Abstract
Submergence-1 (Sub1), a major quantitative trait locus affecting tolerance to complete submergence in lowland rice (Oryza sativa), contains two or three ethylene response factor (ERF)–like genes whose transcripts are regulated by submergence. In the submergence-intolerant japonica cultivar M202, this locus encodes two ERF genes, Sub1B and Sub1C. In the tolerant near-isogenic line containing the Sub1 locus from the indica FR13A, M202(Sub1), the locus additionally encodes the ERF gene Sub1A. During submergence, the tolerant M202(Sub1) displayed restrained leaf and internode elongation, chlorophyll degradation, and carbohydrate consumption, whereas the enzymatic activities of pyruvate decarboxylase and alcohol dehydrogenase were increased significantly compared with the intolerant M202. Transcript levels of genes associated with carbohydrate consumption, ethanolic fermentation, and cell expansion were distinctly regulated in the two lines. Sub1A and Sub1C transcript levels were shown to be upregulated by submergence and ethylene, with the Sub1C allele in M202 also upregulated by treatment with gibberellic acid (GA). These findings demonstrate that the Sub1 region haplotype determines ethylene- and GA-mediated metabolic and developmental responses to submergence through differential expression of Sub1A and Sub1C. Submergence tolerance in lowland rice is conferred by a specific allele variant of Sub1A that dampens ethylene production and GA responsiveness, causing quiescence in growth that correlates with the capacity for regrowth upon desubmergence.
INTRODUCTION
Flooding of croplands is a frequent natural disaster in many regions of the world. The flooding of root systems and partial to complete submergence of aerial organs can dramatically reduce crop productivity. Plant submergence attributable to complete flooding restricts the diffusion of oxygen and carbon dioxide by 104-fold, which has a dramatic impact on biochemical activities, such as aerobic respiration and photosynthesis (Armstrong and Drew, 2002). The inhibition of these processes stimulates a variety of responses that can enhance the survival of transient inundation. Acclimative responses are species and genotype specific and can include altered rates of petiole/internode elongation, altered anatomy and cell ultrastructure in leaves and roots, development of lateral/adventitious roots, formation of aerenchyma, and a switch from aerobic to anaerobic respiration (Drew, 1997; Fukao and Bailey-Serres, 2004). The acclimative responses to submergence stress also engender potentially lethal side effects, such as carbohydrate starvation and the accumulation of toxic end products that acidify the cytosol or damage membrane integrity (Drew, 1997; Gibbs and Greenway, 2003; Fukao and Bailey-Serres, 2004).
Most nonaquatic plants are damaged by transient inundation of aerial tissue for 24 to 48 h. Exceptionally, rice (Oryza sativa) is generally tolerant of submergence. Deepwater rice and the widely cultivated lowland rice overcome submergence stress by antithetical strategies (Fukao and Bailey-Serres, 2004). Deepwater rice avoids the stress by the activation of gibberellic acid (GA)–promoted internode elongation, which allows plants to outgrow submergence and thereby restore gas exchange above the water surface. Lowland rice cultivars are typically cultivated in flooded paddies but are generally intolerant of complete submergence. The submergence-tolerant East Indian accession FR13A restricts leaf and internode elongation during inundation and can recommence the initiation of leaf development upon desubmergence (Singh et al., 2001; Das et al., 2005). Physiological responses to submergence stress in rice have been compared using tolerant and intolerant varieties. However, none of these analyses has been performed on near-isogenic lines. Consequently, the significance and genetic determinants of the observed variation in metabolic and developmental responses have remained unclear.
Submergence-1 (Sub1) is a major quantitative trait locus affecting submergence tolerance in lowland rice, which accounts for 35 to 69% of phenotypic variance in tolerance in diverse backgrounds (Xu and Mackill, 1996; Nandi et al., 1997; Sripongpangkul et al., 2000; Xu et al., 2000; Toojinda et al., 2003). Detailed genetic and physical mapping of Sub1 revealed that this locus contains a variable cluster of two to three genes that encode proteins with the DNA binding domain common to the ethylene response factors (ERFs)/ethylene-responsive element binding proteins/Apetala2-like proteins (Xu et al., 2006). All three Sub1 region genes fall in the B-2 subclass of ERF proteins, which contains a single 58- to 59-residue ERF domain. It has been shown in a few cases to interact with GC-rich cis-acting sequences of target genes and to regulate ethylene-mediated responses (Gutterson and Reuber, 2004). The genes Sub1B and Sub1C are present in a wide range of indica and japonica varieties, whereas Sub1A is limited to a subset of indica varieties. A single amino acid substitution distinguishes the Sub1A-1 allele of tolerant indica varieties, including FR13A. The Sub1A alleles in tolerant and intolerant varieties are highly and poorly induced under submergence, respectively. Moreover, Sub1A is absent from all japonica germplasms studied and some indica germplasms, all of which are submergence-intolerant (Xu et al., 2006). In support of Sub1A as a key determinant of submergence tolerance, the transformation of the intolerant japonica variety Liaogeng with a constitutively expressed Sub1A gene significantly increased tolerance to submergence (Xu et al., 2006).
Submergence promotes the biosynthesis of 1-aminocyclopropane-1-carboxylic acid, which is converted to ethylene in an oxygen-dependent manner (Banga et al., 1996; Kende et al., 1998; Peng et al., 2001). Oxygen produced by photosynthesis or from the surrounding aqueous environment can be used for ethylene synthesis during submergence (Mommer et al., 2004, 2005). Ethylene levels can also increase as a result of physical entrapment by the surrounding aqueous environment, as observed in the semiaquatic eudicot Rumex palustris (Banga et al., 1996). This hormone plays important roles in the acclimative responses to submergence and flooding. It contributes to the programmed cell death that leads to the formation of aerenchyma and adventitious roots under hypoxia (Drew et al., 2000; Mergemann and Sauter, 2000; Gunawardena et al., 2001) and promotes internode and petiole elongation that allows deepwater rice and R. palustris to extend photosynthetic organs above the water surface (Kende et al., 1998; Peeters et al., 2002). Cell elongation under submergence involves increased accumulation of gene transcripts encoding expansins, which mediate cell wall loosening. In R. palustris, Expansin A transcripts increased after submergence and ethylene treatment, and the induction by submergence was suppressed by an ethylene action inhibitor, 1-methylcyclopropene (Vreeburg et al., 2005). Ethylene also triggers an increase in the endogenous content of GA and the sensitivity to this phytohormone in deepwater rice and R. palustris, leading to the conclusion that internode and petiole elongation during submergence results from ethylene-mediated GA responses (Kende et al., 1998; Peeters et al., 2002). Recently, it was shown that the ethylene entrapped during submergence contributed to a decrease in endogenous abscisic acid content in R. palustris (Benschop et al., 2005). The change in both of these hormones was a prerequisite for petiole elongation.
In this study, we compared the acclimative responses to submergence of near-isogenic japonica lines that differ in haplotype of an ERF gene cluster at the Sub1 locus. Our results demonstrate that the ERF domain genes Sub1A and Sub1C are differentially regulated by submergence, ethylene, and GA at the level of transcript accumulation. The presence of Sub1A-1 at the Sub1 locus results in ethylene- and GA-mediated negative regulation of genes associated with carbohydrate catabolism and cell elongation as well as positive regulation of genes involved in ethanolic fermentation during submergence. These data demonstrate that evolutionary divergence in an ERF domain gene cluster is responsible for the positive and negative gene regulation that controls multifaceted cellular and developmental responses to submergence and confers submergence tolerance in lowland rice.
RESULTS
Regulation of Three ERF Genes at the Sub1 Locus under Submergence
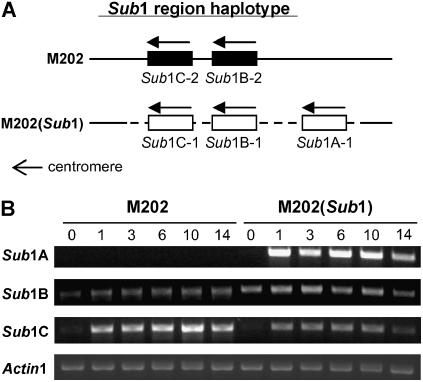
The Sub1 region on rice chromosome 9 contains a cluster of two or three Sub1 genes (Sub1A, Sub1B, and Sub1C), and genotypic variation at this complex locus confers distinctions in submergence tolerance (Xu et al., 2006). Submergence-intolerant japonica rice, including the inbred M202, lacks Sub1A and possesses Sub1B and Sub1C. The near-isogenic line M202(Sub1) was generated by introgression of the Sub1 region from the submergence-tolerant indica cultivar FR13A (Xu et al., 2004). The M202(Sub1) introgression line contains all three Sub1 genes of indica origin in a genomic region of 182 kb based on molecular mapping and genomic sequence analysis (Figure 1A) (Xu et al., 2006). In the submergence stress studies presented here, 14-d-old plants were completely submerged for up to 14 d and leaf tissue was collected at six time points. Semiquantitative RT-PCR analysis confirmed the absence of the Sub1A transcript in leaves of M202, consistent with the absence of this gene from the Nipponbare genome sequence and other japonica varieties studied. The level of Sub1A mRNA increased rapidly in abundance after 1 d of submergence in M202(Sub1) leaves and remained increased for up to 14 d of stress (Figure 1B). The Sub1B transcript was detected in both genotypes and was slightly upregulated by the stress, with higher levels of the transcript consistently detected in M202(Sub1). As observed for Sub1A, the Sub1C transcript was increased dramatically for the duration of the stress period. The level of the Sub1C transcript was higher in M202 compared with M202(Sub1). The distinctions in Sub1B and Sub1C mRNA levels under stress in the two lines may be attributable to the distinct regulation of the japonica versus indica alleles or may reflect the absence versus presence of Sub1A in M202 and M202(Sub1), respectively.
