Abstract
Fruit and seed formation in plants is normally initiated after pollination and fertilization, and, in the absence of fertilization, flowers senesce. In the Arabidopsis thaliana mutant fruit without fertilization, a mutation in AUXIN RESPONSE FACTOR8 (ARF8) results in the uncoupling of fruit development from pollination and fertilization and gives rise to seedless (parthenocarpic) fruit. Parthenocarpy was confirmed in two additional recessive alleles and was caused by mutations within the coding region of ARF8. Genetic experiments indicate that ARF8 acts as an inhibitor to stop further carpel development in the absence of fertilization and the generation of signals required to initiate fruit and seed development. Expression of ARF8 was found to be regulated at multiple levels, and transcriptional autoregulation of ARF8 was observed. Analysis of plants transformed with a transcriptional PARF8:β-glucuronidase (GUS) construct or a translational ARF8:GUS fusion construct displayed distinct developmental regulation of the reporter in floral tissues involved in pollination and fertilization and in the carpel wall. After fertilization, the level of GUS activity declined in the developing seed, while in unfertilized ovules that are destined to senesce, ARF8:GUS expression spread throughout the ovule. This is consistent with a proposed role for ARF8 in restricting signal transduction processes in ovules and growth in pistils until the fruit initiation cue.
INTRODUCTION
Fruit development and seed set in flowering plants normally occur in a coordinated manner following pollination of the stigma and subsequent double fertilization in the ovule of the flower (Gillaspy et al., 1993). When the egg and central cell of the female gametophyte are not fused with sperm cells, they remain in a quiescent state and eventually degrade as the flower undergoes senescence (O'Neill and Nadeau, 1997). This has led to the interpretation that signaling processes are required to activate development of the fertilization products leading to the initiation of seed and fruit development (Raghavan, 2003).
Various phytohormones, including gibberellins, cytokinin, and auxin, are involved in signaling processes following pollination and fertilization as a prerequisite for further growth and development of seeds and fruits (Nitsch, 1952, 1970; Coombe, 1960; Garcia-Martinez and Hedden, 1997; Fos et al., 2000, 2001). Developing seeds appear to be essential for fruit growth and development because they are sources of phytohormones, and there may be a requirement for phytohormones continuously throughout seed and fruit formation (Nitsch, 1970; Eeuwens and Schwabe, 1975; Archbold and Dennis, 1985; Talon et al., 1990a; Garcia-Martinez et al., 1991; Ben-Cheikh et al., 1997; Swain et al., 1997; Ozga et al., 2002).
Fruit development can be uncoupled from fertilization and seed development, and the fruits, termed parthenocarpic, are seedless (Talon et al., 1992; Fos and Nuez, 1996; Robinson and Reiners, 1999; Varoquaux et al., 2002). Parthenocarpy has a genetic basis (Pike and Peterson, 1969; Lin et al., 1984; de Menezes et al., 2005) and has been exploited by farmers and plant breeders for the production of seedless fruits (Sykes and Lewis, 1996). Elevated endogenous phytohormone levels have been observed during parthenocarpic fruit set (George et al., 1984; Talon et al., 1990b, 1992), suggesting that increased supply of phytohormones to fruits from sources other than seeds may be sufficient to induce fruit growth. Accordingly, parthenocarpy can be induced in Arabidopsis thaliana and in diverse agricultural species by the exogenous application of auxins, cytokinins, or gibberellins (Gillaspy et al., 1993; Vivian-Smith and Koltunow, 1999) or by expression of auxin biosynthesis genes in ovaries and ovules (Rotino et al., 1997; Carmi et al., 2003; Mezzetti et al., 2004). Nonetheless, the molecular events directly involved in the initiation of fruit development and their link to plant hormone signal transduction processes remain unknown.
We have isolated the parthenocarpic fruit without fertilization (fwf) mutant from Arabidopsis in which fruit development and growth are uncoupled from pollination and fertilization events (Vivian-Smith et al., 2001). When fertilization is prevented in fwf by removal of floral organs surrounding the carpel, a seedless, dehiscent fruit or silique develops. In this article, we show that the fwf mutant contains a lesion in AUXIN RESPONSE FACTOR8 (ARF8), a member of the ARF transcription factor family. Although ARF transcription factors, including ARF8, have been the focus of many earlier molecular studies, the characterization of the fwf mutant allele of ARF8 together with other T-DNA insertion alleles demonstrates that ARF8 is an important regulator of fruit initiation and that the disruption of its normal function induces parthenocarpy in Arabidopsis. The involvement of ARF8 potentially provides a molecular link between the process of fruit initiation and the auxin signal transduction pathway.
RESULTS
fwf Contains a Lesion in ARF8
Unfertilized Arabidopsis carpels elongate slightly by cellular expansion (Figure 1), but they do not form dehiscent siliques, which is a defining feature of parthenocarpy in fwf (Vivian-Smith et al., 2001). fwf was originally characterized in the Landsberg erecta (Ler) background and was mapped to chromosome five (Vivian-Smith et al., 2001). Further fine mapping (see Methods) positioned fwf in a genomic interval of ∼110 kb between the genetic markers PhyC and SO191 (Figures 2A and 2B). Twenty-four annotated genes were present in this region (Arabidopsis Genome Initiative, 2000; Figure 2C), and the ARF8 gene was a candidate based on the phenotypic data previously obtained (Vivian-Smith et al., 2001; Vivian-Smith, 2001). ARF8 was amplified by PCR from both mutant and wild-type plants in the Ler background and sequenced. We identified a transition from G to A in the predicted translation initiation codon in the PCR product isolated from fwf plants, which converted the predicted start Met ATG to ATA. This base substitution created a cleaved-amplified polymorphic sequence (CAPS) marker (see Methods; Konieczny and Ausubel, 1993) that fully correlated with the fwf phenotype in segregating populations (Vivian-Smith, 2001).
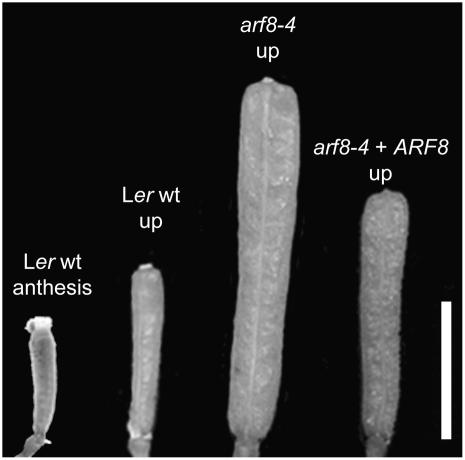
Figure 1.
Pistils from Emasculated Flowers.
Comparison of wild-type anthesis carpels and unpollinated (up) pistils from Ler, arf8-4, and arf8-4+ARF8 plants 7 d after emasculation. Bar = 3 mm.
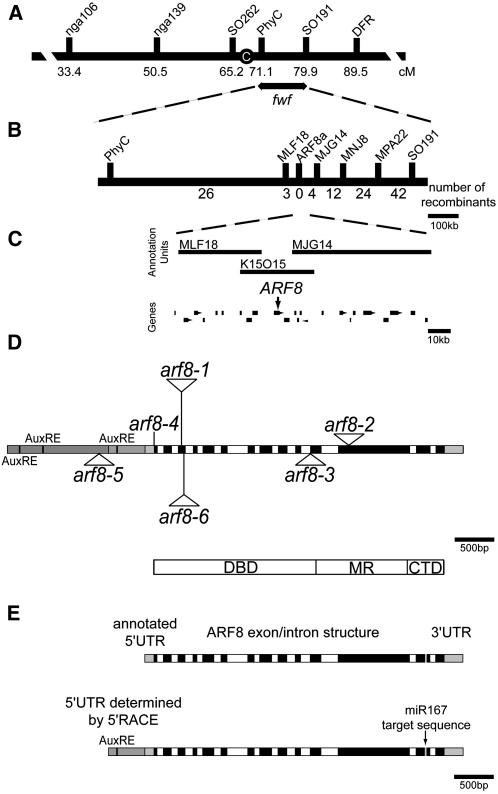
Figure 2.
Physical and Genetic Mapping of fwf.
(A) Genetic map of chromosome 5 showing genetic markers used in cloning fwf and the map position of fwf. C, centromere; cM, centiMorgan
(B) Physical map position of fwf with the number of recombinants between the markers indicated below.
(C) BAC vectors spanning the FWF region and the annotated genes delineated within this region. The position of ARF8 is indicated by the arrow.
(D) Diagram of the ARF8 gene with annotation of mutant alleles used in this study. T-DNA insertion lines are indicated by triangular insertions and the arf8-4 allele as a line, as it is an ethyl methanesulfonate mutation. Alleles and their ecotypes examined in this study were arf8-1 (Ws; Tian et al., 2004), arf8-4 (Ler; Vivian-Smith et al., 2001), arf8-5 (Col; Alonso et al., 2003), and arf8-6 (Col; http://www.hort.wisc.edu/krysan/DS-Lox/). Gray boxes indicate the promoter region and 3′ untranslated region (UTR). Three putative auxin response elements (AuxREs) are found in the promoter region. Black boxes specify exons, and white boxes indicate introns. A schematic representation of the ARF8 protein and its domains is represented below. DBD, DNA binding domain; MR, middle region; CTD, C-terminal domain.
(E) Structure of the ARF8 gene. Top: ARF8 gene with short 5′UTR as annotated in the database. Bottom: ARF8 gene with long 5′UTR as determined by 5′-RACE. The positions of the putative AuxRE present in the transcript leader region and of the microRNA target sequence (miR167) present in exon 13 are indicated.
