 76 d, phenotypes were observed, and leaf count was taken when inflorescence shoots were visible. Error bars show sd.
(F) FT2 transcript was abundant in poplar lines transformed with the Pro35S:FT2 construct, whereas no transcript was detected in wild-type or vector control plants. A downstream gene, AP1, was expressed in the Pro35S:FT2 poplar lines. The poplar UBQ transcript was also amplified to verify that similar amounts of cDNA were used in the RT-PCR. Numbers at left represent the size of the amplified cDNA fragments in base pairs. The RT-PCR data are representative of at least three experiments.
FT2 transcript was abundant in the Pro35S:FT2 poplar trees that flowered and was not detected in control trees (Figure 3F). Unlike in control trees, expression of a potential downstream gene, Pd AP1, was high in Pro35S:FT2 trees (Figure 3F). AP1 was selected as a potential downstream gene based on the Arabidopsis flowering model (Mouradov et al., 2002), recently published data suggesting that AP1 is likely a direct target of FT in Arabidopsis (Abe et al., 2005; Wigge et al., 2005), and unpublished data showing that AP1 is expressed in floral meristems of P. trichocarpa (A. Brunner, personal communication).
FT2 Induces Early Flowering in Arabidopsis
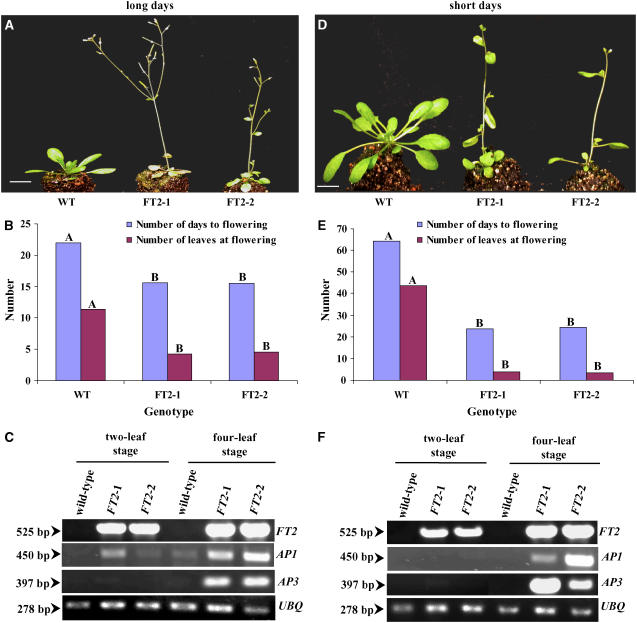
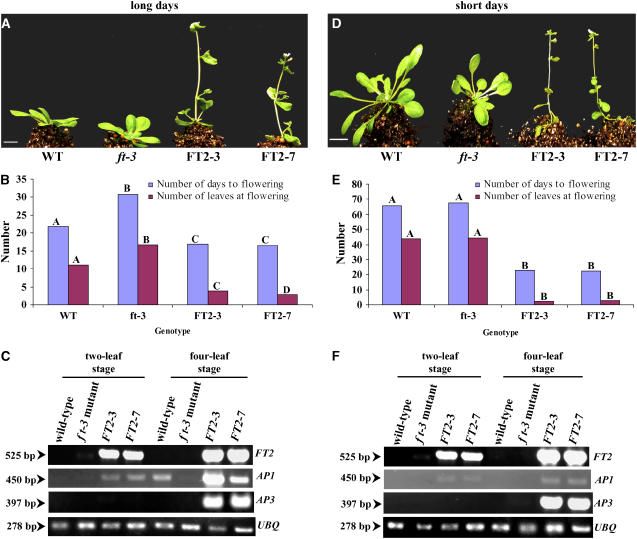
To validate our findings in poplar, we transformed Pro35S:FT2 into wild-type Arabidopsis (ecotype Columbia [Col-0]). Six independent transgenic lines in the third generation were selected for phenotypic observation. All of the lines flowered earlier than their wild-type counterparts. Then, we randomly selected two lines for a detailed phenotypic analysis under long and short days. Regardless of the duration of the light period, transgenic lines induced earlier flowering relative to controls (Figures 4A and 4D). Flowering time, in terms of number of days to flowering and number of leaves at flowering, differed significantly (P < 0.001) between the transgenic lines and controls under both long and short days (Figures 4B and 4E). Transgenic lines flowered within 16 d of seed sowing at the 4-leaf (rosette) stage under long days, whereas wild-type controls flowered within 22 d of seed sowing at the 11-leaf stage. Under short days, transgenic lines flowered within 25 d of seed sowing at the 4-leaf stage, whereas wild-type controls flowered within 64 d of seed sowing at the 44-leaf stage. The abundance of FT2 transcript in each line was examined using RT-PCR at the two- and four-leaf stages. In wild-type control plants, no transcript was detected at any developmental stage or daylength (Figures 4C and 4F). However, the abundance was high in Pro35S:FT2 lines throughout development. These data suggest that the high abundance of FT2 transcript in transgenic lines induced early flowering compared with wild-type controls.
Figure 4.
Ectopic Expression of FT2 Induced Early Flowering under Long Days and Short Days in Wild-Type Arabidopsis (Col-0).
(A) Two randomly selected lines (FT2-1 and FT2-2) that constitutively expressed FT2 exhibited an early-flowering phenotype relative to wild-type controls under long days. The photograph was taken 18 d after planting. Bar = 1 cm.
(B) Number of days to flowering and number of leaves at flowering differed significantly between transgenic lines and controls (Genotype) (n = 15; P < 0.001). Different letters above bars of the same color indicate that the genotypes differ significantly for flowering time.
(C) RT-PCR analysis of the accumulation of FT2, AP1, and AP3 transcripts at the two- and four-leaf stages of Arabidopsis. Numbers at left represent the size of the amplified cDNA fragments in base pairs. The Arabidopsis UBQ transcript was also amplified to verify that similar amounts of cDNA were used in the RT-PCR. The RT-PCR data are representative of at least three experiments.
(D) Two randomly selected lines (FT2-1 and FT2-2) that constitutively expressed FT2 exhibited an early-flowering phenotype relative to wild-type controls under short days. The photograph was taken 30 d after planting. Bar = 1 cm.
(E) Number of days to flowering and number of leaves at flowering differed significantly between transgenic lines and controls (Genotype) (n = 15; P < 0.001). Different letters above bars of the same color indicate that the genotypes differ significantly for flowering time.
(F) RT-PCR analysis of the accumulation of FT2, AP1, and AP3 transcripts at the two- and four-leaf stages of Arabidopsis. Numbers at left represent the size of the amplified cDNA fragments in base pairs. The Arabidopsis UBQ transcript was also amplified to verify that similar amounts of cDNA were used in the RT-PCR. The RT-PCR data are representative of at least three experiments.