Figure 1.
Comparison of the Allele Composition and mRNA Accumulation of Sub1 Region Genes in M202 and M202(Sub1).
(A) Gene and allele composition of ERF domain genes in the Sub1 region of chromosome 9. The Sub1 haplotype in M202 (japonica) consists of Sub1B-2 and Sub1C-2, whereas the genomic region in M202(Sub1) encodes Sub1A-1, Sub1B-1, and Sub1C-1 (Xu et al., 2006). The dashed line indicates the ∼182 kb introgressed from the indica accession FR13A. Arrows represent the direction of transcription for the ERF genes.
(B) Analysis of Sub1 gene transcript accumulation in M202 and M202(Sub1) leaves during submergence. Fourteen-day-old plants were submerged for up to 14 d, and leaf tissue was harvested at specific time points (days 0, 1, 3, 6, 10, and 14). Total RNA was analyzed by semiquantitative RT-PCR using gene-specific primers for Sub1A, Sub1B, and Sub1C. The level of Actin1 mRNA was used as a loading control. The number of cycles for linear amplification was optimized for each primer pair. Representative results from at least three independent biological replicate experiments are shown.
Submergence Recovery and Chlorophyll Maintenance under Submergence Are Influenced by the Sub1 Haplotype
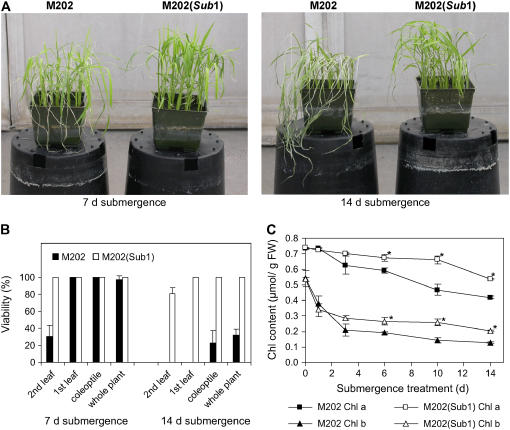
Fourteen-day-old plants of these near-isogenic lines consisted of a coleoptile and two fully expanded leaves with no evident difference in leaf and internode development. The influence of the M202 versus the FR13A Sub1 haplotype on submergence tolerance and the viability of the coleoptile, leaf, and whole plant was evaluated at 7 d after recovery from 7 or 14 d of submergence (Figures 2A and 2B). The two genotypes had a distinct appearance upon desubmergence. M202 leaves were more prostrate, spindly, and chlorotic. After 7 d of submergence, only 30.7% of the second leaves of M202 were intact, whereas all leaves of M202(Sub1) were fully green. The viability of the coleoptile and first leaf was not significantly different in the two genotypes after 7 d of inundation. After a more prolonged submergence (14 d), all of the first and second leaves of M202 appeared brown and withered, whereas all of the first leaves and 81.3% of the second leaves of M202(Sub1) were fully green and appeared undamaged. The coleoptile of M202(Sub1) was also considerably more tolerant to 14 d of submergence. Viability after 14 d of submergence was significantly different in these lines, as judged by the emergence and growth of the third and subsequent leaves (Figure 2B). All of the M202(Sub1) plants produced new leaves during the recovery period, whereas only 32.0% of the M202 plants did so. These data confirm that the introgression of the FR13A Sub1 region into M202 is sufficient to dramatically enhance the viability of fully emerged leaves and to confer the ability to resume apical meristem development upon desubmergence.
Figure 2.
Phenotypes of M202 and M202(Sub1) Plants after Submergence.
(A) Rice plants after 7 d of recovery from submergence. Fourteen-day-old plants were submerged for 7 d (left) or 14 d (right). After submergence, plants were returned to normal growth conditions for 7 d and photographed.
(B) Viability of coleoptile, first leaf, second leaf, and whole plants after desubmergence. The leaf and whole plant viability of each genotype was evaluated in the samples shown in (A). Fully green (nonchlorotic) leaves were scored as viable. Plants were scored as viable if a new leaf appeared during recovery. The data represent means ± sd from three independent biological replicates (n = 75).
(C) Decrease in chlorophyll a/b content in leaves during submergence. Fourteen-day-old plants were submerged for up to 14 d, and leaf tissue was harvested at specific time points (days 0, 1, 3, 6, 10, and 14). Chlorophyll was extracted in 80% (v/v) buffered acetone and quantified by a spectrophotometer. The data represent means ± sd from three independent biological replicates. Asterisks indicate significant differences between the two genotypes (P < 0.05). FW, fresh weight.
In rice, submergence stress increases the transcript level and enzymatic activity of chlorophyllase, which promotes the degradation of chlorophyll (Ella et al., 2003). To determine whether genetic variation at the Sub1 region influences chlorophyll maintenance (biosynthesis and/or breakdown) during submergence, chlorophyll contents were monitored over 14 d of submergence (Figure 2C). Chlorophyll a/b contents declined gradually in both genotypes, but M202(Sub1) maintained significantly higher levels of chlorophyll than did M202 from day 6. Chlorophyll b content decreased more rapidly than chlorophyll a content, reflecting the conversion of chlorophyll b to a in the degradation pathway (Folly and Engel, 1999). These data confirm that the FR13A Sub1 haplotype enhances the maintenance of chlorophyll during submergence.
Differential Regulation of Elongation under Submergence Is Controlled by the Sub1 Haplotype
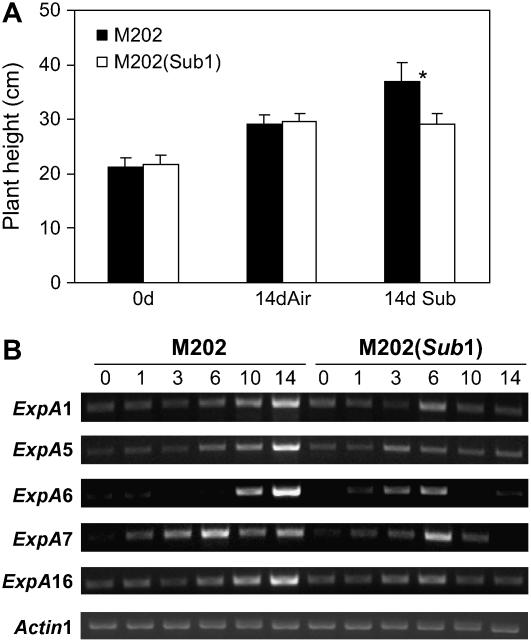
Generally, submergence-sensitive lowland rice cultivars elongate more rapidly than tolerant cultivars under submergence stress (Singh et al., 2001; Das et al., 2005). To examine whether the Sub1 haplotype controls growth rate, plant height was compared in M202 and M202(Sub1) plants grown under normal (aerobic) or submerged conditions for 14 d (Figure 3A). The height of 14-d-old plants of these genotypes was nearly identical, and both lines grew uniformly under normal conditions. In response to 14 d of submergence stress, M202 displayed significantly greater elongation than M202(Sub1). The increase in plant height of the tolerant line was the same under submergence and normal conditions, indicating an acceleration in elongation of the intolerant line under the stress. These data confirm that the FR13A Sub1 haplotype suppresses elongation during submergence but does not influence plant height under normal conditions. At maturity, these two lines were indistinguishable in height (data not shown). The lack of effect of the Sub1 haplotype on normal growth and development is consistent with the observation that the Sub1A gene was submergence-inducible.
Figure 3.
Leaf Elongation in M202 Is Greater Than in M202(Sub1) Plants under Submergence.
(A) Plant height after submergence treatment. Fourteen-day-old plants were grown under aerobic (Air) or submerged (Sub) conditions for 14 d. Plant height was measured at days 0 and 14. The height of plants submerged for 14 d was recorded upon desubmergence. The data represent means ± sd from three independent biological replicates (n = 75). The asterisk indicates that M202 plants after 14 d of submergence were significantly more elongated than other plants (P < 0.01).
(B) Analysis of ExpA gene transcript accumulation in leaves during submergence. Fourteen-day-old plants were submerged for up to 14 d, and leaf tissue was collected at specific time points (days 0, 1, 3, 6, 10, and 14). Total RNA was analyzed by semiquantitative RT-PCR using gene-specific primers for ExpA. The level of Actin1 mRNA was used as a loading control.
Expansins are proteins that mediate cell wall loosening in plants. Increased expansin mRNA and protein levels are correlated with cell elongation (Li et al., 2003). A total of 80 expansin-encoding genes have been recognized in the japonica rice genome and classified into four subgroups by phylogenetic analysis (Li et al., 2003). At least seven Expansin A (ExpA) mRNAs accumulate in leaves of deepwater rice, and their abundance was upregulated by submergence (Lee and Kende, 2002). The levels of five ExpA transcripts, ExpA1, ExpA5, ExpA6, ExpA7, and ExpA16, were assessed by semiquantitative RT-PCR analysis in the two near-isogenic lines (Figure 3B). All five mRNAs, except for ExpA7, were gradually increased in M202 over the 14 d of submergence. By contrast, in M202(Sub1), these transcripts increased slightly until day 3 or 6 and then declined. A higher and more sustained accumulation of ExpA mRNAs during submergence was consistent with the significant promotion of leaf and internode elongation in the intolerant M202.