Other Mutations in ARF8 Can Induce Parthenocarpy
Although other mutations in ARF8 have been identified and published, none have been reported to show parthenocarpic fruit development. arf8-1 (Figure 2D) exhibits altered hypocotyl elongation (Tian et al., 2004), while a phenotype for arf8-2 has not been described in detail so far (Okushima et al., 2005). The arf8-3 mutant has delayed stamen development and decreased fecundity (Nagpal et al., 2005). We will henceforth refer to fwf as arf8-4 (Figure 2D).
The flowers of the arf8-1 mutant together with two previously uncharacterized insertional mutants arf8-5 (SALK_049954) and arf8-6 (WiscDsLox324F09) (Figure 2D) were emasculated to test for parthenocarpy. Silique development in emasculated arf8 mutant flowers was compared with emasculated flowers from the corresponding wild-type ecotype background as controls because there are differences in the degree of carpel expansion in unfertilized wild-type flowers from different ecotypes (Vivian-Smith and Koltunow, 1999; Vivian-Smith et al., 2001).
Floral emasculation of plants homozygous for arf8-1 and arf8-6 produced dehiscent, parthenocarpic siliques (Table 1). Both of these recessive alleles (data not shown) contain a T-DNA insertion that disrupts the coding region (Figure 2D). F1 plants derived from crosses between arf8-4 in a near isogenic Columbia (Col) ecotype background (arf8-4NIL) and arf8-6 showed that these mutations were allelic because parthenocarpy was observed in plants trans-heterozygous for both mutations (data not shown).
Table 1.
Analysis of Elongation, Dehiscence, and ARF8 Expression Ratios in Lines Used in This Study
| Line | Silique Lengtha | Elongationb | Dehiscenceb | mRNA Expression Ratioc |
|---|---|---|---|---|
| Ler | 4.5 mm ± 0.5 mm | − | − | 1.0 |
| arf8-4 | 7.5 mm ± 0.8 mm | +++ | ++ | 1.6 ± 0.2 |
| Col | 3.7 mm ± 0.5 mm | − | − | 1.0 |
| arf8-4NIL | 5.9 mm ± 0.7 mm | ++ | ++ | 1.3 ± 0.1 |
| arf8-5 | 4.0 mm ± 0.6 mm | + | − | 2.4 ± 0.2 |
| arf8-6 | 5.2 mm ± 0.2 mm | ++ | ++ | 0.6 ± 0.2 |
| Ws | 3.8 mm ± 0.3 mm | − | − | 1.0 |
| arf8-1 | 5.0 mm ± 0.1 mm | ++ | ++ | 0.2 ± 0.1 |
| arf8-4 + ARF8 #1 | 5.4 mm ± 0.8 mm | + | ++ | 4.1 ± 1.2 |
| arf8-4 + ARF8 #2 | 5.1 mm ± 0.6 mm | + | ++ | 1.0 ± 0.1 |
| arf8-4 + ARF8 #3 | 4.3 mm ± 0.7 mm | − | ++ | ND |
| arf8-4 + ARF8 #4 | 5.6 mm ± 0.6 mm | ++ | ++ | 1.2 ± 0.2 |
| arf8-4 + ARF8 #5 | 5.0 mm ± 0.7 mm | + | ++ | 3.0 ± 0.2 |
| arf8-4 + ARF8 #6 | 4.9 mm ± 0.6 mm | + | ++ | ND |
| arf8-4 + ARF8 #7 | 5.4 mm ± 0.6 mm | + | ++ | 5.9 ± 0.3 |
| arf8-4 + ARF8 #8 | 5.7 mm ± 0.5 mm | ++ | ++ | ND |
| arf8-4 + ARF8 #9 | 6.0 mm ± 0.8 mm | ++ | ++ | ND |
| Ler + ARF8 #1 | 3.8 mm ± 0.7 mm | − | − | ND |
| Ler + ARF8 #2 | 4.3 mm ± 0.4 mm | − | − | ND |
| Ler + ARF8 #3 | 4.6 mm ± 0.4 mm | − | − | ND |
Pistil lengths measured 7 d after emasculation (±sd; a minimum of 40 flowers was emasculated and measured for each line).
−, no elongation/dehiscence; +, some elongation; +++/++, very good/good elongation/dehiscence.
The expression ratio of ARF8 mRNA was determined from flowers at anthesis. The ratio for each line is determined in relation to the expression in the corresponding wild-type line, which was set to 1.0 as the reference point. ND, not determined.
By contrast, emasculation of flowers from homozygous arf8-5 plants, which contain a T-DNA insertion 942 bp upstream of the ARF8 coding region, resulted in nonparthenocarpic, indehiscent pistils (Table 1). Furthermore, arf8-5 plants did not show the other phenotypes found in arf8-1, arf8-4, and arf8-6, including reduced seed set in proximal silique regions, broader silique shape, and precocious carpel growth prior to flower opening (Vivian-Smith et al., 2001; data not shown). Collectively, these data suggest that parthenocarpic fruit development is caused by loss-of-function mutations due to disruptions within the coding region of ARF8.
The Parthenocarpic Phenotype in arf8-4 Is Partially Rescued by an ARF8 Genomic Fragment
Complementation of the arf8-4 allele (Ler background) was attempted by inserting a wild-type ARF8 genomic fragment (as shown in Figure 2D) by plant transformation. Nine independent lines containing one to three copies of the introduced gene were recovered and analyzed. Both the introduced wild-type and endogenous mutant gene were expressed in all nine lines because a mixture of both mutant and wild-type transcript was detected using the CAPS marker following RT-PCR assays (data not shown). All homozygous transgenic lines produced shorter siliques following floral emasculation when compared with the arf8-4 mutant (Figure 1, Table 1). However, these shorter fruit were genuine siliques because they dehisced (Table 1). Moreover, some of the other features associated with arf8-4 mutants, including reduced seed set in proximal silique regions, broad silique shape, and precocious growth of carpels prior to floral bud opening, were also generally retained in these lines. This indicated that only partial complementation had been achieved. At least two possibilities may account for this result. The introduced genomic fragment may be lacking key sequence information, or the arf8-4 mutant allele is generating an inhibitory or competitive effect at the RNA and/or protein level.
The ARF8 Transcript Is Expressed in a Range of Tissues, and mRNA Levels Increase in the arf8-4 Allele
RNA gel blot analysis of both wild-type and arf8-4 plants detected mRNA of equal size in rosette and cauline leaves, inflorescence stems, flowers, and siliques. The size of the ARF8 mRNA transcript found in all wild-type and arf8-4 plant tissues examined was 3.4 kb in length (Figure 3). This was longer than the predicted 2.8-kb transcripts described in annotated databases. Rapid amplification of cDNA ends (5′-RACE) performed on RNA isolated from arf8-4 and wild-type siliques and subsequent sequencing of the PCR products confirmed the presence of the mutation in the expressed RNA from arf8-4 plant tissue and revealed that the mRNA in wild-type and arf8-4 mutants extends a total of 627 bases 5′ to the predicted translation initiation codon (Figure 2E), accounting for the observed 3.4-kb size. Thus, a stable full-length mRNA containing a mutation in the predicted translation start site is produced in arf8-4 plants.
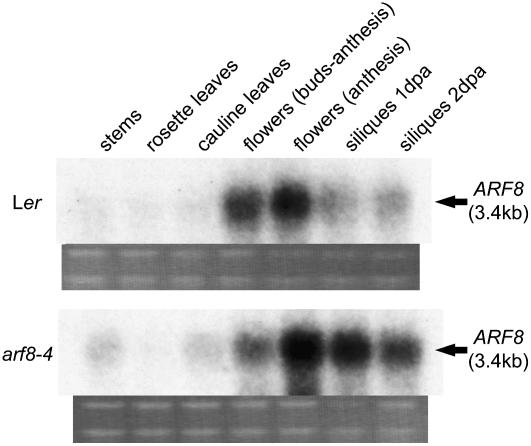
Figure 3.
RNA Gel Blot Analysis.
Comparison of ARF8 mRNA expression in various tissues from Ler and arf8-4 plants. dpa, days postanthesis.
In wild-type plants, RNA gel blot analysis detected the highest levels of steady state ARF8 mRNA in flowers at anthesis (Figure 3). In arf8-4, mRNA levels were higher in most examined tissues compared with wild-type ARF8 levels (Figure 3). Quantitative real-time PCR (qRT-PCR) showed that arf8-4 levels in anthesis flowers were approximately twofold higher than levels of ARF8 mRNA in wild-type anthesis flowers (see Supplemental Table 1 online). The higher level of arf8-4 mRNA persisted in the developing seeded siliques of arf8-4 plants for the 2 d after fertilization where embryos were at early to mid globular stages, respectively (Figure 3).
In situ hybridization analysis showed the expression of ARF8 was unevenly distributed in cells of a particular tissue. Expression varied with respect to the stage of flower development and silique growth (Figures 4A to 4D). Comparable expression patterns were found throughout flower development in both Ler and arf8-4. ARF8 mRNA was detected in sepals, anthers, and carpels prior to anthesis in both Ler and arf8-4 (Figures 4A and 4C). At anthesis, expression was strong within the mesocarp layers of the fruit (Figures 4B and 4D), and ARF8 mRNA was also detected in the septum of the pistil and in the funiculi and integuments of both Ler and arf8-4 ovules (Figures 4B and 4D). After fertilization, ARF8 transcripts remained present in the integuments of both wild-type and arf8-4 ovules 1 to 2 d after fertilization (see Supplemental Figure 1 online). ARF8 mRNA was evident in the mesocarp of the fruit and the carpel septum during growth and was detectable 3 d after fertilization in both Ler and arf8-4 (see Supplemental Figure 1 online). In situ hybridization is not reliable enough to use for the quantification of twofold changes in mRNA level, but we can conclude that the cellular distribution of both mutant and wild-type ARF8 mRNA was diverse and spatially comparable at the light microscopy level in examined tissues.