To support this assertion, we analyzed the abundance of the Arabidopsis AP1 and AP3 transcripts in control and transgenic plants. AP1 is likely a direct target of FT in Arabidopsis (Abe et al., 2005; Wigge et al., 2005); thus, it can serve as a good marker to determine whether plants are at the flowering stage. AP3 is a downstream gene to FT and AP1 and a B-class gene that is involved in the petal/stamen identity program (Jack et al., 1992; Ng and Yanofsky, 2001; Mouradov et al., 2002). Thus, a developing flower expresses AP3. We used AP3 as a marker to show that plants developed flowers. If the constitutive expression of FT2 induces early flowering, the amount of AP1 transcript should be high. In Pro35S:FT2 lines, the abundance of AP1 transcript was at background levels at the two-leaf stage, whereas it increased at the four-leaf stage in long and short days (Figures 4C and 4F). The abundance of AP3 transcript followed a similar pattern. No significant expression of AP1 or AP3 was detected in wild-type controls or the transgenic lines at the two-leaf stage. These findings further support our conclusion that the early-flowering phenotype of poplar was attributable to the expression of FT2.
FT2 Rescues the Arabidopsis Late-Flowering Phenotype
If FT2 and FT are functionally conserved, then FT2 should substitute a mutated form of FT in Arabidopsis. Arabidopsis ft-3 mutant plants (Arg-119→His) exhibit a strong late-flowering phenotype under long days (Kardailsky et al., 1999). We transformed Arabidopsis ft-3 with Pro35S:FT2 to determine whether the wild-type phenotype could be restored. Six independent transgenic lines that were in the third generation were selected for phenotypic observations. All of the lines flowered earlier than the background ft-3 mutant plants. Then, we randomly selected two lines for a detailed phenotypic analysis under long and short days. Regardless of the duration of the light period, transgenic lines induced early flowering relative to ft-3 mutant plants (Figures 5A and 5D). Flowering time, in terms of number of days to flowering and number of leaves at flowering, differed significantly (P < 0.001) between the transgenic lines, ft-3 plants, and wild-type controls under long and short days (Figures 5B and 5E). Transgenic lines flowered within 17 d of seed sowing at the 3/4-leaf (rosette) stage under long days, whereas ft-3 plants and wild-type controls flowered within 31 d (17-leaf stage) and 22 d (11-leaf stage) of seed sowing, respectively. Under short days, transgenic lines flowered within 23 d of seed sowing at the 3-leaf stage, whereas ft-3 plants and wild-type controls flowered within 67 and 66 d of seed sowing, respectively, at the 44-leaf stage.
Figure 5.
Ectopic Expression of FT2 Complemented the Late-Flowering Phenotype of the Arabidopsis ft-3 Mutant under Both Long Days and Short Days.
(A) Two randomly selected lines (FT2-3 and FT2-7) that constitutively expressed FT2 were used. They flowered earlier than the ft-3 mutant and wild-type controls under long days. The photograph was taken 18 d after planting. Bar = 1 cm.
(B) Number of days to flowering and number of leaves at flowering differed significantly between transgenic lines and controls (Genotype) (n = 15; P < 0.001). Different letters above bars of the same color indicate that the genotypes differ significantly for flowering time.
(C) RT-PCR analysis of the accumulation of FT2, AP1, and AP3 transcripts at the two- and four-leaf stages of Arabidopsis. Numbers at left represent the size of the amplified cDNA fragments in base pairs. The Arabidopsis UBQ transcript was also amplified to verify that similar amounts of cDNA were used in the RT-PCR. The RT-PCR data are representative of at least three experiments.
(D) Two randomly selected lines (FT2-3 and FT2-7) that constitutively expressed FT2 flowered earlier than the ft-3 mutant and wild-type controls under short days. The photograph was taken 30 d after planting. Bar = 1 cm.
(E) Number of days to flowering and number of leaves at flowering differed significantly between transgenic lines and controls (Genotype) (n = 15; P < 0.001). Different letters above bars of the same color indicate that the genotypes differ significantly for flowering time.
(F) RT-PCR analysis of the accumulation of FT2, AP1, and AP3 transcripts at the two- and four-leaf stages of Arabidopsis. Numbers at left represent the size of the amplified cDNA fragments in base pairs. The Arabidopsis UBQ transcript was also amplified to verify that similar amounts of cDNA were used in the RT-PCR. The RT-PCR data are representative of at least three experiments.
To support the phenotypic observations, we analyzed the abundance of FT2, AP1, and AP3 transcripts in Pro35S:FT2 and control plants. In Pro35S:FT2 lines, FT2 transcript was abundant at the two- and four-leaf developmental stages in long and short days. However, no transcript was detected in ft-3 and wild-type plants (Figures 5C and 5F). The abundance of AP1 transcript was low at the two-leaf stage in Pro35S:FT2 lines, whereas it increased at the four-leaf stage in long days (Figures 5C and 5F). We detected a background-level expression of AP1 in wild-type and ft-3 plants at the two- and four-leaf stages, except that the abundance of AP1 transcript increased slightly in wild-type plants under long days. In Pro35S:FT2 plants, the AP1 transcript was present at the two- and four-leaf stages in short days, whereas no transcript was detected in wild-type and ft-3 mutant plants. Consistent with the results of FT2 expression, the transcript of AP3 in Pro35S:FT2 plants was abundant in long and short days at the four-leaf stage, at which flowering was observed (Figures 5C and 5F). No significant expression was observed in ft-3 and wild-type plants. Our data showed that FT2 complemented the mutation, indicating that FT2 and FT are functionally conserved.
FT2 Is Involved in the Initiation of Poplar Seasonal Flowers
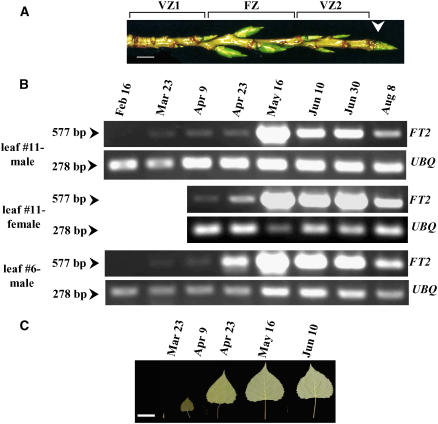
Unlike Arabidopsis, poplar shoots at the reproductive developmental phase initiate axillary early vegetative buds (vegetative zone I), floral buds (floral zone), and late vegetative buds (vegetative zone II) in a sequential manner (Figure 6A) (Yuceer et al., 2003). In wild-type P. deltoides trees, the terminal shoot apex or bud generally does not develop into an inflorescence shoot or a floral bud. Thus, shoots have distinct developmental phases and alternate between vegetative and reproductive growth as part of the seasonal flowering process. We next asked whether FT2 plays a role in these transitions and the initiation of floral meristems. Using RT-PCR, we showed that the abundance of FT2 transcript in preformed leaves 6 and 11 was low during the late winter and early spring, from February to late April (Figure 6B). However, we detected an increase in the transcript in late spring (mid May). The pattern of FT2 expression in leaf 11 was similar in male and female trees. During this time, leaf 11 developed from a primordial preformed leaf to a fully expanded leaf by mid May (Figure 6C).