Carbohydrate Consumption under Submergence Is Modulated by the Sub1 Haplotype
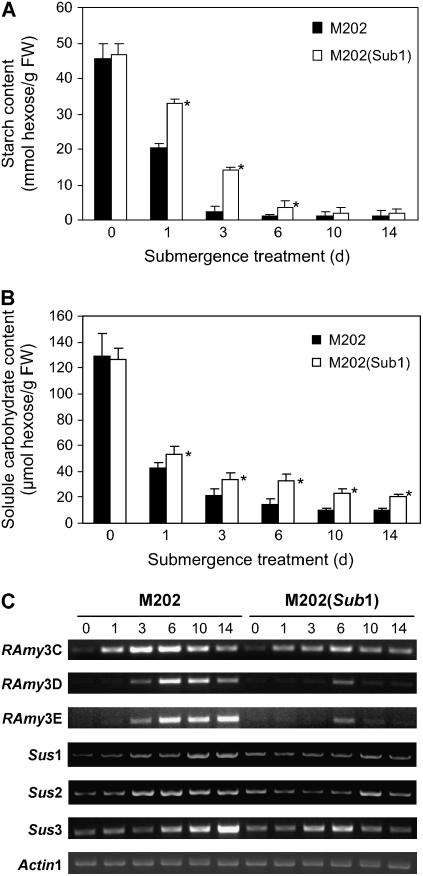
Plant cells consume carbohydrates through energetically inefficient coupling of glycolysis and anaerobic fermentation when oxygen levels are restricted (Drew, 1997). An additional consequence of submergence is that carbohydrate production by photosynthesis is restrained as a result of reduced light intensity, CO2 diffusion, and chlorophyll content. Therefore, long-term submergence causes extensive carbohydrate and energy (nucleotide triphosphate) starvation. To investigate the influence of the Sub1 haplotype on carbohydrate metabolism, starch and total soluble carbohydrate contents in leaves were monitored in the two genotypes over 14 d of submergence (Figures 4A and 4B). Leaves of both genotypes had similar starch content (Figure 4A). In M202, starch was rapidly consumed and decreased to 9.8% of the total starch content at day 0 after 3 d of submergence. The decline in starch was significantly less dramatic in M202(Sub1), which retained 32.0% of the total starch content on day 3. Nonetheless, starch reserves were greatly depleted in both genotypes by day 10. These observations reveal that M202 leaves experience a more rapid and prolonged period of starch starvation during submergence. A similar trend was confirmed for the amount of total soluble carbohydrates, including glucose, fructose, and sucrose, in leaves of the two genotypes (Figure 4B). More than 60% of soluble carbohydrates were consumed within 1 d of submergence in both genotypes. After the initial response, the consumption rate decelerated, with the magnitude of diminution in total soluble carbohydrates significantly less severe in the tolerant M202(Sub1).
Figure 4.
Carbohydrate Consumption Is Accelerated in Leaves of M202 Relative to M202(Sub1) during Submergence.
(A) Starch contents in leaves during submergence. Fourteen-day-old plants were submerged for up to 14 d, and leaf samples were collected at specific time points (days 0, 1, 3, 6, 10, and 14). Leaf starch content was determined by an enzymatic method. The data represent means ± sd from three independent biological replicates. Asterisks indicate significant differences between the two genotypes at that time point (P < 0.05). FW, fresh weight.
(B) Total soluble carbohydrate contents in leaves during submergence. Leaf samples used for the starch assay were analyzed to determine total carbohydrate contents by the anthrone method. The data represent means ± sd from three independent biological replicates. Asterisks indicate significant differences between the two genotypes at that time point (P < 0.05).
(C) Accumulation of gene transcripts associated with carbohydrate catabolism. Leaf samples analyzed for starch and total soluble carbohydrates were used to extract total RNA, which was analyzed by semiquantitative RT-PCR using gene-specific primers for α-amylases (RAmy) and sucrose synthases (Sus). The level of Actin1 mRNA was used as a loading control.
A number of metabolic enzymes are involved in the catabolism of starch and soluble carbohydrates in plants. Anoxia-intolerant species, such as wheat (Triticum aestivum) and barley (Hordeum vulgare), fail to degrade conserved starch under conditions of oxygen deprivation because α-amylases are not synthesized and active in their seeds under anoxia (Guglielminetti et al., 1995b). Exceptionally, transcripts of rice α-amylases accumulate in the seed embryo and aleurone during germination even under anoxia (Hwang et al., 1999). In addition, α-amylase protein levels and activity were shown to be induced by anoxia in rice seedlings (Guglielminetti et al., 1995b). Regulation of leaf α-amylase genes in lowland rice leaves under submergence has not been reported. Semiquantitative RT-PCR detection of the transcripts of three α-amylase genes, Rice Amylase-3C (RAmy3C), RAmy3D, and RAmy3E, revealed that their upregulation was controlled by the Sub1 locus (Figure 4C). The level of RAmy3C mRNA increased immediately under submergence stress, reached a maximum by day 6, and then decreased through day 14 in both genotypes. Overall RAmy3C transcript induction was greater in M202. RAmy3D and RAmy3E transcript increases occurred later than in RAmy3C; the increases in these transcripts were considerably lower in M202(Sub1) leaves.
Sucrose, which is the major energy source and transport form of carbohydrates in rice, can be hydrolyzed via two distinct pathways: the more energy-efficient sucrose synthase pathway and the invertase pathway (Zeng et al., 1999; Geigenberger, 2003; Fukao and Bailey-Serres, 2004). It has been shown that the transcript level and the enzymatic activity of sucrose synthase increase, whereas those of invertase decrease, during oxygen deprivation in maize (Zea mays) roots and potato (Solanum tuberosum) tubers, suggesting that sucrose synthase is the principal enzyme that converts sucrose to phosphorylated hexose sugars under low oxygen stress (Zeng et al., 1999; Geigenberger, 2003). In M202 leaves, the transcript levels of all three sucrose synthase genes, Sus1, Sus2, and Sus3, became increased by the middle of the submergence period and remained increased through day 14 (Figure 4C). By contrast, Sus gene transcripts were only transiently and much less dramatically increased in M202(Sub1). Thus, the Sub1 haplotype controls the regulation of the transcript levels of genes encoding α-amylase and sucrose synthase, which are required for carbohydrate catabolism during submergence.
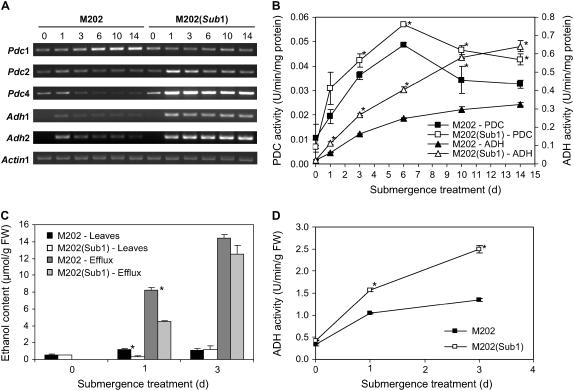
Transcript Levels of Genes Associated with Ethanolic Fermentation under Submergence Are Controlled by the Sub1 Haplotype
The activation of fermentation under conditions of oxygen deprivation is required to recycle NAD+, which is essential for the maintenance of glycolysis. It is well established that oxygen deprivation generally promotes a transient burst in lactate fermentation that is followed by an extended period of increased ethanolic fermentation (Drew, 1997). This increase typically requires increases in pyruvate decarboxylase (PDC) and alcohol dehydrogenase (ADH). In fact, adh and pdc loss-of-function mutants in maize, rice, and Arabidopsis thaliana succumb rapidly to low oxygen and submergence stress, confirming that ethanolic fermentation is necessary for acclimation to the transient stress (Schwartz, 1969; Rahman et al., 2001; Baxter-Burrell et al., 2003; Ismond et al., 2003; Kürsteiner et al., 2003). To examine the role of the Sub1 haplotype in ethanolic fermentation during submergence, transcript levels of the Pdc and Adh genes were evaluated in leaves of the two genotypes (Figure 5A). In M202, Pdc1 mRNA gradually accumulated until day 6 and remained constant until day 14. By contrast, in M202(Sub1), the Pdc1 mRNA increase was limited. The Pdc2 and Pdc4 transcript accumulation was consistent with the Adh1 and Adh2 mRNA levels in both lines. A dramatic increase in these transcripts occurred within 1 d of submergence and remained steady through day 14 in tolerant M202(Sub1); the increase of these transcripts was greatly limited in M202.
Figure 5.
Ethanolic Fermentation in M202 and M202(Sub1) Leaves in Response to Submergence.
(A) Accumulation of gene transcripts associated with ethanolic fermentation during submergence. Fourteen-day-old seedlings were exposed to submergence stress for up to 14 d, and leaf tissue was harvested at specific time points (days 0, 1, 3, 6, 10, and 14). Total RNA extracted from the leaf tissues was analyzed by semiquantitative RT-PCR using gene-specific primers for Pdc and Adh transcripts. The level of Actin1 mRNA was used as a loading control.