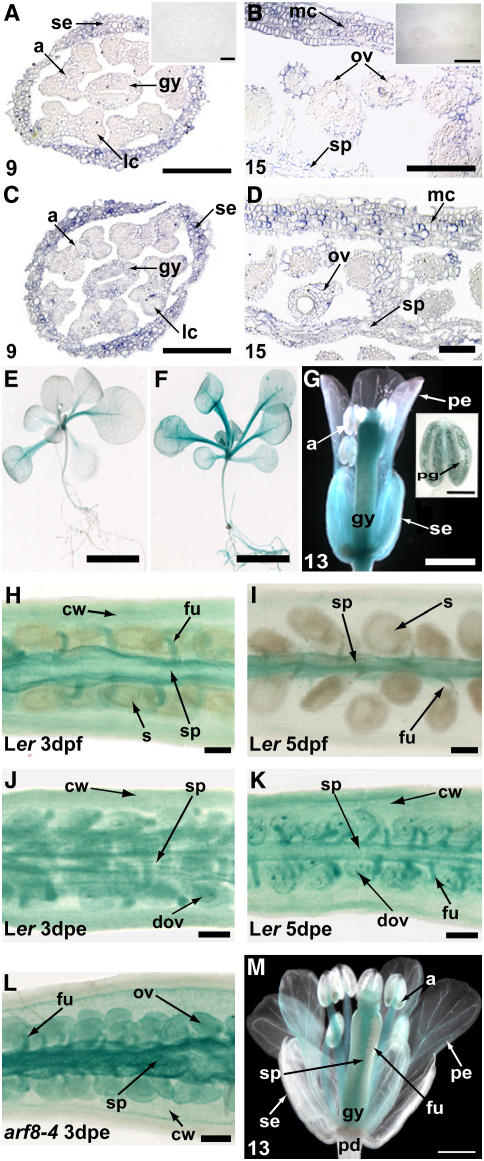
Figure 4.
Expression of ARF8.
(A) to (D) In situ hybridizations on thin sections of developing Ler ([A] and [B]) and arf8-4 ([C] and [D]) flowers ([A] and [C]) and siliques ([B] and [D]) hybridized with antisense-labeled ARF8 RNA. Insets ([A] and [B]) show control hybridizations with sense-labeled ARF8 RNA. The numbers at the bottom left indicate the stages of flower development (Smyth et al., 1990). Bars = 100 μm.
(E) and (F) ARF8 is involved in the modulation of PARF8:GUS expression. Seedlings of Ler (E) and arf8-4 (F) plants transformed with the PARF8:GUS construct at 15 d after germination. Bars = 1 mm.
(G) Ler flower transformed with PARF8:GUS at anthesis (bar = 500 μm). Inset shows an anther with stained pollen (bar = 50 μm).
(H) to (L) PARF8:GUS expression in parthenocarpic pistils resembles that of fertilization-induced silique development. Ler silique 3 d after fertilization (H); Ler silique 5 d after fertilization (I); Ler silique 3 d after emasculation (J); Ler silique 5 d after emasculation (K); arf8-4 silique 3 d after emasculation (L). Bars = 50 μm.
(M) Ler flower transformed with ARF8:GUS at anthesis. Bar = 500 μm.
a, anther; cw, carpel wall; dov, degenerating ovule; dpe, days postemasculation; dpf, days postfertilization; fu, funiculus; gy, gynoecium; ov, ovule; pd, pedicel; pe, petal; pg, pollen grain; s, seed; se, sepal; sp, septum; lc, locule; mc, mesocarp.
ARF8 Shows Transcriptional Autoregulation
The increased levels of arf8-4 mRNA in various tissues was further investigated by transforming Ler and arf8-4 plants with a transcriptional fusion of the ARF8 promoter fused to the β-glucuronidase (GUS) gene (PARF8:GUS; Jefferson et al., 1987). Three independent, homozygous lines containing PARF8:GUS in each background were examined. Histochemical staining showed that the introduced transcriptional fusion was expressed at lower levels in all Ler tissues compared with transgenic arf8-4 tissues at the same stage, although the spatial expression pattern was conserved. This was most obvious in seedlings (Figures 4E and 4F) but was also clearly evident in flowers and developing siliques. Two weeks after germination, arf8-4 seedlings exhibited strong GUS activity throughout the hypocotyl, the inflorescence meristem, and the cotyledon margins. The true leaves had high GUS levels around the veins, the leaf margins, and the trichomes, and the root hairs were also intensely stained (Figure 4F). The GUS activity in transgenic Ler plants was significantly lower but showed the same pattern (Figure 4E). These observations are consistent with arf8-4 mutants having an impaired ability to downregulate expression levels from the ARF8 promoter in the transcriptional GUS fusion. Therefore, in wild-type plants, ARF8 appears to be involved either directly or indirectly in the modulation of transcriptional expression from its own promoter.
To investigate ARF8 regulation further, we examined mRNA levels in the nonparthenocarpic arf8-5 allele (Col background) containing a T-DNA insertion between two putative auxin response elements in the predicted ARF8 promoter (Figure 2D). We detected higher levels of ARF8 mRNA in arf8-5 flowers compared with that in wild-type Col-1 flowers of the same stage using qRT-PCR. The levels observed in arf8-5 were also higher than those found in the arf8-4 allele in the Col near isogenic line (arf8-4NIL; Table 1). The increased ARF8 expression in the arf8-5 plants suggests that the normal levels of expression might be altered by the presence of the T-DNA insertion within the promoter possibly influencing self-regulation.
PARF8:GUS Expression during Parthenocarpic and Fertilization-Induced Silique Development in arf8-4 and Wild-Type Plants
The expression patterns of the PARF8:GUS construct were compared in flowers of wild-type and arf8-4 mutant plants transformed with the PARF8:GUS construct. Throughout flower and fruit development, GUS activity was detected in a similar pattern to the mRNA distribution found in the in situ hybridization experiments. GUS activity was strong in carpels and pollen grains of Ler plants at anthesis, with comparably weaker activity in sepals and petals (Figure 4G). Further comparison of GUS activity in flowers after fertilization and after emasculation showed that 3 d after fertilization, GUS expression was visible within the carpel wall, the carpel septum, and the funiculi but not in the seeds (Figure 4H). Five days after fertilization, expression became restricted to the septum and the base of the funiculi (Figure 4I). After emasculation, there were changes in the observed expression pattern. Three days after emasculation of transgenic Ler plants, GUS activity was evident throughout the ovule in addition to the septum and carpel wall tissue (Figure 4J). Five days after emasculation, GUS activity had decreased in the carpel wall and in the septum but remained strong in the funiculi with some patchy GUS activity within the female gametophyte (Figure 4K).
In arf8-4 mutant plants containing PARF8:GUS, the GUS activity in the flowers at anthesis and in the developing seeds after fertilization was spatially similar to that in pollinated Ler plants (see Supplemental Figure 1 online). However, staining was stronger and was detected in higher levels over the 3 d following fertilization, consistent with the data obtained by RNA gel blot analysis (Figure 3). In parthenocarpic siliques of arf8-4 plants containing PARF8:GUS, GUS activity was observed 3 d after emasculation in the carpel walls, the septum, the funiculi, and within the ovules (Figure 4L). GUS activity became restricted to the septum and the base of the funiculi at 7 d after emasculation (see Supplemental Figure 1 online), coincident with the onset of ovule senescence. The restricted expression of the PARF8:GUS marker in the septum and funiculi during parthenocarpic silique formation in arf8-4 resembles that found in fertilization-induced Ler siliques and is distinct from that in Ler pistils undergoing senescence.
ARF8:GUS Expression Reveals Additional Levels of Regulation
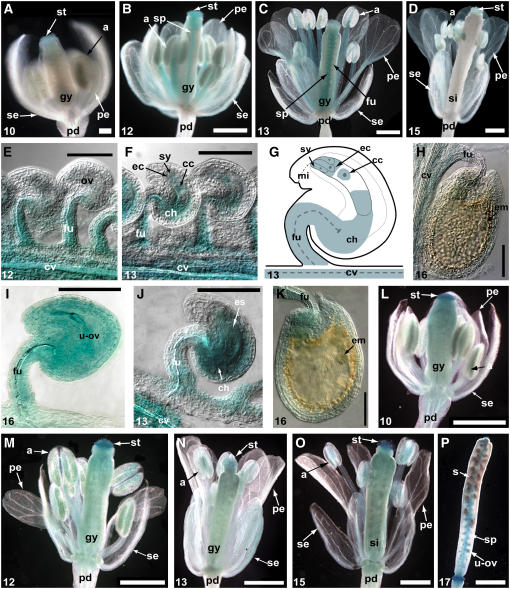
A translational gene fusion was made with the wild-type ARF8 genomic fragment (shown in Figure 2D) and the GUS gene generating ARF8:GUS. Ler plants were transformed with the construct, and 14 independent lines were recovered and analyzed for GUS activity. Even though the introduced translational ARF8:GUS fusion contained the same ARF8 promoter fragment used in PARF8:GUS, a more restricted pattern of expression was observed in flowers and siliques of transgenic wild-type plants at anthesis because expression was not evident in sepals or the pedicel (cf. Figures 4G and 4M).