Figure 6.
Seasonal P. deltoides Shoot and Leaf Development, and the Abundance of FT2 in Leaves.
(A) A poplar shoot at the reproductive developmental stage produces early vegetative buds (vegetative zone 1 [VZ1]), floral buds (floral zone [FZ]), and late vegetative buds (vegetative zone 2 [VZ2]) in a sequential manner. Each bud forms in the axil of a leaf (not shown). Once the shoot ceases annual growth, it forms a vegetative terminal bud (arrowhead), in which the SAM resides. Floral buds are much larger than vegetative buds. Bar = 1 cm.
(B) Accumulation of FT2 transcript increased in preformed leaves, leaf 6 (VZ1) and leaf 11 (FZ), by mid May and then decreased in male and female trees. The poplar UBQ transcript was also amplified to verify that similar amounts of cDNA were used in the RT-PCR. Numbers at left represent the size of the amplified cDNA fragments in base pairs. The RT-PCR data are representative of at least three experiments.
(C) During the time indicated, leaf 11 developed from a primordial preformed leaf to a fully expanded leaf by mid May, after which no leaf expansion was observed. Bar = 2 cm.
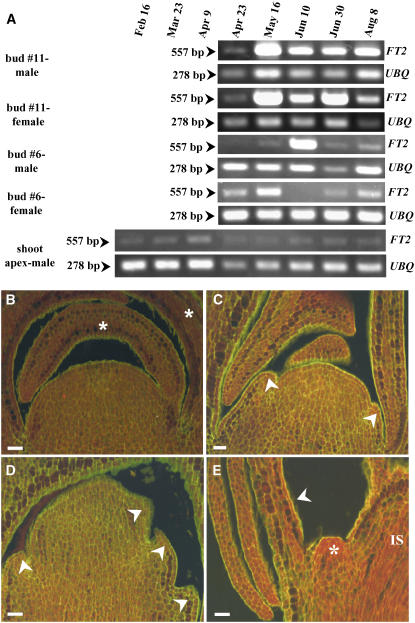
Beginning in mid May, FT2 transcript in male and female trees was abundant in bud 11, which formed an inflorescence shoot (Figure 7A). We only observed scale formation in this bud during late April (Figure 7B). Then, bracts, which are the first sign of floral meristem formation (Yuceer et al., 2003), began forming on flanks of the inflorescence shoot in early May (Figures 7C and 7D). Floral meristems in axils of bracts were beginning to form in early June (Figure 7D). Thus, the timing of the increase of FT2 transcript in preformed leaves and bud 11 is consistent with the morphological changes in bud 11. To further support this observation, we examined the abundance of FT2 transcript in the vegetative zone I bud (bud 6) during the same period. We detected an increase in the abundance of FT2 transcript in male trees only in mid June, whereas FT2 transcript in female trees increased slightly in mid May (Figure 7A). However, at the other sampling dates, its abundance was at background levels. This finding might be attributable to the instability of FT2 transcript in this bud, whose fate was destined to be a vegetative shoot (which produces true leaves), instead of an inflorescence shoot (which produces bracts and floral meristems). In addition, we observed that the abundance of FT2 transcript was at background levels in the shoot apex (Figure 7A). Thus, no change in pattern was detected. When the expression of FT2 transcript was increased experimentally in the transgenic poplar, the normally vegetative terminal shoot was transformed into an inflorescence shoot that produced flowers (Figure 3D). Consequently, the terminal shoot was unable to make the transition back to the vegetative phase. This indicates that the expression of FT2 is repressed at the normally vegetative terminal shoot apex in poplar.
Figure 7.
Temporal Abundance of FT2 Transcript in Buds, and Morphological Changes in Bud 11 of Mature P. deltoides Shoots.
(A) Abundance of FT2 transcript increased in bud 11 (destined to be a floral bud) by mid May and then stayed steady. However, it was at background levels in bud 6 (vegetative or leaf bud) and at the shoot apex (vegetative). The poplar UBQ transcript was also amplified to verify that similar amounts of cDNA were used in the RT-PCR. Numbers at left represent the size of the amplified cDNA fragments in base pairs. The RT-PCR data are representative of at least three experiments.
(B) Bud scales (asterisks) were formed on flanks of the meristem in bud 11 on April 28. Bar = 5 μm.
(C) Bracts (arrowheads) began forming on flanks of the inflorescence meristem on May 9. Bar = 5 μm.
(D) The inflorescence meristem continued growing, and more bracts (arrowheads) were formed on May 23. Bar = 25 μm.
(E) Floral meristems (asterisk) were visible in axils of bracts (arrowhead) on flanks of the inflorescence shoot (IS) on June 13. Bar = 25 μm.
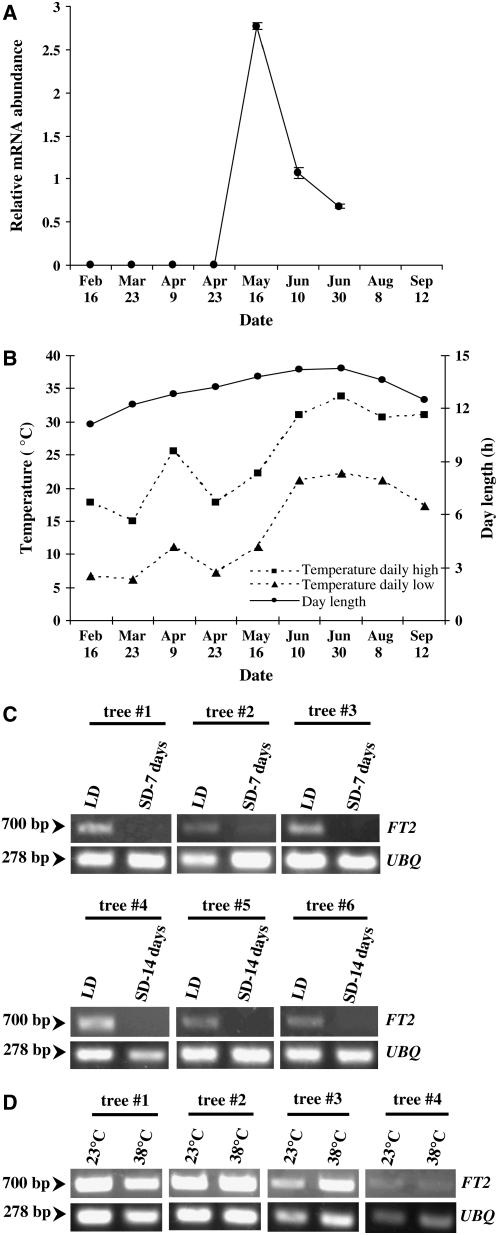
We confirmed the increase in FT2 transcript in mid May using real-time RT-PCR (Figure 8A). Potential factors involved in this increase include development, daylength, and temperature. We have already shown that tree development from the juvenile to the mature stage is likely a factor (Figures 2C, 2D, and 3). Our data also suggest that on seasonal shoots, the development of leaf 11 from a primordial preformed leaf to a fully expanded, mature leaf by mid May correlates with the increase in FT2 transcription (Figure 6C). During this period, daylength increases by ∼3 h, and then it decreases beginning in June (Figure 8B). Could this increase in daylength be the basis of the increase in the abundance of Pd FT1 transcript? To answer this question, 717-1B4 (P. alba × P. tremula) trees were grown under long (14 h) and short (8 h) days for 14 d. The FT2 transcript was abundant under long days throughout the experiment, whereas it was either at background levels (first 7 d) or undetectable after 14 d under short days (Figure 8C). Poplar trees grown in short days ceased shoot growth. These data suggest that long days promote the abundance of FT2 transcript. There is also an increasing trend in temperature from February to July during the flower bud initiation period (Figure 8B). Thus, poplar trees were treated under two temperature regimes (23 and 38°C) to determine whether temperature would play a role in the abundance of FT2 transcript. We did not observe a change in the expression pattern of FT2 under either temperature regime (Figure 8D), suggesting that temperature is not a factor in the seasonal expression pattern of FT2.