(B) PDC and ADH activities in leaves during submergence. Specific activities of PDC and ADH were assayed for the leaf tissue used for RT-PCR analysis of Pdc and Adh mRNAs. The data represent means ± sd from three independent biological replicates. Asterisks indicate significant differences between the two genotypes at that time point (P < 0.05).
(C) Ethanol content of leaves and surrounding medium of submerged plants. Ten-day-old plants were submerged in water in test tubes for up to 3 d. Leaf tissues and the surrounding medium were collected at days 0, 1, and 3, and their ethanol contents were quantified enzymatically. The data represent means ± sd from three independent biological replicates. Asterisks indicate significant differences between the two genotypes at that time point (P < 0.01). FW, fresh weight.
(D) ADH activity in leaves used for the ethanol assay. ADH enzymatic activity was assayed in the leaf tissues used for the analysis of ethanol content. ADH values are presented on a fresh weight basis. The data represent means ± sd from three independent biological replicates. Asterisks indicate significant differences between the two genotypes at that time point (P < 0.01).
The enzymatic activities of PDC and ADH were also monitored in leaves of the two genotypes under submergence stress (Figure 5B). PDC activities increased at most 4.9- and 8.1-fold in M202 and M202(Sub1), respectively, compared with 14-d-old plants grown in air. The increase in specific activity of PDC was significantly greater in M202(Sub1) after day 3. PDC activity increased to a maximum by day 6 and then decreased through day 14 in both genotypes. ADH activities increased 14.8- and 30.6-fold at the maximum in M202 and M202(Sub1), respectively. In both genotypes, ADH activity increased continuously for 14 d of submergence, although the rate of increase was gradually lessened. These data suggest that the Sub1 haplotype controls the magnitude but not the temporal pattern of PDC and ADH activities during submergence.
The limitation of carbohydrate consumption in the tolerant M202(Sub1) could in fact restrict ethanolic fermentation, despite the higher capacity for this process in this line. Ten-day-old seedlings grown on agar medium in test tubes were fully submerged, and ethanol content and ADH activity were monitored at days 0, 1, and 3 (Figures 5C and 5D). Higher ADH activity induction was observed in M202(Sub1) but was not reflected by leaf and efflux ethanol content. In M202, the ethanol contents in both the leaves and the surrounding water were significantly higher at day 1 (Figure 5C). At day 3, these values were similar for both genotypes. These data further demonstrate that the Sub1 haplotype influences both carbohydrate utilization and the capacity for ethanolic fermentation.
Gene Expression Responsive to Ethylene and GA Is Influenced by the Sub1 Haplotype
An increase in the production and accumulation of ethylene is essential for a variety of acclimative responses to submergence (Fukao and Bailey-Serres, 2004). The observation that the intolerant M202 showed increased leaf and internode elongation under submergence led to the hypothesis that the Sub1 haplotype might influence production or sensitivity to ethylene. To evaluate this possibility, ethylene released during submergence was measured in the two genotypes (Figure 6A). Ethylene level was enhanced fivefold to sixfold after 1 d of submergence in both genotypes. After 3 d, ethylene evolution continued, but it was 1.6-fold higher in M202 than in M202(Sub1).
Figure 6.
Ethylene Sensitivity Is Greater in Leaves of M202 Relative to M202(Sub1).
(A) Ethylene production during submergence. Ten-day-old plants were submerged in water in test tubes for up to 3 d. Ethylene gas that accumulated in the headspace of the test tube was quantified by gas chromatography. The data represent means ± se from five independent biological replicates. Asterisks indicate significant differences between the two genotypes at that time point (P < 0.05). FW, fresh weight.
(B) Analysis of Sub1 region gene transcript accumulation in response to ethylene. Fourteen-day-old plants were treated with 1 or 100 ppm ethylene for 6 h in the light. Total RNA was analyzed by semiquantitative RT-PCR using gene-specific primers for Sub1A, Sub1B, and Sub1C, as described for Figure 1B.
(C) Analysis of ExpA gene transcript accumulation in response to ethylene. Total RNA extracted from ethylene-treated leaves was analyzed by semiquantitative RT-PCR using gene-specific primers for ExpA, as described for Figure 3.
(D) Analysis of transcript levels for genes associated with carbohydrate metabolism in response to ethylene. Total RNA extracted from ethylene-treated leaves was analyzed by semiquantitative RT-PCR using gene-specific primers for α-amylases (RAmy) and sucrose synthases (Sus), as described for Figure 4.
(E) Analysis of transcript levels for genes associated with ethanolic fermentation in response to ethylene. Total RNA extracted from ethylene-treated leaves was analyzed by semiquantitative RT-PCR using gene-specific primers for Pdc and Adh, as described for Figure 5.
The level of Actin1 mRNA was used as a loading control for (B) to (E).
To discern whether Sub1A, Sub1B, or Sub1C influences the transcript accumulation of ethylene-responsive genes, the transcript levels of these ERF genes and Sub1-regulated genes were evaluated in 14-d-old plants after treatment with 1 or 100 ppm ethylene (Figures 6B to 6E). Sub1A mRNA, the gene present only in the tolerant line, was increased dramatically in M202(Sub1) in response to 1 and 100 ppm ethylene (Figure 6B). Exposure to high levels of ethylene also promoted an increase in the Sub1C transcript in both genotypes, but the transcript level was slightly higher in M202. By contrast, Sub1B transcript accumulation was not ethylene-responsive. The levels of all expansin transcripts tested increased in response to ethylene in M202 (Figure 6C). However, these genes were either nonresponsive or less responsive to ethylene in M202(Sub1), generally consistent with the haplotype-specific differential regulation of these transcripts under submergence. These results suggest that the ethylene- and submergence-responsive induction of Sub1A may suppress the expression of the expansin genes.
The ethylene response of genes related to carbohydrate catabolism was also surveyed (Figure 6D). RAmy3C and RAmy3D mRNA accumulation was responsive to ethylene at similar levels in both genotypes, whereas RAmy3E transcript abundance was not affected by ethylene. This result contrasts somewhat with the submergence-promoted increase in RAmy3D and RAmy3E, which was considerably more pronounced in the M202 genotype. These results indicate an involvement of ethylene that is not repressed by the Sub1 haplotype under aerobic conditions. Sucrose synthase gene transcripts were generally increased in response to 1 ppm ethylene, with the exception of Sus3 mRNA, which remained at the threshold of detection in M202(Sub1). Thus, ethylene treatment did not recapitulate the genotype-specific increases in Sus transcript levels under submergence. Ethylene treatment also altered Adh and Pdc transcript levels (Figure 6E). Low and high concentrations induced an obvious accumulation of Pdc1 transcript in M202 but not in M202(Sub1). Conversely, the ethylene-mediated increase in the other mRNAs associated with ethanolic fermentation, such as Pdc2, Pdc4, Adh1, and Adh2, was restrained in M202 compared with M202(Sub1). The distinct ethylene-responsiveness of the genes related to ethanolic fermentation in the two genotypes corresponded directly with expression patterns observed during submergence (Figure 5A).
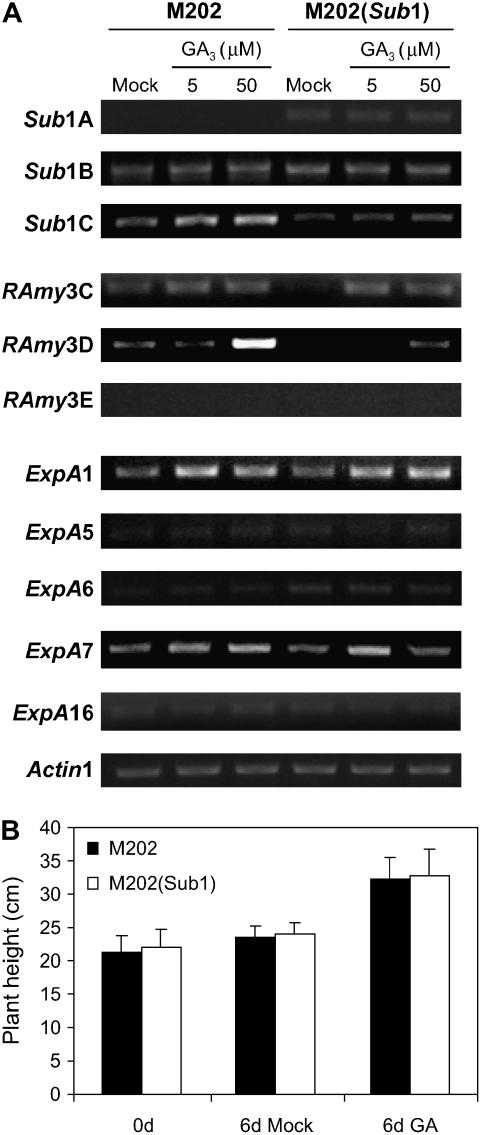
GA is also implicated in the acclimative response to submergence, particularly the elongation of internodes and petioles during submergence in deepwater rice and R. palustris, respectively (Kende et al., 1998; Peeters et al., 2002). A number of expansin genes are upregulated by GA in the internodes of deepwater rice (Cho and Kende, 1997; Lee and Kende, 2002). GA also induces the mRNA accumulation of some α-amylase genes in the embryo and aleurone layers of cereal seeds (Bethke et al., 1997; Gibson, 2004). To discern any effect of the Sub1 haplotype on the response to GA in leaves, we evaluated the expression of the three Sub1 ERFs and the GA-responsive genes associated with acclimation to submergence stress (Figure 7A). The transcript levels of Sub1A and Sub1B were unaltered by 5 and 50 μM GA3, whereas Sub1C mRNA accumulation was induced by GA treatment, specifically in M202. This GA-responsiveness correlated with higher levels of Sub1C under submergence in M202. RAmy3C mRNA, which is responsive to GA, accumulated similarly in both genotypes after GA treatment. By contrast, the induction of RAmy3D mRNA was much greater in M202 with 50 μM GA3. Of the five expansins analyzed here, only ExpA1 and ExpA7 transcripts accumulated after GA treatment, but a clear difference was not observed between the two genotypes (Figure 7A). GA-promoted elongation under normal growth conditions was similar in 14-d-old seedlings of the two genotypes (Figure 7B), consistent with the similar accumulation of ExpA transcripts in response to GA3.