Analysis of the expression of ARF8:GUS during floral development showed that it was first detected in flowers at stage 10 (Smyth et al., 1990) in the stigma (Figure 5A). During flower stages 11 and 12, GUS staining was also evident in the carpel septum, the anther filaments, and then in developing pollen at the tetrad stage after meiosis (Figure 5B). At pollination (flower stage 13), ARF8:GUS expression was evident in the ovule funiculi, the carpel wall, and in the petals (Figure 5C). After fertilization (Figure 5D, stage 15), the level of GUS activity declined and was barely visible in the stigmatic papillae, pollen, and silique walls. The altered pattern of expression of the ARF8 translational fusion relative to the mRNA localization and the expression pattern described previously for plants containing the transcriptional fusion (PARF8:GUS) indicates that ARF8 might be regulated at the posttranscriptional level.
Figure 5.
Expression of Translational ARF8:GUS Fusion Protein.
(A) to (D) GUS staining patterns in Ler flowers and siliques transformed with ARF8:GUS at different developmental stages. Ler flower bud at stage 10 (A); Ler flower around stage 12 (B); Ler flower at anthesis (stage 13) (C); Ler silique after fertilization (stage 15) (D).
(E) to (I) GUS staining patterns in Ler and arf8-4 ovules between anthesis and globular embryo stage.
(E) Staining in septum tissue and funiculi before anthesis.
(F) After pollination, GUS staining is evident in the chalazal region and in the mature embryo sac around the fused polar nuclei and the egg apparatus.
(G) Schematic representation of GUS staining in ovules after pollination.
(H) At the globular embryo stage, no staining is visible in fertilized ovules.
(I) Unfertilized ovules show strong GUS expression throughout the ovule.
(J) GUS expression was observed earlier and in a general distribution pattern in the embryo sac of arf8-4 ovules transformed with ARF8:GUS.
(K) In the arf8-4 background, staining in the chalazal region is still visible at the globular embryo stage in fertilized ovules.
(L) to (P) GUS staining patterns in arf8-4 flowers and siliques transformed with ARF8:GUS at different developmental stages.
(L) arf8-4 flower at stage 10.
(M) arf8-4 flower at stage 12. Expression is visible in the septum and within the ovules.
(N) arf8-4 flower at anthesis (stage 13).
(O) After fertilization (stage 15), staining remains visible in the carpel walls, the septum, and the ovules.
(P) In growing siliques (stage 17), ARF8:GUS is expressed in the septum and in unfertilized ovules.
The numbers at the bottom left indicate the stages of flower development (Smyth et al., 1990). a, anther; cc, central cell; ch, chalazal end; cv, carpel vasculature; ec, egg cell; em, embryo; es, embryo sac; fu, funiculus; gy, gynoecium; mi, micropylar end; ov, ovule; pd, pedicel; pe, petals; se, sepals; sp, septum; si, silique; st, stigma; sy, synergids; u-ov, unfertilized ovule. Bars = 1 mm in (A) to (D) and (L) to (P) and 100 μm in (E) to (K).
ARF8 Is Expressed in the Ovule and the Embryo Sac
Before pollination (stage 12), expression of ARF8:GUS in the carpel was detected primarily in the septum tissue around the vasculature and also in the funiculus of the ovule (Figure 5E). After pollination, but before fertilization had occurred (stage 13), there was a lack of staining between the funiculus and carpel vasculature. GUS activity in the ovule was also evident in the chalazal region of the integuments adjacent to the embryo sac (Figure 5F). Expression in the mature embryo sac was evident in three discrete zones: one in the chalazal cytoplasm of the embryo sac, a second in the fused polar nuclei and surrounding cytoplasm, and the third in the egg apparatus comprising synergids and the egg cell (Figures 5F and 5G). After fertilization, the level of GUS activity in the embryo and endosperm compartments decreased relative to that seen in the embryo sac and by the globular embryo stage of seed development GUS was barely detectable (Figure 5H).
In unfertilized wild-type ovules that are destined to senesce, ARF8:GUS expression was evident throughout the ovule (Figure 5I), resembling the expression pattern observed in ovules of emasculated Ler siliques transformed with the PARF8:GUS construct. The difference in expression pattern between fertilized and senescing ovules reveals that ARF8 expression is regulated differently in the ovule depending on whether or not fertilization occurs.
ARF8:GUS Expression Is Spatially and Temporally Altered in the arf8-4 Background
The ARF8:GUS construct was inserted into arf8-4 mutant plants by plant transformation, and 14 independent lines were obtained. Analysis of the resulting expression pattern showed significant differences in the expression of ARF8:GUS in the arf8-4 background compared with that observed in Ler. Expression was observed much earlier in the developing carpel and was evident in flower buds as early as stage 5 (see Supplemental Figure 1 online). Expression continued throughout flower development, silique elongation, and early embryo development (Figures 5J to 5P). Expression in carpel and silique wall tissues was much higher than that found in the Ler background and somewhat masked the underlying expression in septum and funiculi (cf. Figures 5C and 5N). Septum and funiculi expression began earlier, at stage 9, and septum expression did not decline after fertilization but continued throughout silique growth in the arf8-4 background until siliques turned yellow (Figures 5L to 5P). Expression of ARF8:GUS was also observed earlier in the embryo sac of the ovules, and a general distribution pattern was observed instead of the three discrete zones seen in the Ler background (cf. Figures 5F and 5J). GUS activity declined after fertilization, but in contrast with the Ler background, where it had disappeared by the early globular embryo stage, staining was still visible in the chalazal region and the funiculus of the ovule when the embryo was at the globular stage of development in the arf8-4 background (cf. Figures 5H and 5K). GUS activity continued to decrease and was not visible at the early heart stage of embryo development (see Supplemental Figure 1 online). There were also differences in staining intensities in some tissues between the Ler and arf8-4 backgrounds, with decreased ARF8:GUS activity in petals and filaments in arf8-4 (Figures 5L to 5N) compared with Ler (Figures 5A to 5C). These data indicate that ARF8:GUS is being regulated differently in the arf8-4 mutant. The temporally extended expression agrees with the different level of expression of PARF8:GUS in the arf8-4 background and the higher levels of mRNA evident in developing arf8-4 siliques.
ARF8:GUS Induces the Formation of the Dehiscence Zone in Transgenic Ler Plants
We tested Ler and arf8-4 plants containing ARF8:GUS for parthenocarpy by floral emasculation. Surprisingly, the transgenic Ler plants developed genuine siliques that dehisced but did not elongate significantly over that observed in unfertilized, expanded carpels (Table 2). This contrasted with the indehiscent carpels (Table 1) observed previously in Ler plants containing the identical genomic fragment lacking the GUS gene fused in frame to the 3′ end of the ARF8 gene. This indicates that the introduced ARF8:GUS is compromising the activity of the endogenous ARF8 gene to prohibit dehiscence zone formation in the absence of fertilization. Introduction of ARF8:GUS into the arf8-4 background resulted in partial complementation of the parthenocarpic phenotype because elongation was reduced but the dehiscence zone still formed (Table 2). This was consistent with the inability to complement the arf8-4 mutant with the genomic fragment (Table 1), further supporting the possibility that the arf8-4 allele can interfere with the activity of the endogenous ARF8 gene.
Table 2.
Analysis of Elongation and Dehiscence in the Translational ARF8:GUS Lines
| Line | Silique Lengtha | Elongationb | Dehiscenceb |
|---|---|---|---|
| Ler | 4.1 mm ± 0.4 mm | − | − |
| Ler + ARF8:GUS | 3.9 mm ± 0.8 mm | − | ++ |
| arf8-4 | 7.2 mm ± 0.6 mm | +++ | ++ |
| arf8-4 + ARF8:GUS | 4.9 mm ± 0.7 mm | + | ++ |
Pistil lengths measured 7 d after emasculation (±sd; a minimum of 37 flowers was emasculated and measured).
−, no elongation/dehiscence; +, some elongation; +++/++, very good/good elongation/dehiscence.
DISCUSSION
ARF8 Is a Negative Regulator of Fruit Initiation and Growth
The parthenocarpic fwf phenotype (Vivian-Smith et al., 2001) is caused by a lesion in ARF8. ARF8 is a member of a family of 23 transcription factors in Arabidopsis (Guilfoyle and Hagen, 2001; Liscum and Reed, 2002; Remington et al., 2004) and has previously been linked to light-stimulated hypocotyl elongation, root growth, and auxin homeostasis (Tian et al., 2004). ARF8 has also been shown to be involved in stamen filament elongation and anther dehiscence (Nagpal et al., 2005). Our analysis demonstrates that ARF8 is also a negative regulator of fruit initiation because loss-of-function arf8 alleles allow parthenocarpy and alter carpel mesocarp cell division (Vivian-Smith et al., 2001). The degree of carpel elongation in unfertilized flowers and the extent of parthenocarpic fruit growth was found to be dependent on the ecotype background, with the strongest silique elongation observed in the Ler background and significantly weaker elongation found in mutants in the Col and Wassilewskija (Ws) ecotypes (Vivian-Smith et al., 2001; this study).
Partial Complementation of arf8-4 May Be Due to Inhibitory or Competitive Effects
Our attempt to complement the arf8-4 mutation by the introduction of a wild-type genomic ARF8 fragment only reduced the extent of parthenocarpic silique length, while other arf8-4 characteristics, including the formation of the dehiscence zone without seed set, were still observed. Similarly, full complementation was not achieved in arf8-1, as the defective elongation in light-grown hypocotyls was incompletely restored (Tian et al., 2004). One possibility is that other genomic elements necessary for full ARF8 function were absent in the fragments used for complementation. Alternatively, the arf8-1 and arf8-4 alleles may produce aberrant transcripts or proteins that prevent full complementation by the introduced wild-type copy of the gene. Consistent with this suggestion, both the arf8-1 (Tian et al., 2004; data not shown) and arf8-4 alleles produce transcripts with the potential to encode truncated ARF8 proteins that might interfere with wild-type ARF8 protein function. While further experiments are necessary to determine if a protein fragment can indeed be produced in these mutants, the ability of the ARF8:GUS transgene to induce parthenocarpic phenotypes in wild-type plants (albeit very short but dehiscent siliques) suggests that altered ARF8 proteins can have dominant-negative effects.