Figure 8.
Environmental Factors Affecting the Abundance of FT2 Transcript in Poplar.
(A) In leaf 11, RT-PCR results were confirmed using quantitative real-time PCR (n = 3). Error bars show sd.
(B) Trends in daylength and temperature during the flower bud initiation and development period in 2005 (north-central Mississippi).
(C) RT-PCR analysis of the accumulation of FT2 transcript under long (LD) and short (SD) days. Numbers at left represent the size of the amplified cDNA fragments in base pairs. The poplar UBQ transcript was also amplified to verify that similar amounts of cDNA were used in the RT-PCR. The RT-PCR data are representative of at least three experiments.
(D) Abundance of FT2 transcripts under two temperature regimes. Numbers at left represent the size of the amplified cDNA fragments in base pairs. The poplar UBQ transcript was also amplified to verify that similar amounts of cDNA were used in the RT-PCR. The RT-PCR data are representative of at least three experiments.
DISCUSSION
First-Time Flower Initiation in Poplar
Although a few foreign genes (e.g., Arabidopsis LEAFY and AP1 genes) have been reported to induce flowering in several woody perennial species such as citrus and poplar (Weigel and Nilsson, 1995; Pena et al., 2001), native genes involved in the flowering of perennial woody species have not been identified. Here, we report that FT2 is involved in first-time and seasonal flowering in the woody perennial poplar. Concurrent with final revisions of this article, Bohlenius et al. (2006) reported that another poplar FT family member, Pt FT1, is involved in the regulation of flowering time in poplar. FT2 is 91% similar to FT1 in coding regions at the amino acid level (Figure 1). Although similar environmental factors are present at the juvenile and reproductive developmental stages, poplar trees do not normally flower until the age of 7 to 10 years (Braatne et al., 1996). Our results showed at least a threefold increase in the abundance of FT2 transcript from the juvenile to the reproductive developmental stage in poplar, suggesting that a critical level of FT2 expression is needed to initiate flowering. Consistent with this finding, FT levels in wild-type Arabidopsis plants are low at the juvenile developmental stage and gradually increase from the juvenile to the reproductive phase under both long and short days (Kardailsky et al., 1999; Kobayashi et al., 1999; Takada and Goto, 2003). Consequently, overexpression of FT induces flowering at an early developmental stage in Arabidopsis. We postulate that low abundance of FT2 transcript during the lengthy juvenile period might be a major constraint to flower initiation in juvenile poplar. When FT2 transcript (Pro35S:FT2) levels were increased in juvenile poplar trees, rapid shoot-to-flower or shoot-to-inflorescence conversion was observed. Bohlenius et al. (2006) observed similar phenotypes when FT1 was overexpressed in a male hybrid poplar (P. tremula × P. tremuloides). The observation that ectopic expression of FT2 was sufficient to promote the flowering of wild-type Arabidopsis plants in an early developmental stage is also consistent with a role of FT2 in phase transition. These results suggest that the regulation of FT2 transcript has a direct and/or systemic role in the transition to the reproductive phase.
Why is the abundance of FT2 transcript low during the juvenile period? Are juvenile poplar trees incompetent to perceive floral stimuli, such as photoperiod? The only changing variables from the juvenile to the reproductive developmental stage are chronological age and the size of the trees. One hypothesis is that the tree develops the capacity for the first-time flowering as a result of either attaining a certain size or undergoing a certain number of cycles of growth and dormancy (Chalupka and Cecich, 1997). Evidence from birch (Betula verrucosa) supports the former factor (Longman and Wareing, 1959). Periodic growth (cycles between active growth and dormancy) did not induce flowering after six cycles, whereas continuous growth resulted in taller trees with flowers at very early ages after 1 year. However, how this size factor plays a role in the abundance of FT2 transcript and first-time flowering remains unknown. Perhaps it is related to development.
An alternative hypothesis is that juvenility acts to reduce the expression of FT2 transcription or a systemic signal upstream of FT2 in the floral pathway. Potentially, chromatin-based repression of FT2 could play a central role in poplar. In eukaryotes, chromatin structure causes a restrictive ground state, preventing proteins (e.g., RNA polymerases and transcriptional regulators) from binding to DNA (Workman and Kingston, 1998). It is becoming clear in Arabidopsis that modification of chromatin structure, by disruption of Polycomb group protein complexes at target loci, plays a major role in the floral transition. For example, the Polycomb group proteins EMBRYONIC FLOWER2 and FERTILIZATION-INDEPENDENT ENDOSPERM act as repressors of the floral homoeotic genes during embryo and vegetative development (Kinoshita et al., 2001; Yoshida et al., 2001; Moon et al., 2003). Specifically, the plant homeodomain finger protein, EARLY BOLTING IN SHORT DAYS, represses FT expression (Pineiro et al., 2003). A homolog of HETEROCHROMATIN PROTEIN1, TFL2, counteracts the promotion effects of CO on FT expression to ensure photoperiodic regulation of flowering (Takada and Goto, 2003). TFL2 particularly silences flowering-related genes within euchromatin (Nakahigashi et al., 2005). Two homologs of TFL2 are present in the poplar genome (poplar database accession numbers eugene3.00130688 and eugene3.00190501), but it is unknown whether they repress the expression of FT2 transcription or a systemic signal upstream of FT2. We do not dismiss the possibility that other poplar-specific proteins are involved in the repression mechanism. Perhaps the repression mechanisms in effect during the juvenile period of poplar are overcome by environmental cues, such as photoperiod or vernalization, through a pathway of genes as trees grow larger.