Figure 7.
GA3 Sensitivity of M202 and M202(Sub1) Leaves.
(A) Analysis of transcript levels for Sub1 region, α-amylase (RAmy), and ExpA genes in response to GA3. Leaves of 14-d-old plants were treated with 5 or 50 μM GA3 for 24 h. Total RNA extracted from GA3-treated leaves was analyzed by semiquantitative RT-PCR using gene-specific primers for Sub1 genes, α-amylase, and ExpA. The level of Actin1 mRNA was used as a loading control.
(B) Response of plant growth to GA3. Five- or 14-d-old plants were treated with mock solution (0.1% [v/v] DMSO) or 100 μM GA3 solution in 0.1% (v/v) DMSO for 3 d. Plant height in the two genotypes was measured after 3 d of treatment. The data represent means ± sd from three independent biological replicates (n = 75).
DISCUSSION
The results presented here demonstrate that the Sub1 haplotype regulates diverse acclimative responses to submergence, including chlorophyll breakdown, leaf and internode elongation, and carbohydrate catabolism, as well as the induction of enzymes required for ethanolic fermentation. Nearly 100% of M202(Sub1) plants with the Sub1 haplotype from FR13A (Sub1A-1, Sub1B-1, and Sub1C-1) recovered from 14 d of submergence compared with <40% of M202 plants (Sub1B-2 and Sub1C-2). The management of carbohydrate and thereby energy economy is critical to surviving oxygen deprivation, as the consequential ATP shortfall leads to a reduction in cytosolic pH and a loss of membrane integrity (Greenway and Gibbs, 2003). In general, the amount of stored carbohydrates in plant organs is positively correlated with the level of submergence tolerance (Jackson and Ram, 2003). Consistently, starch and soluble carbohydrates were better preserved in M202(Sub1) leaves during submergence (Figures 4A and 4B). The observed distinctions in α-amylase and sucrose synthase mRNA accumulation are likely to be responsible for the repression of carbohydrate consumption in M202(Sub1) leaves (Figure 4C). The better maintenance of chlorophyll in M202(Sub1) leaves would lessen carbohydrate starvation, because photosynthesis may continue even under submerged conditions, albeit at low levels as a result of the reduced light intensity and CO2 availability (Figure 2C). Indeed, anatomical and biochemical adjustments to submergence, such as reduction of gas diffusion resistance by thinning of the cell wall and cuticle, restriction of photorespiration, and attenuation of excitation pressure on the chloroplast electron transport system in leaves, contribute to the continued photosynthesis during submergence in submergence-tolerant R. palustris (Mommer et al., 2005). The greater tolerance of the coleoptile to damage than the first and second leaves is in keeping with the observation that the coleoptile emerges from the seed of rice and Echinochloa species during germination under anaerobic conditions as a result of the ability to use starch reserves of the seed, whereas the emergence of leaves is more sensitive to the availability of oxygen (Kennedy et al., 1980).
A major function of ethanolic fermentation during submergence is to recycle NAD+ from NADH, which is required for anaerobic energy production by glycolysis (Fukao and Bailey-Serres, 2004). Paradoxically, the FR13A Sub1 haplotype limited starch and soluble carbohydrate depletion during submergence but increased PDC and ADH activities, which enhance the capacity for ethanolic fermentation (Figures 4 and 5). Interestingly, PDC and ADH activities were not positively correlated with the depletion of carbohydrate reserves or ethanol production during submergence. The amount of endogenous and effluxed ethanol was higher for M202 at day 1 of submergence, even though the enzymatic activity of ADH was lower (Figures 5C and 5D). Despite the dichotomy in gene regulation, the production of ethanol was not markedly different by day 3 of submergence in the two lines. The benefit of increased ADH might extend beyond the regeneration of NAD+. It is thought that the accumulation of acetaldehyde results in serious cellular injury during oxygen deficiency (Drew, 1997; Gibbs and Greenway, 2003). Indeed, leaves of submergence-tolerant rice varieties contained lower levels of aldehydes during submergence (Singh et al., 2001). It is possible that the more dramatic induction of ADH during submergence in M202(Sub1) is critical for the timely conversion of the toxic by-product, acetaldehyde, to neutral and diffusable ethanol.
Submergence stimulates ethylene production and accumulation within plant tissues (Banga et al., 1996; Kende et al., 1998). Ethylene evolution increased considerably in the two rice genotypes studied, with significantly greater levels in M202 than in M202(Sub1) after 3 d of submergence, indicating another role of the Sub1 haplotype (Figure 6A). Although ethylene plays a central role in adaptive responses to submergence, excessive ethylene accumulation may cause negative responses that compromise submergence tolerance. In Citrus fruit, ethylene promotes gene expression, de novo protein synthesis, and enzymatic activity of chlorophyllase (Trebitsh et al., 1993; Jacob-Wilk et al., 1999). Application of an ethylene biosynthesis/signaling inhibitor, 1-methylcyclopropene, reduced chlorophyllase gene expression, enzymatic activity, and chlorophyll breakdown in rice leaves during submergence (Ella et al., 2003). This treatment also suppressed the consumption of starch and soluble sugars during the stress and enhanced recovery. Thus, ethylene enhances the enzymatic degradation of chlorophyll and the consumption of carbohydrate reserves. The finding that the Sub1 haplotype influences ethylene production and the expression of ethylene-responsive genes strongly suggests that the Sub1 locus regulates production and sensitivity to ethylene, which finely modulates myriad acclimative responses to submergence, including leaf and internode elongation, carbohydrate consumption, and ethanolic fermentation.
The Sub1 locus is polymorphic and can encode two or three submergence-inducible ERF genes, of which all or a subset could regulate the expression of genes associated with acclimation to submergence. The accumulation of Sub1A and Sub1B transcripts was more pronounced in M202(Sub1), whereas Sub1C was more highly induced in M202, throughout the period of submergence (Figure 1B). Several lines of evidence indicate that Sub1A confers submergence tolerance (Xu et al., 2006). First, this gene is absent in all japonica and some indica varieties, all of which are submergence-intolerant. Second, indica lines carrying a Sub1A allele with a Ser-to-Pro substitution at residue 186 and showing a lower level of induction are intolerant to submergence. Finally, transformation of an intolerant japonica rice variety with a maize ubiquitin1 promoter:Sub1A-1 (Ubi1:Sub1A-1) displayed increased submergence tolerance. The demonstration here of the differential regulation of Sub1 region genes by ethylene and GA provides further evidence that Sub1A-1 is the critical determinant in submergence tolerance provided by the FR13A haplotype. The low level of expression of Sub1C-1 in response to submergence, ethylene, and GA could be attributable to distinctions in the promoter of this allele and/or negative regulation of Sub1C-1 by Sub1A-1. Sub1A and Sub1C transcripts are increased by the application of ethylene, with slightly higher induction of Sub1C mRNA in M202 (Figure 6B). The distinctions in ethylene-induced transcript levels in the two lines show that Sub1A directly or indirectly affects the activation and repression of a subset of the ethylene-responsive genes evaluated here. We favor the conclusion that SUB1A-1 negatively regulates Sub1C expression because of two independent observations. First, the level of Sub1A transcript is extremely low during submergence, but Sub1C mRNA is highly accumulated under the stress in intolerant indica lines. Second, transgenic lines that ectopically express Sub1A-1 (Ubi1:Sub1A-1) in japonica rice display limited induction of Sub1C-2 under submergence (Xu et al., 2006). These transgenic lines are significantly reduced in stature at the seedling and adult stages (Xu et al., 2006; T. Fukao and K. Xu, unpublished data). In Arabidopsis, overexpression of Arabidopsis ERF1 and tomato (Solanum lycopersicum) Pti4 also decreases plant size in a manner similar to the constitutive ethylene response mutant constitutive triple response1 and Ethylene-Insensitive3/Ethylene-Insensitive-like1–overexpressing plants (Solano et al., 1998; Gu et al., 2002; Wu et al., 2002). These observations further support the notion that SUB1A, a putative DNA binding protein, regulates ethylene-responsive gene expression.