ARF8 Expression Is Regulated at Multiple Levels
Our analysis in both the wild-type and arf8-4 backgrounds has shown that the expression of ARF8 is regulated at multiple levels. RNA gel blot analysis, RNA in situ hybridizations, and the observed expression pattern of a transcriptional PARF8:GUS construct showed that the expression of both mutant and wild-type mRNA was spatially comparable in developing flowers and during fertilization-induced silique growth. However, differences were found in the expression patterns of PARF8:GUS during parthenocarpic silique formation in arf8-4 compared with that in Ler pistils undergoing senescence after emasculation. The parthenocarpic expression pattern resembled more that found in fertilization-induced Ler siliques, indicating that initiation of fruit development alters the expression of ARF8. Furthermore, both RNA gel blot analysis and qRT-PCR found higher levels of mRNA in the arf8-4 plants, which also persisted in developing siliques. Experiments using the transcriptional PARF8:GUS fusion construct suggest that ARF8 is involved in transcriptional autoregulation. The transcriptional regulation of ARF8 and genes under control of the ARF8 promoter is disturbed in the arf8-4 mutant, leading to higher expression levels of those genes.
The analysis of a translational ARF8:GUS construct revealed an additional level of regulation. Expression of the translational ARF8:GUS fusion protein was altered in the arf8-4 mutant in several ways compared with the wild-type pattern. ARF8:GUS levels in arf8-4 appeared to be reduced in petals and especially anther filaments in the mutant. ARF8 activity in these tissues may be connected to the inhibitory effects of outer floral whorls on silique elongation (Vivian-Smith et al., 2001), and a role for ARF8 in stamen filament elongation has been demonstrated in the arf6-2 arf8-3 mutant (Nagpal et al., 2005). Moreover, a temporal change in expression was also observed, especially in the carpel walls, the septum, the funiculi, and the ovules, where GUS activity was detected earlier and persisted in the mutant.
ARF8 translation may also be regulated via the 5′-transcript leader region. The ARF8 mRNA is 629 bp longer than previously reported, and the translational leader region contains 10 upstream AUGs that the ribosomal translational complex can potentially act on before reaching the presumed ARF8 initiation codon. These upstream open reading frames (uORFs) may regulate ARF8 translation, as similar uORFs have been identified in nine other ARF genes (Nishimura et al., 2004, 2005) where they have been shown to affect translation of the main ORF. Short uORFs in the 5′-transcript leader region of ETTIN (ARF3) and MONOPTEROS (ARF5) repress the expression of the downstream main ORFs at the translational level (Nishimura et al., 2005). Short upstream ORFs have also been reported in other genes where they significantly regulate translation of the main ORF (Geballe and Sachs, 2000; Morris and Geballe, 2000; Kawaguchi and Bailey-Serres, 2002).
Potential also exists for regulation of ARF8 by short-interfering RNAs, as recent research has shown that many ARFs are the target of microRNAs (miRNAs; Mallory et al., 2005) and transacting silence-inducing RNAs (Allen et al., 2005; Williams et al., 2005). The ARF8 gene contains a miRNA target sequence (miR167; see Figure 2E) also present in ARF6 (Rhoades et al., 2002; Bartel and Bartel, 2003; Kasschau et al., 2003). The levels of miR167, which are not regulated by auxin, appear to set a homeostatic level of ARF translation (Mallory et al., 2005) and might therefore be involved in the proposed posttranscriptional regulation or transcriptional autoregulation of ARF8. However, the role of the miRNA in the modulation of ARF8 expression and in the regulation of hypocotyl and root growth, flower development, or fruit initiation has not yet been investigated.
Signal Transduction Events in the Ovule May Regulate Fruit Growth
Fertilization is critical for initiating wild-type fruit formation. The expression pattern of ARF8 in the ovule and female gametophyte around fertilization suggests that ARF8 might act in these tissues to regulate fruit initiation.
ARF8:GUS expression is switched off soon after fertilization has occurred in wild-type plants, indicating that a fertilization signal deactivates ARF8. The removal of ARF8 activity after fertilization might abolish a developmental block that represses fruit growth and allows initiation of seed and fruit developmental programs.
ARF8:GUS expression is spatially and temporally altered in the parthenocarpic arf8-4 background, indicating a disturbed regulation of the introduced gene in the arf8-4 mutant. This may result from an interaction between the ARF8:GUS gene and a mutant arf8-4 protein product, as we have observed transcriptional autoregulation. However, we have yet to examine arf8-4:GUS expression and determine if a mutant protein is produced. We speculate that all of the examined arf8 mutant alleles are unlikely to produce a functional ARF8 protein, and parthenocarpy may simply occur because fruit development is not blocked.
In unfertilized ovules, expression of the ARF8:GUS marker persists and staining is observed throughout the ovule, indicating that the negative regulation through ARF8 is kept active. These expression patterns are therefore consistent with ARF8 acting as a negative regulator of fruit initiation, and collectively they indicate a central role for the ovules in mediating positive and negative signals involved in fruit development.
We observed previously that the ats mutation, which changes ovule integument structure, enhances the parthenocarpy phenotype when combined with arf8-4 (Vivian-Smith et al., 2001). In emasculated ats arf8-4 plants, seedless siliques formed that are comparable in length to those induced after fertilization. The ats mutation also counteracts the inhibitory effects of surrounding floral whorls on silique elongation (Vivian-Smith et al., 2001). ATS is KANADI4, and it is specifically expressed in the initiating ovule integument (McAbee et al., 2006), suggesting that modifications to the ovule integument influence parthenocarpic fruit growth in ats arf8-4 plants. The importance and contribution of different structural components of the ovule to parthenocarpic fruit development can be examined genetically using a suite of ovule mutants in the arf8-4 background. This should provide further information concerning the relationship of signal transduction events in the ovule and fruit growth and the role of ARF8 in these processes.
Model for the Role of ARF8 in Fruit Development
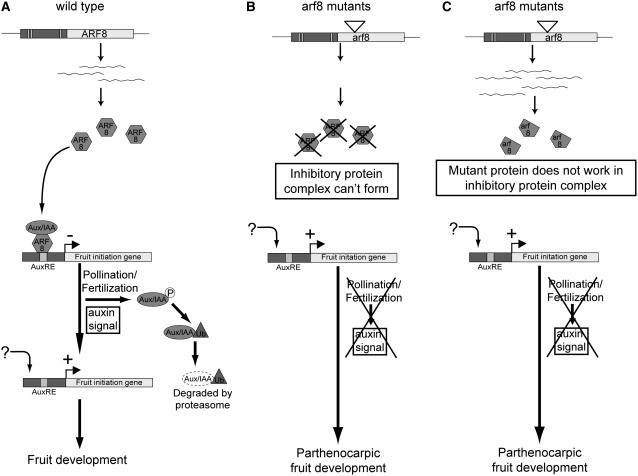
Figure 6 shows a model for the role of ARF8 during the transition from carpel to fruit growth. Our data suggest that ARF8 represses fruit development, and in the simplest model, ARF8 may do so by directly activating genes that themselves repress fruit development. Alternatively, ARF8 may invoke repression by being a member of a complex of proteins. Protoplast transformation experiments support the concept that the transcriptional activity of the ARF8 protein is regulated by heterodimerization with auxin/indole-3-acetic acid (Aux/IAA) proteins that inhibit this activity (Guilfoyle et al., 1998; Ulmasov et al., 1999a, 1999b; Guilfoyle and Hagen, 2001; Rogg and Bartel, 2001; Liscum and Reed, 2002; Tiwari et al., 2003). Physical interactions between ARF8 and members of both the Aux/IAA repressor and ARF protein families have been demonstrated (Hardtke et al., 2004; Tatematsu et al., 2004). While studies have shown that ARF8 transcription is not regulated by auxin per se (Ulmasov et al., 1999a; Pufky et al., 2003; Okushima et al., 2005), auxin indirectly regulates ARF activity by promoting turnover of Aux/IAA proteins (Ulmasov et al., 1999b; Gray et al., 2001; Tiwari et al., 2001; Zenser et al., 2001). This allows the ARFs to become active and impose their regulatory influence on the expression of auxin-responsive genes (Gray et al., 2001; Kepinski and Leyser, 2004). Consistent with an essential role for the closely related ARF6 and ARF8 proteins in flower development, several studies have demonstrated that ARF8 can mediate auxin-induced gene activation (Ulmasov et al., 1999b; Tiwari et al., 2003; Nagpal et al., 2005), and several auxin-responsive genes have been identified as candidates for direct regulation by these two ARFs in microarray experiments (Nagpal et al., 2005). An increase in total auxin levels within flower buds during various developmental stages was not observed (Nagpal et al., 2005), suggesting that localized changes in auxin levels may be more important in regulating ARF8 function.
Figure 6.
Model for the Role of ARF8 in Fruit Development.
(A) Model showing ARF8 action in restricting the expression of auxin-responsive genes in a complex together with an Aux/IAA repressor protein. The auxin signal induced by pollination leads to phosphorylation and subsequent proteolytic degradation of the Aux/IAA protein, and unknown factors subsequently activate expression of the fruit initiation genes.