Seasonal Flower Initiation and Shoot Architecture in Poplar
Our data indicate that FT2 plays an additional role in the initiation of seasonal flowers in poplar. The shoot architecture of poplar has evolved to accommodate both vegetative and reproductive growth, with no detrimental interference with growth at either stage. The juvenile poplar shoots form only vegetative buds that contain vegetative shoots with true leaves. However, upon the transition from the juvenile to the reproductive phase, poplar shoots begin producing flower buds that contain inflorescence shoots with bracts and floral meristems. Thus, shoots develop in a defined pattern that has specific vegetative and floral bud locations. Shoots initiate early vegetative buds, floral buds, and late vegetative buds in a sequential manner (Yuceer et al., 2003). Although FT2 transcripts were rare in early developmental stages of preformed leaves (e.g., leaves 6 and 11 from the base of shoots), they were abundant in fully expanded leaves in mid May. Then, there was a decrease in transcript abundance. What is the basis of the increase and decrease in the amount of FT2 transcript? In Arabidopsis, FT transcript becomes abundant under long days and promotes flowering (Kardailsky et al., 1999; Kobayashi et al., 1999). A similar, but reverse, molecular mechanism is present in rice to control flowering time in response to daylength. The expression of Hd3a, a homolog of FT, is induced specifically under short days and promotes flowering in rice (Kojima et al., 2002). Because daylength increases by ∼3 h from February to late May and then decreases beginning in June, we hypothesized that the change in daylength is the basis of the increase and decrease in the abundance of FT2 transcript. Our data show that the abundance of FT2 transcript decreased when poplar trees were transferred from long days to short days, suggesting that long days promote FT2 transcription. In addition, our data indicate that temperature does not appear to play a role in the abundance of FT2 transcripts. The genetic mechanism controlling FT2-induced flowering in poplar is possibly related to photoperiodic flowering in Arabidopsis and rice. However, how daylength-induced FT2 transcription interacts with leaf development and maturation remains elusive. Perhaps leaves become sensitive to light at a particular developmental stage, or FT2 is also controlled by the developmental pathway. The latter is the case in Arabidopsis, in which photoperiod and developmental pathways converge on FT (Samach et al., 2000).
The terminal shoot bud or apical meristem normally does not become a flower bud or an inflorescence shoot in poplar. We observed that the abundance of FT2 transcript was at background levels in the shoot apex of normally growing mature trees throughout the growing season. This finding indicates that the expression of FT2 is repressed at the normally vegetative terminal shoot apex in poplar. However, when poplar was transformed with Pro35S:FT2, in addition to axillary flowers in leaf axils, the terminal shoots of Pro35S:FT2 poplar trees were transformed into inflorescence shoots that produced flowers in their flanks. Then, the inflorescence shoot formed a terminal flower, which terminated the growth. Pro35S:FT2 poplar trees express FT2 in every cell, because FT2 is under the control of the constitutive CaMV 35S promoter. Thus, axillary and terminal meristems are converted to flowers/inflorescence shoots. Interestingly, the transition of the terminal shoots of Pro35S:FT2 poplar trees to inflorescence shoots resembles the normal Arabidopsis flowering process, in which the vegetative shoot makes a transition to an inflorescence shoot that generally does not revert to the vegetative phase (Boss et al., 2004). These data further support our expression data indicating that FT2 is suppressed at the shoot apex of wild-type poplar trees.
In the Arabidopsis model, FT is activated in the phloem companion cells of mature leaves (Takada and Goto, 2003), its mRNA moves out of the phloem to the SAM (Huang et al., 2005), and then FT protein interacts with FD protein at the SAM to induce floral development through AP1 (Abe et al., 2005; Wigge et al., 2005). FT mRNA appears to be part of a systemic signal. This model supports the classical studies showing that a photoinduced systemic signal, florigen, is synthesized in leaves and transported to the SAM, where floral development is induced (Zeevaart, 1976). Surprisingly, we observed an increase in the abundance of FT2 transcript in bud 11 but not in bud 6 and the shoot apex in poplar. Bud 11 subsequently formed bracts and floral meristems in flanks of the inflorescence shoot, whereas bud 6 formed a vegetative shoot with true leaves. Unlike in Arabidopsis, is the systemic floral signal transported directly from leaves to the axillary meristems in poplar? It is possible that the FT2 mRNA/protein, along with other florigenic compounds produced in preformed leaves, are transported to the developing buds in the floral zone through direct vascular connections. Evidence suggests that leaves along the axis of a poplar shoot are interconnected by vascular tissues in a specific, repeating pattern (Larson and Pizzolato, 1977; Pizzolato and Larson, 1977; Dickson, 1986). This observation can be evolutionarily and developmentally important in the perennial plant poplar. Poplar grows vertically via the terminal bud/shoot tip for 100 to 200 years. Conversion of the terminal shoot to an inflorescence shoot, as we observed in Pro35S:FT2 poplar trees, would affect tree identity, growth, and wood quality. Thus, poplar has perhaps developed a different signaling mechanism, through the regulation of FT2 transcription among various components of a shoot, that reflects the present architecture, in which both vegetative and reproductive growth are accommodated.
METHODS
Isolation of Full-Length FT2 Transcript from Poplar
A combination of PCR-based genome walking and rapid amplification of cDNA ends (RACE) was used to clone FT2 from Populus deltoides. FT cDNA and genomic DNA (Arabidopsis thaliana) were aligned using Lasergene (DNASTAR). The primers 3FT-1 (primary) and 3FT-5 (secondary) were designed based on the FT genomic DNA. All of the primer sequences used in this research can be found in Supplemental Table 1 online. The BD GenomeWalker universal kit (BD Biosciences) was used for genome walking on P. deltoides DNA isolated from leaves using the DNeasy plant maxi kit (Qiagen). A 600-bp genomic sequence was amplified, cloned into pGEM-T Easy vector (Promega), and cycle-sequenced using the CEQ 8000 genetic analysis system (Beckman Coulter). The poplar DNA sequence was aligned with FT cDNA. A 300-bp overlapping region was identified. Then, primer 5FT2-E was designed in the overlapping region for 5′ RACE using the SMART RACE cDNA amplification kit (BD Biosciences). Primer FT2-5E3 was designed to amplify the full-length FT2 cDNA by conducting 3′ RACE. Primers FT2-5E3 (forward) and FT2-E4 (reverse) and Pfu DNA polymerase (Stratagene) were used for PCR amplification of FT2 cDNA (GenBank accession number AY515152).
Phylogenetic Analysis
To identify the related sequences in the GenBank (http://www.ncbi.nlm.nih.gov/BLAST) and poplar (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html) databases, protein–protein BLAST was conducted using the deduced amino acid sequence for FT2. To construct the phylogeny, we included sequences with E values of ≤10−5. Shorter sequences were not included. ClustalX was used to create multiple alignments (Thompson et al., 1997) (see Supplemental Figure 2 online). The resulting alignment was used to generate the phylogenetic tree by the neighbor-joining method (Saitou and Nei, 1987). TreeView was used to draw the tree (Page, 1996). We conducted bootstrap analysis to estimate nodal support based on 1000 replicates.