The Sub1 haplotype also influences GA-mediated gene expression. Application of GA3 promoted the accumulation of Sub1C transcript in leaves of M202 but not in M202(Sub1) (Figure 7). GA treatment increased the levels of RAmy3D transcript only in M202. This finding indicates that in the absence of Sub1A-1, Sub1C may function to promote RAmy3D mRNA accumulation in response to GA. The transcription and transcript stability of genes associated with carbohydrate catabolism are regulated by sugar availability. Sucrose starvation prolonged the half-lives of RAmy1A, RAmy3D, and RAmy3E in rice suspension cells (Sheu et al., 1996). Application of exogenous sugars, such as glucose, fructose, and sucrose, suppresses α-amylase expression at the transcriptional and posttranscriptional levels in cereal seeds (Loreti et al., 2000; Gibson, 2004). In maize root tips, the level of hypoxia-inducible sucrose synthase transcript is repressed by glucose (Koch, 2004). Interestingly, exogenous glucose and sucrose repress GA-mediated α-amylase expression in barley embryos (Perata et al., 1997; Loreti et al., 2000). These data suggest that the alteration of carbohydrate content during submergence would affect the expression of genes responsive to sugar and GA. Distinctions in levels of α-amylase and sucrose synthase transcripts in the two genotypes during submergence could reflect significant differences in soluble carbohydrate reserves in leaves. The Sub1 haplotype might reduce carbohydrate responsiveness, because soluble carbohydrates in M202(Sub1) decrease to levels lower than those that induce α-amylase and sucrose synthase transcripts in M202 during submergence.
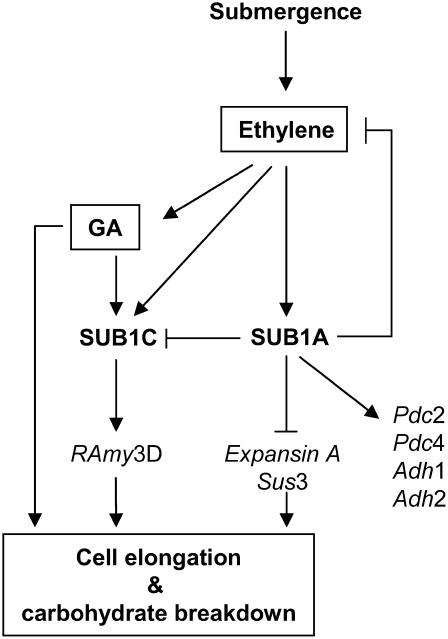
We propose a model for the ethylene- and GA-mediated transcriptional regulation of genes involved in acclimation to submergence by SUB1A and SUB1C (Figure 8). Submergence-promoted ethylene production or entrapment stimulates the accumulation of the Sub1A transcript in leaves of submergence-tolerant rice. SUB1A of tolerant indica enhances the levels of mRNAs associated with ethanolic fermentation and represses the accumulation of mRNAs responsible for cell elongation and carbohydrate catabolism. SUB1A also restricts ethylene production during submergence, possibly by feedback regulation. It has been shown that ethylene increases the endogenous GA content and sensitivity to this phytohormone in deepwater rice and R. palustris (Kende et al., 1998; Peeters et al., 2002). Although the level of Sub1C mRNA is increased by both ethylene and GA, the presence of Sub1A is correlated with limited accumulation of Sub1C mRNA. This suppression of Sub1C mRNA accumulation is also observed during submergence in M202(Sub1) and transgenic lines overexpressing Sub1A (Xu et al., 2006). It follows that through limitation of ethylene-mediated GA production and subsequent Sub1C mRNA accumulation, SUB1A represses GA-mediated responses in cell elongation and carbohydrate breakdown. The level of soluble carbohydrates in M202(Sub1) may be maintained above the threshold that promotes the transcription of genes responsive to sugar starvation, including α-amylases and sucrose synthases. Thus, the Sub1A-1 allele of tolerant indica is responsible for the repression of ethylene- and GA-mediated gene expression involved in carbohydrate consumption and cell elongation as well as the enhancement of fermentation capacity during submergence. The coordination of carbohydrate consumption and energy economy by Sub1A influences the enhancement of submergence tolerance in lowland rice. The analysis of the molecular functions of the three SUB1 proteins is under investigation.
Figure 8.
Model for Ethylene- and GA-Mediated Regulation of Gene Transcripts Associated with Acclimative Responses to Submergence by SUB1A and SUB1C.
Submergence triggers ethylene production and accumulation within plant cells, which promotes the accumulation of Sub1A and Sub1C transcripts. SUB1A of submergence-tolerant indica activates the expression of genes associated with ethanolic fermentation and represses the expression of genes involved in cell elongation and carbohydrate breakage. SUB1A also limits the production of ethylene during submergence, which restricts GA production and sensitivity. Repression of ethylene-mediated GA production and response results in the restriction of Sub1C mRNA accumulation as well as GA-dependent cell elongation and carbohydrate consumption. SUB1A also suppresses the accumulation of Sub1C transcript during submergence. A limitation of GA-dependent carbohydrate consumption by SUB1A may suppress α-amylase and sucrose synthase mRNA accumulation, because transcription of these genes is enhanced by sugar starvation. Consequently, SUB1A of submergence-tolerant indica regulates the ethylene- and GA-mediated gene expression responsible for carbohydrate consumption, cell elongation, and ethanolic fermentation and thereby confers submergence tolerance in lowland rice.
METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa) cv M202 and the Sub1 introgression line M202(Sub1) (accession number DX236-17-2-4) were kindly provided by Abdelbagi Ismail and David Mackill of the International Rice Research Institute (Xu et al., 2004). The seeds were sterilized in 1% (v/v) sodium hypochlorite for 1 h and rinsed thoroughly with deionized water. Seeds were placed on wet filter paper for 3 d at 25°C in the dark, and pregerminated seeds were transplanted into soil-containing pots and grown for 14 d in a greenhouse (30°C day, 20°C night) under natural light conditions.
Submergence, Ethylene, and GA Treatments
All submergence and hormonal treatments were replicated in at least three independent biological experiments. Light gray plastic tanks (65 × 65 × 95 cm) were filled with 90 cm of water, which was left standing for 1 d in the greenhouse (30°C day, 20°C night) before placement of the potted plants in the tank. Fourteen-day-old seedlings in soil-containing pots were completely submerged for up to 14 d. The tank water was not circulated or refreshed during the treatment. The turbidity of the tank water did not visibly increase during the submergence period. All leaves of each plant were harvested at 3 pm on the day of treatment specified, immediately frozen in liquid nitrogen, and stored at −80°C until use. Whole plant and leaf viability was evaluated after 7 d of recovery under normal growth conditions. Plants were scored as viable when one or more new leaves appeared during the recovery period. The viability of the coleoptile, first leaf, and second leaf was also recorded after recovery. Fully green (nonchlorotic) leaves were scored as viable.
Fourteen-day-old plants were treated with ethylene gas or GA3 solution before gene expression analysis. For ethylene treatment, seedlings were placed in a closed Lucite chamber filled with 1 or 100 ppm ethylene in air for 6 h in the light (50 μmol·m−2·s−1). For GA treatment, leaves from 14-d-old plants were floated on mock solution (0.05% [v/v] DMSO) or GA3 solution containing 5 or 50 μM GA3 (Sigma-Aldrich) and 0.05% (v/v) DMSO for 24 h in the light (50 μmol·m−2·s−1). After treatment, leaf samples were frozen immediately in liquid nitrogen and stored at −80°C until use.
To observe the response of plant growth to GA, sterilized seeds were incubated on wet filter paper for 5 d in the light (50 μmol·m−2·s−1) and the seedlings were transferred to mock solution (0.1% [v/v] DMSO) or 100 μM GA3 solution in 0.1% (v/v) DMSO. Fourteen-day-old seedlings (25 plants) were sprayed with mock solution or 200 mL of GA3 solution (100 μM) once per day for 3 d in the greenhouse. Plant height of each seedling (8 or 17 d old) was recorded after 3 d of treatment.
Ethylene Measurement
The relative rate of ethylene production was measured during submergence. Dehulled seeds were sterilized in 70% (v/v) ethanol for 10 min and in 2% (v/v) sodium hypochlorite for 20 min. After rinsing with sterilized deionized water, each seed was placed on 1× Murashige and Skoog medium (3 mL) in a 16-mm × 15-cm test tube at 20°C for 10 d (16 h light/8 h dark; light level, 50 μmol·m−2·s−1). For submergence stress, the test tube was filled with 18 mL of sterilized deionized water, closed with a loose plastic cap, and incubated for up to 3 d at 25°C in the light (50 μmol·m−2·s−1). Two hours before the submergence treatment was complete, the test tube was tightly closed with a rubber serum stopper, and 2 h later, the accumulated gas sample (0.9 mL) was withdrawn from the headspace of the test tube with a 1-mL syringe and analyzed using a gas chromatograph (6850 series; Hewlett Packard) equipped with an alumina-based capillary column (Agilent Technologies) (Larsen and Cancel, 2004).
Semiquantitative RT-PCR
Total RNA was extracted from 100 mg of tissue using the RNeasy plant mini kit (Qiagen). Single-stranded cDNA was synthesized from 2 μg of total RNA using SuperScript II RNase H− reverse transcriptase (Invitrogen) according to the manufacturer's protocol. RT-PCR was performed in a reaction mixture (50 μL) containing 2 μL of cDNA solution, 5 μL of 10× PCR buffer, 0.2 μM primers, 0.2 mM deoxynucleotide triphosphates, and 1.25 units of Taq DNA polymerase (Qiagen). Sequences of primer pairs, PCR conditions, and product sizes are listed in Supplemental Table 1 online. The number of cycles used for amplification with each primer pair was adjusted to be in the linear range. RT-PCR products were confirmed by DNA sequence analysis. All RT-PCR data are representative of at least three independent experiments replicated over the course of several months.