(B) In arf8 mutants in which no ARF8 protein can be formed (null mutants), the inhibitory complex of ARF8 with the Aux/IAA protein cannot form. Therefore, the expression of the fruit initiation genes can be activated in the absence of pollination and fertilization, leading to parthenocarpic fruit development.
(C) In arf8 mutants that can produce a mutated version of the ARF8 protein (arf8), this mutant protein fragment can either not bind to the Aux/IAA protein or does not bind the promoter of the fruit initiation genes. Therefore, the inhibitory complex is not correctly formed, and the expression of the fruit initiation genes can be activated in the absence of pollination and fertilization, leading to parthenocarpic fruit development.
In addition to its role with ARF6 in flower development, we propose that before pollination and fertilization, ARF8 is bound to the promoters of a range of primary auxin-responsive genes that play an essential role in fruit initiation and development. Transcription of these fruit initiation genes is repressed at this stage by a protein complex that includes ARF8 bound to Aux/IAA proteins, which functions as a repressor (Figure 6A). This extends the model described above to include an active repressor role of the ARF8-Aux/IAA complex rather than merely a lack of gene activation by ARF8. The existence of a functional repressor function of the ARF8-Aux/IAA complex is required to explain the observation that reduced ARF8 function allows fruit initiation. The identity of the Aux/IAA protein(s) proposed to act with ARF8 is not known, but a likely candidate is the protein encoded by the Arabidopsis ortholog of tomato (Solanum lycopersicum) IAA9 (Wang et al., 2005). As described above, this repressor function of ARF8 may occur in ovules.
Pollination is known to induce increases in both auxin and ethylene levels in floral organs, and this correlates with subsequent growth or senescence of these organs (O'Neill, 1997). In wild-type flowers, a fertilization-induced auxin burst could induce the degradation of the Aux/IAA protein through a proteolytic pathway (Gray et al., 1999, 2001; Rogg and Bartel, 2001; Hellmann and Estelle, 2002; Dharmasiri and Estelle, 2004; Jenik and Barton, 2005). This would abolish the repression of crucial auxin-responsive fruit initiation genes by the ARF8-Aux/IAA protein complex (Figure 6A).
In the arf8 mutant lines, this mechanism of repression is impaired. If no ARF8 protein is made, the inhibitory complex cannot form or is unable to bind to the promoters of the auxin-responsive fruit initiation genes (Figure 6B). Alternatively, the formation of a functional inhibitory complex can be prevented by a mutant version of the ARF8 protein that cannot bind to either the Aux/IAA protein or other components of the protein complex, or to the promoters of fruit development genes (Figure 6C). As a result, repression of fruit initiation gene transcription does not occur, and parthenocarpy results by allowing fruit initiation to occur in the absence of fertilization. Once the proposed ARF8-Aux/IAA complex is removed, either ARF8 is replaced by other transcription factors, presumably other ARFs, or ARF8 itself functions as a transcriptional activator. If the latter hypothesis is correct, the capacity to initiate fruit development in arf8 mutants implies that ARF8 is functionally redundant in this activation process, presumably with ARF6.
The notion that biological processes actively restrict the carpel from forming a fruit was first proposed by Nitsch (1952), who from various studies held the view that auxin was a key component of this process. Many subsequent studies have suggested that developing seeds are important sources for phytohormones, such as auxins, needed for fruit development (Nitsch, 1970; Eeuwens and Schwabe, 1975; Archbold and Dennis, 1985; Talon et al., 1990a; Garcia-Martinez et al., 1991; Ben-Cheikh et al., 1997; Swain et al., 1997; Ozga et al., 2002). The discovery that FWF is ARF8 supports these suggestions and means that we can now begin to dissect the developmental pathways controlling this crucial process.
METHODS
Plant Growth and Scoring Parthenocarpy
Arabidopsis thaliana seeds were surface-sterilized and grown as described before (Vivian-Smith and Koltunow, 1999). Parthenocarpy was assessed by flower emasculation (Vivian-Smith and Koltunow, 1999; Vivian-Smith et al., 2001). Only siliques above flower position 20 were used, collected, and photographed, and their lengths were determined using the Scion Image Beta 4.02 program (http://www.scioncorp.com/frames/fr_scion_products.htm). Plants producing siliques that significantly and reliably elongated more than the corresponding wild-type ecotype plants and that formed a dehiscence zone were scored as parthenocarpic.
Fine Mapping of the fwf Mutation and Map-Based Cloning of fwf
The map position of the fwf lesion was described on chromosome V between the markers AthPhyC and AthSO191 (Vivian-Smith et al., 2001). Fine mapping was performed using recombinant populations from crosses between a Col-4 female parent and the fwf mutant (Ler ecotype) as pollen donor together with a population that consisted of F2 individuals from a cross between Col-4 and the recessive ats fwf double mutant (Ler ecotype) that was linked in coupling phase (Vivian-Smith et al., 2001). Recombinants between fwf and ats facilitated mapping, and both populations were used to generate informative recombinants in the AthPhyC and AthSO191 interval through a PCR screening method. Plants were assessed by the absence or presence of siliques exhibiting parthenocarpy when emasculated.
Plant DNA was extracted for PCR screening in microtitre plates essentially as described by Langridge et al. (1991). Mutliplex PCR reactions were made with primers to the AthPhyC and AthSO191 CAPS markers. A total of 121 recombinant chromatids were identified from 2442 plants (Vivian-Smith, 2001). Each recombinant plant was transferred to soil, grown, and scored for parthenocarpic silique elongation. Genomic DNA was extracted from leaf tissue (Edwards et al., 1991) and AthPhyC and AthSO191 checked. New simple sequence length polymorphisms (Bell and Ecker, 1994) and new CAPS (Konieczny and Ausubel, 1993) molecular markers were created for the region spanning AthPhyC and AthSO191 (Vivian-Smith, 2001). Markers surrounding the ARF8a CAPS marker had the least number of recombinants. This marker is PCR amplified with ARF8+1478F (5′-GAGCTCCTTTAAGACAGCAGTTTGT-3′) and ARF8+2817R (5′-CCTAGGAAAGTTTAGTTACCCTGAGAC-3′) and cut with AccI (New England Biolabs). The nearest centromeric marker was MLF18 (#2), a marker dominant in Col-1, amplified by MLF18#2F (5′-TTTGTCAATGTTGGGTTCCG-3′) and MLF18#2R (5′-GGGAGACGGGTGAGACAAAT-3′). The nearest telomeric marker was MJG14 (#2) amplified by MJG14#2F (5′-GGATCGTTAGGCAATGGGAT-3′) and MJG14#2R (5′-TGGATACGATGGGGACAAAA-3′) and cut with VspI (New England Biolabs). The latter two markers defined 110 kb spanning part of the P1 vector MLF18 on the left and MJG14 on the right and the whole TAC vector K15O15.
ARF8 T-DNA Mutants
The arf8-1 mutant was published as a hypocotyl elongation mutant allele (Tian et al., 2004) in the Ws background. Seeds were kindly donated by Kotaro Yamamoto (Hokkaido University, Sapporo, Japan).
The Col plant line SALK_049954 was designated arf8-5. The line was identified from the collection of SALK lines (Alonso et al., 2003) and obtained from the ABRC. The position of the T-DNA insertion was verified by sequencing, and it was found to be 431 bp 5′ to the annotation published in The Arabidopsis Information Resource database (Figure 2D). The Col plant line WiscDsLox324F09 (University of Wisconsin T-DNA lines; http://www.hort.wisc.edu/krysan/DS-Lox/) was designated arf8-6 and was identical in phenotype to the fwf allele in Col-1 (arf8-4NIL; Vivian-Smith, 2001).
Cloning of ARF8 and Complementation of fwf
The ARF8 gene, including 1.8 kb of 5′ upstream promoter sequence and the complete coding region including the 3′UTR, was amplified from Ler wild-type and fwf mutant plants via PCR using the eLONGase amplification system (Invitrogen) according to the manufacturer's instructions with the primers ARF8-1830F (5′-CTCGAGTGAGAAGTCTATGATG-3′) and ARF8+2817R (5′-CCTAGGAAAGTTTAGTTACCCTGAGAC-3′). The PCR products were cloned using the pGEM-T Easy Vector System I (Promega), and sequencing revealed that the fwf fragment contained the transition from G to A in the predicted translation initiation codon (Vivian-Smith, 2001). The identified mutation in ARF8 from fwf plants creates a CAPS marker. Amplification with the primers ARF8-143F (5′-AGGAGATGGAGAAAGACGAG-3′) and ARF8+48R (5′-CTCTCCTTCATGACCCTGTTG-3′) and subsequent digest with Hsp92II (Promega) resulted in bands of 142 bp + 41 bp + 8 bp from Ler wild-type plants, whereas 183 bp + 8 bp bands were present in fwf plants.
The wild-type version of ARF8 was subcloned into the pBIN19 vector (Bevan, 1984) and transformed into Agrobacterium tumefaciens strain LBA4404. Ler and fwf plants were separately transformed with the construct via the floral dip method (Clough and Bent, 1998).
RNA Preparation and RNA Gel Blot Analysis
Total RNA was extracted from Arabidopsis plant tissues using TRIZOL LS reagent (Invitrogen) or the RNeasy plant mini kit (Qiagen). RNA gel blot analysis was performed with 10 μg of total RNA as described (Goetz et al., 2001) using a radioactive labeled ARF8-specific probe. The probe was labeled using the random primer Rediprime II DNA labeling system (Amersham Biosciences).