Tissue, Developmental, and Daily Analysis of FT2 Transcript in Poplar
Various tissues (e.g., leaf 11, root, shoot apex [<2 mm in size with primordial leaves and scales removed], and bud 11) from three male and three female wild-type mature P. deltoides trees (11 years old) in central Mississippi were used to identify the tissue specificity and determine the mRNA abundance of FT2. We sampled these tissues on May 16, because FT2 mRNA was most abundant in leaves on this date. For the analysis of FT2 mRNA expression through P. deltoides developmental stages, leaf 11 from the base of the shoot was collected at the juvenile (1 and 2 years old) and mature (11 years old) developmental stages. We collected samples from three 1-year-old, three 2-year-old, and three 11-year-old trees in 2004. Mature trees (11 years old) were male, but we did not know the gender of juvenile trees. Juvenile and mature trees were genetically unrelated. We also collected samples from three juvenile trees (1 year old) and three mature trees (11 years old, male) in 2003 and 2005. Although juvenile and mature trees were genetically unrelated and the gender of juvenile trees was not known in 2005, juvenile and mature trees were male and came from the same clone (genetically identical) in 2003. A lift-truck was used to collect samples from ∼30-m-tall trees. To identify the daily expression pattern of FT2, leaf 11 in the floral zone was sampled from mature trees at six different time points: 5:30 am, 7:30 am, 12:30 pm, 6:30 pm, 9:30 pm, and 12:30 am. Light fluence rate (μmol·m−2·s−1) was measured at each time point using a light meter (Li-Cor model LI-189). Total RNA from samples was isolated using the RNeasy plant mini kit (Qiagen) with in-column DNase I digestion (Qiagen). The expression pattern of FT2 transcript was analyzed using RT-PCR. One microgram of total RNA from each sample was reverse-transcribed using Moloney murine leukemia virus reverse transcriptase (Promega). Gene-specific primers FT2-PT1 (forward) and FT2-E4 (reverse) were used to detect FT2 transcript. To detect the P. deltoides ubiquitin transcript (UBQ), the gene-specific primers UBQ-1 (forward) and UBQ-2 (reverse) were used. By following the manufacturer's instructions, quantitative real-time PCR was prepared using the LightCycler FasterStart DNA Master SYBR Green I kit (Roche) and analyzed with the LightCycler 2.0 system (Roche). The standard curve was generated by log [cDNA] (represented by the amount of total RNA used in the real-time reaction) versus the crossing point value using a series of dilutions of the first-strand cDNA. The ratio between the expression levels of FT2 and UBQ for each sample was calculated using the relative quantitative analysis method. Each assay was repeated at least three times.
Binary Vector Construction and Arabidopsis and Poplar Transformation
The protein-coding region of FT2 cDNA was PCR-amplified with Pfu DNA polymerase using the primers FT2-PT1 and FT2-PT2. To develop the construct Pro35S:FT2, the fragment was subsequently cloned into the pBI121 binary vector (BD Biosciences) to replace the β-glucuronidase reporter gene, which was downstream of the CaMV 35S promoter. Agrobacterium tumefaciens strain C58 was transformed with the binary vector harboring Pro35S:FT2, which was then used to transform both Arabidopsis and poplar.
The ft-3 mutant and wild-type (Col-0) Arabidopsis plants were transformed with the Pro35S:FT2 construct using the floral dipping transformation method (Clough and Bent, 1998). The transformed lines were selected on medium containing half-strength Murashige and Skoog salts and kanamycin (50 μg/mL). The third generation of transgenic lines was used for phenotypic assessment of flowering time. The female poplar clone 717-1B4 (P. alba × P. tremula) was transformed with the Pro35S:FT2 construct and the vector control (pBI101; BD Biosciences) using the established poplar transformation procedure (Han et al., 2000). Poplar plants (30 to 60 d old) were grown in vitro as transformation material. Leaf discs (1 × 1 cm2) were used to generate explants. All of the cultures were maintained in an incubator (Percival Scientific) at 25°C with a 16-h-light photoperiod under cool-white fluorescent lamps. Under our experimental conditions, regeneration of roots from preparation of leaf discs and preculturing took 210 to 231 d (7 to 8 months).
Analysis of Flowering Response in Transgenic Arabidopsis Lines
The third generation of Pro35S:FT2 transgenic lines, including wild-type (Col-0) and ft-3 mutant plants, was grown at 22/19°C (day/night) under short days (8-h photoperiod) and long days (16-h photoperiod). Plants were arranged in a randomized complete block design with three blocks. Each genotype within a block was represented by five plants. Thus, 15 plants from each genotype were observed for each daylength condition. Flowering time was measured by counting the number of rosette leaves and the number of days from sowing when the first flower bud was seen. Analysis of variance was performed with the SAS software package (SAS Institute) for the number of leaves at flowering and the number of days to flowering to determine whether there were significant differences among genotypes (α = 0.05). Means were separated by Fisher's protected LSD procedure in SAS.
To support flowering-time data, transcript analysis of FT2, AP1, and AP3 was conducted. Total RNA was extracted from (1) the Pro35S:FT2 transgenic lines both in the wild-type and ft-3 mutant backgrounds, (2) wild-type (Col-0) plants, and (3) ft-3 plants at the two- and four-leaf stages. Under long days, tissues from the two- and four-leaf stages of wild-type and ft-3 plants were sampled at 12 and 15 d after planting, respectively. Tissues from the Pro35S:FT2 transgenic lines produced in wild-type and ft-3 mutant backgrounds were sampled at 12 and 16 d after planting for the two- and four-leaf stages, respectively. Under short days, tissues from the two- and four-leaf stages of wild-type and ft-3 plants were collected at 14 and 21 d after planting, respectively. Tissues from the Pro35S:FT2 transgenic lines produced in wild-type and ft-3 backgrounds were sampled at 14 and 16 d after planting for the two- and four-leaf stages, respectively. The RNeasy plant mini kit with in-column DNase I digestion (Qiagen) was used to extract total RNA. RT-PCR was used to analyze FT2, AP1, and AP3 expression in Arabidopsis. One microgram of total RNA from each sample was reverse-transcribed using Moloney murine leukemia virus reverse transcriptase (Promega). Gene-specific primers FT2-PT1 (forward) and FT2-PT2 (reverse), TAP1-1 (forward) and TAP1-2 (reverse), and AP3-1 (forward) and AP3-2 (reverse) were used to detect FT2, AP1, and AP3, respectively. Gene-specific primers UBQ-1 (forward) and UBQ-2 (reverse) were used to detect the Arabidopsis ubiquitin transcript (UBQ). The amplified FT2, AP1, AP3, and UBQ fragments were cloned and sequenced for confirmation.