Chlorophyll Assay
Chlorophyll a/b contents were assayed as described by Porra et al. (1989). Chlorophyll was extracted from 50 mg of tissue in 1 mL of 80% (v/v) acetone containing 2.5 mM sodium phosphate buffer, pH 7.8, on ice. After centrifugation at 4°C for 20 min at 16,000g, A663.6, A646.6, and A750 were measured with a spectrophotometer (DU800; Beckman).
Carbohydrate Assay
Leaf tissue (50 mg) was homogenized in 1 mL of 80% (v/v) ethanol and incubated at 80°C for 20 min. After centrifugation at 16,000g for 10 min, the supernatant was collected in a new tube. The tissue extraction procedure was repeated twice. The three extracts were combined, dried under vacuum, and dissolved in 0.5 mL of water. Total soluble carbohydrate content was quantified by the anthrone method, with glucose as the standard (Yem and Willis, 1954). The sample solution (100 μL) was mixed with 1.0 mL of 0.14% (w/v) anthrone solution in 100% H2SO4 and incubated in boiling water for 20 min. After cooling, A620 was determined. Starch content was measured according to the method of Walters et al. (2004). The ethanol-insoluble fraction was washed with water, resuspended in 0.5 mL of water, and autoclaved at 121°C for 3 h. After cooling, the suspension was adjusted to 25 mM sodium citrate, pH 4.8, and hydrolyzed with 9 units of α-amylase and 3 units of amyloglucosidase for 16 h at 37°C. The glucose content was assayed enzymatically by the method of Guglielminetti et al. (1995a).
Ethanol Assay
Ten-day-old seedlings were submerged in test tubes for up to 3 d as described for the ethylene measurements. Ethanol content in leaves and the water used for seedling submergence was quantified by the method of Rumpho and Kennedy (1981). Leaf tissue (20 mg) was homogenized in 0.2 mL of 5 M ice-cold HClO4, and the extract was centrifuged at 4°C for 20 min at 16,000g. The supernatant (100 μL) was neutralized with 0.25 mL of 1 M K2CO3, and the precipitates were removed by centrifugation. The obtained supernatant and the water used for submergence were analyzed for ethanol in an assay mixture (1 mL) containing 100 μL of sample solution, 100 mM Tris-HCl, pH 9.0, 0.6 mM NAD+, and 40 units of ADH. The samples were incubated at 25°C for 1 h, and ethanol content was determined by measuring the reduction of NAD+ at A340. Ethanol was used as the standard.
Enzyme Activity Assay
PDC (EC 4.1.1.1) activity was assayed as described by Fukao et al. (2003). Crude protein was extracted from 50 mg of tissue in ice-cold extraction buffer (0.4 mL) containing 50 mM MEPS-NaOH, pH 6.2, 1 mM MgCl2, 5 mM DTT, and 1 mM thiamine pyrophosphate chloride. The extract was centrifuged at 4°C for 20 min at 16,000g. The reaction mixture (1 mL) contained 100 μL of extract, 50 mM MEPS-NaOH, pH 6.2, 0.5 mM MgCl2, 0.1 mM thiamine pyrophosphate chloride, 10 mM oxamate, 0.12 mM NADH, and 660 nkat of yeast ADH. To initiate the reaction, 50 μL of 400 mM pyruvate was added, and the coupled NADH oxidation was monitored at A340 at 25°C for 2 min. ADH (EC 1.1.1.1) activity was assayed by the method of Fukao et al. (2003). Leaf tissue (50 mg) was homogenized in cold extraction buffer (0.4 mL) containing 100 mM Tris-HCl, pH 9.0, 20 mM MgCl2, and 0.1% (v/v) 2-mercaptoethanol on ice. After centrifugation, the supernatant was used for the activity assay. The reaction mixture (1 mL) contained 50 μL of extract, 50 mM Tris-HCl, pH 9.0, and 1 mM NAD+. Ethanol (50 μL) was added to initiate the reaction, and NAD+ reduction was monitored at A340 at 25°C for 2 min. Protein was quantified by the method of Bradford (1976), with BSA as the standard.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ011598 (Sub1A), AP005705 (Sub1B), and AP006758 (Sub1C).
Supplemental Data
The following material is available in the online version of this article.
Supplemental Table 1. Sequences of Primers and PCR Conditions Applied for Semiquantitative RT-PCR.
Supplementary Material
Acknowledgments
We thank David Mackill, Abdelbagi Ismail, and Sigrid Heuer for valuable comments and discussion. We are also grateful to Paul Larsen for technical assistance with the ethylene treatments and measurements. This work was supported by the USDA National Research Initiative Competitive Grants Program (Grant 2002-35101-13359) and by USAID for International Development Linkage Project funds from the International Rice Research Institute to J.B.-S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Julia Bailey-Serres (serres@ucr.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.106.043000.
References
- Armstrong, W., and Drew, M.C. (2002). Root growth and metabolism under oxygen deficiency. In Plant Roots: The Hidden Half, 3rd ed. Y. Waisel, A. Eshel, and U. Kafkafi, eds (New York: Marcel Dekker), pp. 729–761.
- Banga, M., Slaa, E.J., Blom, C.W., and Voesenek, L.A. (1996). Ethylene biosynthesis and accumulation under drained and submerged conditions. Plant Physiol. 112 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter-Burrell, A., Chang, R., Springer, P., and Bailey-Serres, J. (2003). Gene and enhancer trap transposable elements reveal oxygen deprivation-regulated genes and their complex patterns of expression in Arabidopsis. Ann. Bot. (Lond.) 91 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop, J.J., Jackson, M.B., Gühl, K., Vreeburg, R.A.M., Croker, S.J., Peeters, A.J.M., and Voesenek, A.C.J. (2005). Contrasting interactions between ethylene and abscisic acid in Rumex species differing in submergence tolerance. Plant J. 44 756–768. [DOI] [PubMed] [Google Scholar]
- Bethke, P.C., Schuurink, R., and Jones, R.L. (1997). Hormonal signaling in cereal aleurone. J. Exp. Bot. 48 1337–1356. [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Cho, H.-T., and Kende, H. (1997). Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell 9 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, K.K., Sarkar, R.K., and Ismail, A.M. (2005). Elongation ability and non-structural carbohydrate levels in relation to submergence tolerance in rice. Plant Sci. 168 131–136. [Google Scholar]
- Drew, M.C. (1997). Oxygen deficiency and root metabolism: Injury and acclimation under hypoxia and anoxia. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 223–250. [DOI] [PubMed] [Google Scholar]
- Drew, M.C., He, C.J., and Morgan, P.W. (2000). Programmed cell death and aeronchyma formation in roots. Trends Plant Sci. 5 123–127. [DOI] [PubMed] [Google Scholar]
- Ella, E.S., Kawano, N., Yamauchi, Y., Tanaka, K., and Ismail, A.M. (2003). Blocking ethylene perception enhances flooding tolerance in rice seedlings. Funct. Plant Biol. 30 813–819. [DOI] [PubMed] [Google Scholar]
- Folly, P., and Engel, N. (1999). Chlorophyll b to chlorophyll a conversion precedes chlorophyll degradation in Hordeum vulgare L. J. Biol. Chem. 274 21811–21816. [DOI] [PubMed] [Google Scholar]
- Fukao, T., and Bailey-Serres, J. (2004). Plant responses to hypoxia—Is survival a balancing act? Trends Plant Sci. 9 449–456. [DOI] [PubMed] [Google Scholar]
- Fukao, T., Kennedy, R.A., Yamasue, Y., and Rumpho, M.E. (2003). Genetic and biochemical analysis of anaerobically-induced enzymes during seed germination of Echinochloa crus-galli varieties tolerant and intolerant of anoxia. J. Exp. Bot. 54 1421–1429. [DOI] [PubMed] [Google Scholar]
- Geigenberger, P. (2003). Response of plant metabolism to too little oxygen. Curr. Opin. Plant Biol. 6 223–250. [DOI] [PubMed] [Google Scholar]
- Gibbs, J., and Greenway, H. (2003). Mechanisms of anoxia tolerance in plants. I. Growth, survival and anaerobic catabolism. Funct. Plant Biol. 30 1–47. [DOI] [PubMed] [Google Scholar]
- Gibson, S.I. (2004). Sugar and phytohormone response pathways: Navigating a signaling network. J. Exp. Bot. 55 253–264. [DOI] [PubMed] [Google Scholar]
- Greenway, H., and Gibbs, J. (2003). Mechanisms of anoxia tolerance in plants. II. Energy requirements for maintenance and energy distribution to essential processes. Funct. Plant Biol. 30 999–1036. [DOI] [PubMed] [Google Scholar]
- Gu, Y.-Q., Wildermuth, M.C., Chakravarthy, S., Loh, Y.-T., Yang, C., He, X., Han, Y., and Martin, G.B. (2002). Tomato transcription factors Pti4, Pti5, and Pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 14 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielminetti, L., Perata, P., and Alpi, A. (1995. a). Effect of anoxia on carbohydrate metabolism in rice seedlings. Plant Physiol. 108 735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielminetti, L., Yamaguchi, J., Perata, P., and Alpi, A. (1995. b). Amylolytic activities in cereal seeds under aerobic and anaerobic conditions. Plant Physiol. 109 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena, A.H., Pearce, D.M., Jackson, M.B., Hawes, C.R., and Evans, D.E. (2001). Characterization of programmed cell death during aeronchyma formation induced by ethylene or hypoxia in roots of maize (Zea mays L.). Planta 212 205–214. [DOI] [PubMed] [Google Scholar]