DNA Preparation and DNA Gel Blot Analysis
Genomic DNA was extracted from Arabidopsis plant tissues using a CTAB extraction protocol (Goetz et al., 2001). Ten micrograms of genomic DNA were digested with different restriction enzymes for 24 h and then subjected to electrophoresis in a 0.8% agarose gel. DNA was transferred to a nylon membrane (Biodyne B/Plus; PALL Gelman Laboratory), and hybridization and autoradiography were performed as for RNA gel blot analysis.
RT-PCR and qRT-PCR
One microgram of total RNA extracted from Arabidopsis plant tissues with the RNeasy plant mini kit and treated with an on-column RNase-free DNase protocol was used as template for cDNA synthesis with the ThermoScript RT-PCR system (Invitrogen). One microliter of the cDNA was then used for a standard PCR reaction using the primer pair NKE11F (5′-GCGGCCGCGGTACCTTTCCTATGTATCCA-3′) and ARF8+3956R (5′-GTCGACCTAGAGATGGGTCGGGTTTTGC-3′) to amplify ARF8 and the primer pair b-tubF (5′-GGGTGCTGGAAACAATTGGGCTAA-3′) and b-tubR (5′-ACTGCTCACTCACGCGCCTAA-3′) to amplify β-tubulin as a control.
For qRT-PCR, 3 μL of one-fifth diluted cDNA were used as a template in a 15 μL reaction with 300 nM of each primer and 7.5 μL of 2× Absolute QPCR SYBR Green ROX mix (ABgene, Integrated Sciences). Reactions were performed using the Rotor-Gene 3000 (Corbett Research, Adelab Scientific) and analyzed with Rotor-Gene 6.0 software (Corbett Research, Adelab Scientific). Primer pairs used for amplification of ARF8 fragments during qRT-PCR were as follows: Q-ARF8-122F (5′-TTGTACTTCCGGAGCTAAAGAGTT-3′) and Q-ARF8-15R (5′-CAAGAAACCAAACTTTGAAAACC-3′), Q-ARF8+296F (5′-AACCATTGACACCGGAGGAG-3′) and Q-ARF8+481R (5′-GCTGCAGTGTGTAATCCAATGG-3′), or Q-ARF8 +2495F (5′-TGAAGTCGTTTCCACTCATCTTT-3′) and Q-ARF8+2601R (5′-AAGTTTCAGGACCCATACTCACA-3′). The primer pair Q-UbceF (5′-AGGTACAGCGAGAGAAAGTAGCAGA-3′) and Q-UbceR (5′-AACAGAAAAAGCAAGCTGAAAAACA-3′) was used for amplification of a ubiquitin conjugating enzyme (At5g25760) fragment as a control and to normalize cDNA starting amounts.
GUS Constructs, Staining, and Microscopy
To create the transcriptional fusion construct PARF8:GUS, 1.8 kb of the 5′ upstream promoter sequence of ARF8 was amplified from Ler wild-type plants via PCR with the primers ARF8-1830F (5′-CTCGAGTGAGAAGTCTATGATG-3′) and ARF8-1R (5′-CTCGAGTTCAACTTCAAGAA-3′). The PCR fragment was cloned into the vector pART7NAPX, which contains a promoterless GUS gene (Gleave, 1992). The expression cassette, containing the amplified ARF8 promoter, the GUS gene, and the OCS terminator sequence, was excised from the vector with NotI and cloned into the binary vector pART27 (Gleave, 1992). This was transformed via Agrobacterium strain LBA4404 as described above (Clough and Bent, 1998).
The translational fusion constructs consisted of 5.8 kb of genomic sequence, including 1.8 kb of the 5′ upstream promoter sequence and the entire ARF8 coding region excluding the stop codon. This was amplified from wild-type plants and arf8-4 plants with the primers A8-1823SalIF (5′-GTCGACTGAGAAGTCTATGATGAG-3′) and ARF8:G:g:Rev (5′-CATCCCTAGGGAGATGGGTCGGGTTTTGCGGGAA-3′). The PCR fragment was cloned into the vector pGEM T-easy (Promega). SalI and AvrII fragments were released, allowing the ARF8 fragment to be subcloned in frame with a promoterless GUS gene in the vector pCAMBIA1381Xa (GenBank accession number AF234303). The constructs were transformed via floral dip as above (Clough and Bent, 1998).
Histological analysis of GUS enzyme activity in Arabidopsis tissue was done essentially as described by Jefferson et al. (1987). The stained tissues were postfixed in FAA (4% formaldehyde [v/v], 5% acetic acid [v/v], and 50% ethanol [v/v]) and cleared in 70% ethanol before being photographed. Whole-mount tissue samples were viewed with Stemi2000C or Axioplan microscopes (Carl Zeiss). Digital images were captured using a Spot II camera (Diagnostic Instruments Inc). Image processing and reproduction were performed with Auto Montage Essentials (Syncroscopy) and Photoshop 7.0 (Adobe Systems).
In Situ Hybridization
A 328-bp fragment from the 3′UTR of ARF8 was amplified with primers ARF8+2481F (5′-GGTACCAGAAGATGTGCATCAAATGGG-3′) and ARF8+2718R (5′-CCTAGGAAAGTTTAGTTACCCTGAGAC-3′) and cloned into pGEM-T Easy. Plasmids were sequenced to verify identity and orientation of inserts. Probe preparation and in situ hybridizations were performed as described previously (Tucker et al., 2003), except that once probes were added to the formamide-based hybridization solution and cover slips applied, the slides were heated to 80°C for 2 min prior to hybridization overnight at 42°C.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AT5G37020 (ARF8) and AT1G30330 (ARF6).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Expression Ratio of ARF8 mRNA in Various Tissues.
Supplemental Figure 1. Expression of ARF8 and GUS Marker Constructs in Ler and arf8-4 Plants.
Supplementary Material
Acknowledgments
We thank Steve Swain for critical reading of this manuscript, Kotaro Yamamoto for arf8-1, the ABRC for arf8-5, and the University of Wisconsin (Madison, WI) for supplying seeds for arf8-6. We also thank Lauren Hooper, Miva Splawinski, Sandra Protopsaltis, Melissa Pickering, and Carol Horsman for excellent technical assistance. This work was funded by an Australian postgraduate award (A.V.-S.) and a Horticulture Australia grant (A.M.K.) as part of the Key Genes for Horticultural Markets project.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Anna M. Koltunow (anna.koltunow@csiro.au).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.037192.
References
- Allen, E., Xie, Z., Gustafson, A.M., and Carrington, J.C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Archbold, D.D., and Dennis, F.G. (1985). Strawberry receptacle growth and endogenous IAA content as affected by growth regulator application and achene removal. J. Am. Soc. Hortic. Sci. 110 816–820. [Google Scholar]
- Bartel, B., and Bartel, D.P. (2003). MicroRNAs: At the root of plant development? Plant Physiol. 132 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. [DOI] [PubMed] [Google Scholar]
- Ben-Cheikh, W., Perez-Botella, J., Tadeo, F.R., Talon, M., and Primo-Millo, E. (1997). Pollination increases gibberellin levels in developing ovaries of seeded varieties of citrus. Plant Physiol. 114 557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan, M. (1984). Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmi, N., Salts, Y., Dedicova, B., Shabtai, S., and Barg, R. (2003). Induction of parthenocarpy in tomato via specific expression of the rolB gene in the ovary. Planta 217 726–735. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Coombe, B. (1960). Relationship of growth and development to changes in sugars, auxins, and gibberellins in fruit of seeded and seedless varieties of Vitis vinifera. Plant Physiol. 35 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Menezes, C.B., Maluf, W.R., de Azevedo, S.M., Faria, M.V., Nascimento, I.R., Nogueira, D.W., Gomes, L.A.A., and Bearzoti, E. (2005). Inheritance of parthenocarpy in summer squash (Cucurbita pepo L.). Genet. Mol. Res. 4 39–46. [PubMed] [Google Scholar]
- Dharmasiri, N., and Estelle, M. (2004). Auxin signaling and regulated protein degradation. Trends Plant Sci. 9 302–308. [DOI] [PubMed] [Google Scholar]
- Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeuwens, C.J., and Schwabe, W.W. (1975). Seed and pod wall development in Pisum sativum L. in relation to extracted and applied hormones. J. Exp. Bot. 26 1–14. [Google Scholar]
- Fos, M., and Nuez, F. (1996). Molecular expression of genes involved in parthenocarpic fruit set in tomato. Physiol. Plant 98 165–171. [Google Scholar]
- Fos, M., Nuez, F., and Garcia-Martinez, J.L. (2000). The gene pat-2, which induces natural parthenocarpy, alters the gibberellin content in unpollinated tomato ovaries. Plant Physiol. 122 471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fos, M., Proaño, K., Nuez, F., and García-Martínez, J.L. (2001). Role of gibberellins in parthenocarpic fruit development induced by the genetic system pat-3/pat-4 in tomato. Physiol. Plant 111 545–550. [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez, J.L., and Hedden, P. (1997). Gibberellins and fruit development. In Phytochemistry of Fruit and Vegetables, F.A. Tomás-Barberán and R.J. Robins, eds (Oxford, UK: Clarendon Press), pp. 263–286.
- Garcia-Martinez, J.L., Marti, M., Sabater, T., Maldonado, A., and Vercher, Y. (1991). Development of fertilized ovules and their role in the growth of the pea pod. Physiol. Plant 83 411–416. [Google Scholar]
- Geballe, A.P., and Sachs, M.S. (2000). Translational control by upstream open reading frames. In Translational Control of Gene Expression, N. Sonenberg, J.W.B. Hershey, and M.B. Mathews, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 595–614.