Analysis of Poplar Trees in Response to Pro35S:FT2
Poplar plants were initially grown in a biological incubator (Percival Scientific) at 25°C under a 16-h photoperiod from cool-white fluorescent lamps. After root formation, they were transplanted to soil and placed under short (8-h photoperiod) and long (16-h photoperiod) days at 22/19°C (day/night) in the ArabiSun Lighting System (Lehle Seeds). Light intensity at the plant canopy was 135 μmol·m−2·s−1 under short days and 180 μmol·m−2·s−1 under long days. Flowering response and leaf count were taken on each plant. The flowering time of transformed poplar lines was measured by counting the number of days from the inoculation of explants with A. tumefaciens that contained Pro35S:FT2. To analyze the abundance of FT2 and AP1 (a potential downstream gene of FT2) transcripts, total RNA was isolated from controls (wild-type and vector control [pBI101] plants) and the Pro35S:FT2 lines. First-strand cDNA was synthesized as described previously. Gene-specific primers FT2-PT1 and FT2-PT2 and DAP1-1 (forward) and DAP1-2 (reverse) were used to detect the FT2 and AP1 transcripts, respectively. Gene-specific primers UBQ-1 and UBQ-2 were used to detect the poplar UBQ mRNA.
Analysis of Seasonal Flowering in Poplar
For temporal expression analysis of FT2 mRNA during shoot development, leaf 6, leaf 11, bud 6, bud 11 (from the base of the shoot), and the shoot apex (<2 mm in size with primordial leaves and scales removed) from mature male and female P. deltoides trees (described above) were collected from February to August for two successive years. Daylength and temperature (daily high and daily low) data for the year 2005 at each sample collection date were obtained from two Internet resources (http://www.sunrisesunset.com and http://ext.msstate.edu/anr/drec/weather.cgi). Total RNA from each sample was extracted, and first-strand cDNA was synthesized as described above. In addition to conventional RT-PCR, quantitative real-time PCR (as described above) was used to confirm the former. Each assay was repeated at least three times. To analyze the morphological changes in axillary bud meristems (bud 11; floral zone), half-strength Karnovsky's fixative (2% paraformaldehyde and 2.5% glutaraldehyde) with phosphate buffer (0.1 M, pH 7.2) was used to fix the tissues for 48 h at 4°C (Yuceer et al., 2002). The tissues were dehydrated in an ethanol series, cleared in tertiary butyl alcohol, embedded in Paraplast, sectioned at 8 μm, and stained with safranin-fast green. A Leica TCS NT confocal system (Leica Microsystems) was used to examine the stained sections.
To determine the photoperiodic regulation of FT2 transcript, poplar trees (female clone 717-1B4; P. alba × P. tremula) were grown in a growth chamber (Environmental Growth Chambers) at 23°C under long days (14 h of light) at a light intensity of 160 μmol·m−2·s−1 for 1 month. Six trees were then transferred to short days (8 h of light) at 23°C for 2 weeks. Leaf samples were collected 2 h after the beginning of the light period under both long and short days. To determine the temperature regulation of FT2 transcript, poplar trees (clone 717-1B4) were initially grown under long days as described above. Four trees were then transferred to the growth chamber with a temperature regime of 38/23°C (day/night) under long days (14 h) for 2 weeks. This temperature regime was used to mimic the natural environment during summer in Mississippi. Leaf samples were collected 2 h after the beginning of the light period from the trees grown in 23/23°C and 38/23°C temperature regimes. Total RNA from each sample was isolated, and first-strand cDNA was synthesized as described previously. The gene-specific primers FT2-1 (forward) and FT-717-2 (reverse) were used to detect the FT2, whereas the gene-specific primers UBQ-1 and UBQ-2 were used to detect the UBQ mRNA as an internal control.
Accession Numbers
The accession numbers of the sequences used are as follows: At MFT (AF147721), At ATC (AB024715), At BFT (Q9FIT4), TSF (AB027506), At TFL1 (CAB85504), At FT (AB027504), At AP3 (NM_115294), and At UBQ (AY057500) from Arabidopsis thaliana; Sl SP (AAC26161), Sl SP2G (AY186734), Sl SP3D (AY186735), Sl SP5G (AY186736), Sl SP6A (AY186737), and Sl SP9D (AAO31795) from Solanum lycopersicum; Md TFL1-1 (BAD06418), Md TFL1-2 (BAD10967), and Md FT (AB161112) from Malus × domestica; Cs TFL1 (AAR04684) and Cs FT (AB027456) from Citrus sinensis; Vv TFL1 (AAM46142) from Vitis vinifera; Am CEN (CAC21563) from Antirrhinum majus; Os Hd3a (AB052944) from Oryza sativa; Pt CEN-1 (AAQ88444), Pt FT1 (poplar database number LG_VII000284), Pt FT2 (poplar database number eugene3.14090001), Pt FT3 (poplar database number LG_XV001090), Pt FT4 (poplar database number LG_IX0251), Pt AP1 (poplar database number grail3.0006057701), and Pd UBQ (poplar database number eugene3.00111099) from Populus trichocarpa; and Pd FT2 (AY515152) from Populus deltoides.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of the Deduced Amino Acids of Pd FT2 and Pt FT2.
Supplemental Figure 2. Amino Acid Alignment of Proteins Used in the Phylogenetic Analysis.
Supplemental Table 1. Primer Sequences That Were Used in PCR Experiments.
Supplementary Material
Acknowledgments
We thank B. Monroe at the Mississippi State University Electron Microscopy Center for help with tissue sectioning and microscopy. We also thank O. Nilsson, A. Brunner, and R. Meilan for comments on the manuscript. The Arabidopsis ft-3 mutant seeds (CS185) were obtained from the ABRC (Ohio State University). This work was supported by the National Science Foundation and the Mississippi State University Life Sciences and Biotechnology Institute. Approved for publication as Journal Article No. FO313 of the Forest and Wildlife Research Center, Mississippi State University.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Cetin Yuceer (mcy1@ra.msstate.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.106.041038.
References
- Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., Ichinoki, H., Notaguchi, M., Goto, K., and Araki, T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 1052–1056. [DOI] [PubMed] [Google Scholar]
- Alvarez, J., Guli, C.L., Yu, X.-H., and Smyth, D.R. (1992). TERMINAL FLOWER. A gene affecting inflorescence development in Arabidopsis thaliana. Plant J. 2 103–116. [Google Scholar]
- An, H., Roussot, C., Suarez-Lopez, P., Corbesier, L., Vincent, C., Pineiro, M., Hepworth, S., Mouradov, A., Justin, S., Turnbull, C., and Coupland, G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131 3615–3626. [DOI] [PubMed] [Google Scholar]
- Ayre, B.G., and Turgeon, R. (2004). Graft transmission of a floral stimulant derived from CONSTANS. Plant Physiol. 135 2271–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlenius, H., Huang, T., Charbonnel-Campaa, L., Brunner, A.M., Jansson, S., Strauss, S.H., and Nilsson, O. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312 1040–1043. [DOI] [PubMed] [Google Scholar]
- Boss, P.K., Bastow, R.M., Mylne, J.S., and Dean, C. (2004). Multiple pathways in the decision to flower: Enabling, promoting, and resetting. Plant Cell 16 (suppl.), S18–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braatne, J.H., Rood, S.B., and Heilman, P.E. (1996). Life history, ecology, and conservation of riparian cottonwoods in North America. In Biology of Populus, R.F. Stettler, H.D. Bradshaw, Jr., P.E. Heilman, and T.M. Hincley, eds (Ottawa, Canada: NRC Research Press), pp. 57–85.