- Gutterson, N., and Reuber, T.L. (2004). Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 7 465–471. [DOI] [PubMed] [Google Scholar]
- Hwang, Y.-S., Thomas, B.R., and Rodriguez, R.L. (1999). Differential expression of rice α-amylase genes during seedling development under anoxia. Plant Mol. Biol. 40 911–920. [DOI] [PubMed] [Google Scholar]
- Ismond, K.P., Dolferus, R., de Pauw, M., Dennis, E.S., and Good, A.G. (2003). Enhanced low oxygen survival in Arabidopsis through increased metabolic flux in the fermentative pathway. Plant Physiol. 132 1292–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, M.B., and Ram, P.C. (2003). Physiological and molecular basis of susceptibility and tolerance of rice plants to complete submergence. Ann. Bot. (Lond.) 91 227–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob-Wilk, D., Holland, D., Goldschmidt, E.E., Riov, J., and Eyal, Y. (1999). Chlorophyll breakdown by chlorophyllase: Isolation and functional expression of the Chlase1 gene from ethylene-treated Citrus fruit and its regulation during development. Plant J. 20 653–661. [DOI] [PubMed] [Google Scholar]
- Kende, H., van der Knaap, E., and Cho, H.-T. (1998). Deepwater rice: A model plant to study stem elongation. Plant Physiol. 118 1105–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, R.A., Barrett, S.C., Vander Zee, D., and Rumpho, M. (1980). Germination and seedling growth under anaerobic conditions in Echinochloa crus-galli (barnyard grass). Plant Cell Environ. 3 243–248. [Google Scholar]
- Koch, K. (2004). Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 7 235–246. [DOI] [PubMed] [Google Scholar]
- Kürsteiner, O., Dupuis, I., and Kuhlemeier, C. (2003). The pyruvate decarboxylase1 gene of Arabidopsis is required during anoxia but not other environmental stresses. Plant Physiol. 132 968–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, P.B., and Cancel, J.D. (2004). A recessive mutation in the RUB1-conjugating enzyme, RCE1, reveals a requirement for RUB modification for control of ethylene biosynthesis and proper induction of basic chitinase and PDF1.2 in Arabidopsis. Plant J. 38 626–638. [DOI] [PubMed] [Google Scholar]
- Lee, Y., and Kende, H. (2002). Expression of α-expansin and expansin-like genes in deepwater rice. Plant Physiol. 130 1396–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Jones, L., and McQueen-Mason, S. (2003). Expansins and cell growth. Curr. Opin. Plant Biol. 6 603–610. [DOI] [PubMed] [Google Scholar]
- Loreti, E., Alpi, A., and Perata, P. (2000). Glucose and disaccharide-sensing mechanisms modulate the expression of α-amylase in barley embryos. Plant Physiol. 123 939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergemann, H., and Sauter, M. (2000). Ethylene induces epidermal cell death at the site of adventitious root emergence in rice. Plant Physiol. 124 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mommer, L., Pedersen, O., and Visser, E.J.W. (2004). Acclimation of a terrestrial plant to submergence facilitates gas exchange under water. Plant Cell Environ. 27 1281–1287. [Google Scholar]
- Mommer, L., Pons, T.L., Wolters-Arts, M., Venema, J.H., and Visser, E.J.W. (2005). Submergence-induced morphological, anatomical, and biochemical responses in a terrestrial species affect gas diffusion resistance and photosynthetic performance. Plant Physiol. 139 497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi, S., Subudhi, P.K., Senadhira, D., Manigbas, N.L., Sen-Mandi, S., and Huang, N. (1997). Mapping QTL for submergence tolerance in rice by AFLP analysis and selective genotyping. Mol. Gen. Genet. 255 1–8. [DOI] [PubMed] [Google Scholar]
- Peeters, A.J., Cox, M.C., Benschop, J.J., Vreeburg, R.A., Bou, J., and Voesenek, A.C. (2002). Submergence research using Rumex palustris as a model: Looking back and going forward. J. Exp. Bot. 53 391–398. [DOI] [PubMed] [Google Scholar]
- Peng, H.-P., Chan, C.-S., Shih, M.-C., and Yang, S.F. (2001). Signaling events in the hypoxic induction of alcohol dehydrogenase in Arabidopsis. Plant Physiol. 126 742–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perata, P., Matsukura, C., Vernieri, P., and Yamaguchi, J. (1997). Sugar repression of a gibberellin-dependent signaling pathway in barley embryos. Plant Cell 9 2197–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra, R.J., Thompson, W.A., and Kriedemann, P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975 384–394. [Google Scholar]
- Rahman, M., Grover, A., Peacock, W.J., Dennis, E.S., and Ellis, M.H. (2001). Effects of manipulation of pyruvate decarboxylase and alcohol dehydrogenase levels on the submergence tolerance of rice. Aust. J. Plant Physiol. 28 1231–1241. [Google Scholar]
- Rumpho, M.E., and Kennedy, R.A. (1981). Anaerobic metabolism in germinating seeds of Echinochloa crus-galli (barnyard grass): Metabolite and enzyme studies. Plant Physiol. 68 165–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, D. (1969). An example of gene fixation resulting from selective advantage in suboptimal conditions. Am. Nat. 103 479–481. [Google Scholar]
- Sheu, J.-J., Yu, T.-S., Tong, W.-F., and Yu, S.-M. (1996). Carbohydrate starvation stimulates differential expression of rice α-amylase genes that is modulated through complicated transcriptional and posttranscriptional processes. J. Biol. Chem. 271 26998–27004. [DOI] [PubMed] [Google Scholar]
- Singh, H.P., Singh, B.B., and Ram, P.C. (2001). Submergence tolerance of rainfed lowland rice: Search for physiological marker traits. J. Plant Physiol. 158 883–889. [Google Scholar]
- Solano, R., Stepanova, A., Chao, Q., and Ecker, J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSIVE-FACTOR1. Genes Dev. 12 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripongpangkul, K., Posa, G.B., Senadhira, D.W., Brar, D., Huang, N., Khush, G.S., and Li, Z.K. (2000). Genes/QTLs affecting flood tolerance in rice. Theor. Appl. Genet. 101 1074–1081. [Google Scholar]
- Toojinda, T., Siangliw, M., Tragoonrung, S., and Vanavichit, A. (2003). Molecular genetics of submergence tolerance in rice: QTL analysis of key traits. Ann. Bot. (Lond.) 91 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebitsh, T., Goldschmidt, E.E., and Riov, J. (1993). Ethylene induces de novo synthesis of chlorophyllase, a chlorophyll degrading enzyme, in Citrus fruit peel. Proc. Natl. Acad. Sci. USA 90 9441–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreeburg, R.A., Benschop, J.J., Peeters, A.J., Colmer, T.D., Ammerlaan, A.H., Staal, M., Elzenga, T.M., Staals, R.H., Darley, C.P., McQueen-Mason, S.J., and Voesenek, L.A. (2005). Ethylene regulates fast apoplastic acidification and expansin A transcription during submergence-induced petiole elongation in Rumex palustris. Plant J. 43 597–610. [DOI] [PubMed] [Google Scholar]
- Walters, R.G., Ibrahim, D.G., Horton, P., and Kruger, N.J. (2004). A mutant of Arabidopsis lacking the triose-phosphate/phosphate translocator reveals metabolic regulation of starch breakdown in the light. Plant Physiol. 135 891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K., Tian, L., Hollingworth, J., Brown, D.C., and Miki, B. (2002). Functional analysis of tomato Pti4 in Arabidopsis. Plant Physiol. 128 30–37. [PMC free article] [PubMed] [Google Scholar]
- Xu, K., Deb, R., and Mackill, D.J. (2004). A microsatellite marker and a codominant PCR-based marker for marker-assisted selection of submergence tolerance in rice. Crop Sci. 44 248–253. [Google Scholar]
- Xu, K., and Mackill, D.J. (1996). A major locus for submergence tolerance mapped on rice chromosome 9. Mol. Breed. 2 219–224. [Google Scholar]
- Xu, K., Xu, X., Fukao, T., Canlas, P., Maghirang-Rodriguez, R., Heuer, S., Ismail, A.M., Bailey-Serres, J., Ronald, P.C., and Mackill, D.J. (2006). Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature, in press. [DOI] [PubMed]
- Xu, K., Xu, X., Ronald, P.C., and Mackill, D.J. (2000). A high-resolution linkage map of the vicinity of the rice submergence tolerance locus Sub1. Mol. Gen. Genet. 263 681–689. [DOI] [PubMed] [Google Scholar]
- Yem, E.W., and Willis, A.J. (1954). The estimation of carbohydrates in plant extracts by anthrone. Biochem. J. 57 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Y., Wu, Y., Avigne, W.T., and Koch, K.E. (1999). Rapid repression of maize invertases by low oxygen. Invertase/sucrose synthase balance, sugar signaling potential, and seedling survival. Plant Physiol. 121 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.