- George, W.L., Scott, J.W., and Splittstoesser, W.E. (1984). Parthenocarpy in tomato. Hortic. Rev. (Am. Soc. Hortic. Sci.) 6 65–84. [Google Scholar]
- Gillaspy, G., Ben-David, H., and Gruissem, W. (1993). Fruits: A developmental perspective. Plant Cell 5 1439–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave, A. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20 1203–1207. [DOI] [PubMed] [Google Scholar]
- Goetz, M., Godt, D.E., Guivarc'h, A., Kahmann, U., Chriqui, D., and Roitsch, T. (2001). Induction of male sterility in plants by metabolic engineering of the carbohydrate supply. Proc. Natl. Acad. Sci USA 98 6522–6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414 271–276. [DOI] [PubMed] [Google Scholar]
- Guilfoyle, T., Hagen, G., Ulmasov, T., and Murfett, J. (1998). How does auxin turn on genes? Plant Physiol. 118 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle, T.J., and Hagen, G. (2001). Auxin response factors. J. Plant Growth Regul. 20 281–291. [Google Scholar]
- Hardtke, C.S., Ckurshumova, W., Vidaurre, D.P., Singh, S.A., Stamatiou, G., Tiwari, S.B., Hagen, G., Guilfoyle, T.J., and Berleth, T. (2004). Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131 1089–1100. [DOI] [PubMed] [Google Scholar]
- Hellmann, H., and Estelle, M. (2002). Plant development: Regulation by protein degradation. Science 297 793–797. [DOI] [PubMed] [Google Scholar]
- Jefferson, R., Kavanagh, T., and Bevan, M. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik, P.D., and Barton, M.K. (2005). Surge and destroy: The role of auxin in plant embryogenesis. Development 132 3577–3585. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. (2003). P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4 205–217. [DOI] [PubMed] [Google Scholar]
- Kawaguchi, R., and Bailey-Serres, J. (2002). Regulation of translational initiation in plants. Curr. Opin. Plant Biol. 5 460–465. [DOI] [PubMed] [Google Scholar]
- Kepinski, S., and Leyser, O. (2004). Auxin-induced SCFTIR1-Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc. Natl. Acad. Sci. USA 101 12381–12386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using codominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Langridge, U., Schwall, M., and Langridge, P. (1991). Squashes of plant tissue as substrate for PCR. Nucleic Acids Res. 19 6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S., George, W.L., and Splittstoesser, W.E. (1984). Expression and inheritance of parthenocarpy in “Severianin” tomato. J. Hered. 75 62–66. [Google Scholar]
- Liscum, E., and Reed, J.W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49 387–400. [PubMed] [Google Scholar]
- Mallory, A.C., Bartel, D.P., and Bartel, B. (2005). MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17 1360–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAbee, J.M., Hill, T.A., Skinner, D.J., Izhaki, A., Hauser, B.A., Meister, R.J., Venugopala Reddy, G., Meyerowitz, E.M., Bowman, J.L., and Gasser, C.S. (2006). ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. Plant J. 46 522–531. [DOI] [PubMed] [Google Scholar]
- Mezzetti, B., Landi, L., Pandolfini, T., and Spena, A. (2004). The defH9-iaaM auxin-synthesizing gene increases plant fecundity and fruit production in strawberry and raspberry. BMC Biotechnol. 4 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, D.R., and Geballe, A.P. (2000). Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20 8635–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal, P., Ellis, C.M., Weber, H., Ploense, S.E., Barkawi, L.S., Guilfoyle, T.J., Hagen, G., Alonso, J.M., Cohen, J.D., Farmer, E.E., Ecker, J.R., and Reed, J.W. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132 4107–4118. [DOI] [PubMed] [Google Scholar]
- Nishimura, T., Wada, T., and Okada, K. (2004). A key factor of translation reinitiation, ribosomal protein L24, is involved in gynoecium development in Arabidopsis. Biochem. Soc. Trans. 32 611–613. [DOI] [PubMed] [Google Scholar]
- Nishimura, T., Wada, T., Yamamoto, K.T., and Okada, K. (2005). The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell 17 2940–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsch, J.P. (1952). Plant hormones in the development of fruits. Q. Rev. Biol. 27 33–57. [DOI] [PubMed] [Google Scholar]
- Nitsch, J.P. (1970). Hormonal factors in growth and development. In The Biochemistry of Fruits and Their Products, A.C. Hulme, ed (New York: Academic Press), pp. 427–472.
- O'Neill, S.D. (1997). Pollination regulation of flower development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 547–574. [DOI] [PubMed] [Google Scholar]
- O'Neill, S.D., and Nadeau, J.A. (1997). Post-pollination flower development. Hortic. Rev. (Am. Soc. Hortic. Sci.) 19 1–58. [Google Scholar]
- Okushima, Y., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga, J.A., van Huizen, R., and Reinecke, D.M. (2002). Hormone and seed-specific regulation of pea fruit growth. Plant Physiol. 128 1379–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike, L.M., and Peterson, C.R. (1969). Inheritance of parthenocarpy in the cucumber (Cucumis sativus L.). Euphytica 18 101–105. [Google Scholar]
- Pufky, J., Qiu, Y., Rao, M., Hurban, P., and Jones, A. (2003). The auxin-induced transcriptome for etiolated Arabidopsis seedlings using a structure/function approach. Funct. Integr. Genomics 3 135–143. [DOI] [PubMed] [Google Scholar]
- Raghavan, V. (2003). Some reflections on double fertilization, from its discovery to the present. New Phytol. 159 565–583. [DOI] [PubMed] [Google Scholar]
- Remington, D.L., Vision, T.J., Guilfoyle, T.J., and Reed, J.W. (2004). Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 135 1738–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. (2002). Prediction of plant microRNA targets. Cell 110 513–520. [DOI] [PubMed] [Google Scholar]
- Robinson, R.W., and Reiners, S. (1999). Parthenocarpy in summer squash. HortScience 34 715–717. [Google Scholar]
- Rogg, L.E., and Bartel, B. (2001). Auxin signaling: Derepression through regulated proteolysis. Dev. Cell 1 595–604. [DOI] [PubMed] [Google Scholar]
- Rotino, G.L., Perri, E., Zottini, M., Sommer, H., and Spena, A. (1997). Genetic engineering of parthenocarpic plants. Nat. Biotechnol. 15 1398–1401. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain, S.M., Reid, J.B., and Kamiya, Y. (1997). Gibberellins are required for embryo growth and seed development in pea. Plant J. 12 1329–1338. [Google Scholar]
- Sykes, S.R., and Lewis, S. (1996). Comparing Imperial mandarin and Silverhill satsuma mandarins as seed parents in a breeding program aimed at developing new seedless citrus cultivars for Australia. Aust. J. Exp. Agric. 36 731–738. [Google Scholar]
- Talon, M., Hedden, P., and Primo-Millo, E. (1990. b). Gibberellins in Citrus sinensis: A comparison between seeded and seedless varieties. J. Plant Growth Regul. 9 201–206. [Google Scholar]
- Talon, M., Zacarias, L., and Primo-Millo, E. (1990. a). Hormonal changes associated with fruit set and development in mandarins differing in their parthenocarpic ability. Physiol. Plant 79 400–406. [Google Scholar]
- Talon, M., Zacarias, L., and Primomillo, E. (1992). Gibberellins and parthenocarpic ability in developing ovaries of seedless mandarins. Plant Physiol. 99 1575–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu, K., Kumagai, S., Muto, H., Sato, A., Watahiki, M.K., Harper, R.M., Liscum, E., and Yamamoto, K.T. (2004). MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, C., Muto, H., Higuchi, K., Matamura, T., Tatematsu, K., Koshiba, T., and Yamamoto, K.T. (2004). Disruption and overexpression of Auxin Response Factor8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J. 40 333–343. [DOI] [PubMed] [Google Scholar]
- Tiwari, S.B., Hagen, G., and Guilfoyle, T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, S.B., Wang, X.-J., Hagen, G., and Guilfoyle, T.J. (2001). AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, M.R., Araujo, A.-C.G., Paech, N.A., Hecht, V., Schmidt, E.D.L., Rossell, J.-B., de Vries, S.C., and Koltunow, A.M.G. (2003). Sexual and apomictic reproduction in Hieracium subgenus Pilosella are closely interrelated developmental pathways. Plant Cell 15 1524–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999. a). Dimerization and DNA binding of auxin response factors. Plant J. 19 309–319. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999. b). Activation and repression of transcription by auxin response factors. Proc. Natl. Acad. Sci. USA 96 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoquaux, F., Blanvillain, R., Delseny, M., and Gallois, P. (2002). Less is better: New approaches for seedless fruit production. Trends Biotechnol. 18 233–242. [DOI] [PubMed] [Google Scholar]
- Vivian-Smith, A. (2001). The Molecular Basis for the Initiation of Fruit Development and Parthenocarpy. PhD dissertation (Adelaide, Australia: University of Adelaide).
- Vivian-Smith, A., and Koltunow, A.M. (1999). Genetic analysis of growth-regulator-induced parthenocarpy in Arabidopsis. Plant Physiol. 121 437–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian-Smith, A., Luo, M., Chaudhury, A., and Koltunow, A. (2001). Fruit development is actively restricted in the absence of fertilization in Arabidopsis. Development 128 2321–2331. [DOI] [PubMed] [Google Scholar]
- Wang, H., Jones, B., Li, Z., Frasse, P., Delalande, C., Regad, F., Chaabouni, S., Latche, A., Pech, J.-C., and Bouzayen, M. (2005). The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17 2676–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, L., Carles, C.C., Osmont, K.S., and Fletcher, J.C. (2005). A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc. Natl. Acad. Sci. USA 102 9703–9708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenser, N., Ellsmore, A., Leasure, C., and Callis, J. (2001). Auxin modulates the degradation rate of Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 98 11795–11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.