- Bradley, D., Carpenter, R., Copsey, L., Vincent, C., Rothstein, S., and Coen, E.S. (1996. a). Control of inflorescence architecture in Antirrhinum. Nature 379 791–797. [DOI] [PubMed] [Google Scholar]
- Bradley, D., Ratchliffe, O., Vincent, C., Carpenter, R., and Coen, E. (1997). Inflorescence commitment and architecture in Arabidopsis. Science 275 80–83. [DOI] [PubMed] [Google Scholar]
- Bradley, D., Vincent, C., Carpenter, R., and Coen, E. (1996. b). Pathways for inflorescence and floral induction in Antirrhinum. Development 122 1535–1544. [DOI] [PubMed] [Google Scholar]
- Chalupka, W., and Cecich, R.A. (1997). Control of first flowering in forest trees. Scand. J. For. Res. 12 102–111. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Dickson, R.E. (1986). Carbon fixation and distribution in young Populus trees. In Proceedings: Crown and Canopy Structure in Relation to Productivity, T. Fujimori and D. Whitehead, eds, (Ibaraki, Japan: Forestry and Forest Products Research Institute), pp. 409–426.
- Han, K., Meilan, R., Ma, C., and Strauss, S. (2000). An Agrobacterium tumefaciens transformation protocol effective on a variety of cottonwood hybrids (genus Populus). Plant Cell Rep. 19 315–320. [DOI] [PubMed] [Google Scholar]
- Huang, T., Bohlenius, H., Eriksson, S., Parcy, F., and Nilsson, O. (2005). The mRNA of the Arabidopsis gene FT moves from leaf to shoot apex and induces flowering. Science 309 1694–1696. [DOI] [PubMed] [Google Scholar]
- Jack, T., Brockman, L.L., and Meyerowitz, E.M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68 683–697. [DOI] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V.K., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. (1999). Activation tagging of the floral inducer FT. Science 286 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (2001). Polycomb repression of flowering during early plant development. Proc. Natl. Acad. Sci. USA 98 14156–14161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286 1960–1962. [DOI] [PubMed] [Google Scholar]
- Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T., and Yano, M. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43 1096–1105. [DOI] [PubMed] [Google Scholar]
- Larson, P.R., and Pizzolato, T.D. (1977). Axillary bud development in Populus deltoides. I. Origin and early ontogeny. Am. J. Bot. 64 835–848. [Google Scholar]
- Longman, K.A., and Wareing, P.F. (1959). Early induction of flowering in birch seedlings. Nature 184 2037–2038. [Google Scholar]
- Moon, Y.-H., Chen, C., Pan, R.L., Chang, H.-S., Zhu, T., Maffeo, D.M., and Sung, Z.R. (2003). EMF genes maintain vegetative development by repressing the flower program in Arabidopsis. Plant Cell 15 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov, A., Cremer, F., and Coupland, G. (2002). Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell 14 (suppl.), S111–S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahigashi, K., Jasencakova, Z., Schubert, I., and Goto, K. (2005). The Arabidopsis heterochromatin protein1 homolog (TERMINAL FLOWER2) silences genes within euchromatic region but not genes positioned in heterochromatin. Plant Cell Physiol. 46 1747–1756. [DOI] [PubMed] [Google Scholar]
- Ng, M., and Yanofsky, M.F. (2001). Activation of the Arabidopsis B class homeotic genes by APETALA1. Plant Cell 13 739–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, R.D.M. (1996). TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12 357–358. [DOI] [PubMed] [Google Scholar]
- Pena, L., Martin-Trillo, M., Juarez, J., Pina, J.A., Navarro, L., and Martinez-Zapater, J.M. (2001). Constitutive expression of Arabidopsis LEAFY or APETALA1 genes in citrus reduces their generation time. Nat. Biotechnol. 19 263–267. [DOI] [PubMed] [Google Scholar]
- Pineiro, M., Gomez-Mena, C., Schaffer, R., Martinez-Zapater, J.M., and Coupland, G. (2003). EARLY BOLTING IN SHORT DAYS is related to chromatin remodeling factors and regulates flowering in Arabidopsis by repressing FT. Plant Cell 15 1552–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzolato, T.D., and Larson, P.R. (1977). Axillary bud development in Populus deltoides. II. Late ontogeny and vascularization. Am. J. Bot. 64 849–860. [Google Scholar]
- Pnueli, L., Carmel-Goren, L., Hareven, D., Gutfinger, T., Alvarez, J., Ganal, M., Zamir, D., and Lifschitz, E. (1998). The SELFPRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125 1979–1989. [DOI] [PubMed] [Google Scholar]
- Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80 847–857. [DOI] [PubMed] [Google Scholar]
- Reinhardt, D., and Kuhlemeier, C. (2002). Plant architecture. EMBO Rep. 3 846–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwartz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288 1613–1616. [DOI] [PubMed] [Google Scholar]
- Soltis, P.S., Soltis, D.E., and Chase, M.W. (1999). Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature 402 402–404. [DOI] [PubMed] [Google Scholar]
- Somerville, C., and Koornneef, M. (2002). A fortunate choice: The history of Arabidopsis as a model plant. Nature 3 883–889. [DOI] [PubMed] [Google Scholar]
- Takada, S., and Goto, K. (2003). TERMINAL FLOWER 2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN 1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15 2856–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., and Nilsson, O. (1995). A developmental switch sufficient for flower initiation in diverse plants. Nature 377 495–500. [DOI] [PubMed] [Google Scholar]
- Wigge, P.A., Kim, M.C., Jaeger, K.E., Busch, W., Schmid, M., Lohmann, J.U., and Weigel, D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309 1056–1059. [DOI] [PubMed] [Google Scholar]
- Wikstrom, N., Savolainen, V., and Chase, M.W. (2001). Evolution of the angiosperms: Calibrating the family tree. Proc. R. Soc. Lond. B. Biol. Sci. 268 2211–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman, J.L., and Kingston, R.E. (1998). Alterations of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67 545–579. [DOI] [PubMed] [Google Scholar]
- Yoshida, N., Yanai, Y., Chen, L., Kato, Y., Hiratsuka, J., Miwa, T., Sung, Z.R., and Takahashi, S. (2001). EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13 2471–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuceer, C., Harkess, R.L., Land, S.B., Jr., and Luthe, D.S. (2002). Structure and developmental regulation of CONSTANS-LIKE genes isolated from Populus deltoides. Plant Sci. 163 615–625. [Google Scholar]
- Yuceer, C., Land, S.B., Jr., Kubiske, M.E., and Harkess, R.L. (2003). Shoot morphogenesis associated with flowering in Populus deltoides (Salicaceae). Am. J. Bot. 90 194–204. [DOI] [PubMed] [Google Scholar]
- Zeevaart, J.A.D. (1976). Physiology of flower formation. Annu. Rev. Plant Physiol. 27 321–348. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.