Abstract
The floral homeotic APETALA3 (AP3) gene in Arabidopsis thaliana encodes a MADS box transcription factor required for specifying petal and stamen identities. AP3 is a member of the euAP3 lineage, which arose by gene duplication coincident with radiation of the core eudicots. Although Arabidopsis lacks genes in the paralogous Tomato MADS box gene 6 (TM6) lineage, tomato (Solanum lycopersicum) possesses both euAP3 and TM6 genes, which have functionally diversified. A loss-of-function mutation in Tomato AP3 (TAP3) resulted in homeotic transformations of both petals and stamens, whereas RNA interference–induced reduction in TM6 function resulted in flowers with homeotic defects primarily in stamens. The functional differences between these genes can be ascribed partly to different expression domains. When overexpressed in an equivalent domain, both genes can partially rescue the tap3 mutant, indicating that relative levels as well as spatial patterns of expression contribute to functional differences. Our results also indicate that the two proteins have differing biochemical capabilities. Together, these results suggest that TM6 and TAP3 play qualitatively different roles in floral development; they also support the ideas that the ancestral role of AP3 lineage genes was in specifying stamen development and that duplication and divergence in the AP3 lineage allowed for the acquisition of a role in petal specification in the core eudicots.
INTRODUCTION
Gene duplication can provide the raw material for the evolution of new functions. This can occur by subfunctionalization (partitioning of the original gene function into two parts) or by neofunctionalization (acquisition of a new role by one of the duplicates), and such changes are thought to contribute to the retention of both duplicated genes in the genome (Ohno, 1970; Force et al., 1999; Lynch and Conery, 2000; Lynch and Force, 2000). Traditionally, researchers have examined the adaptive significance of the retention of duplicate gene pairs by characterizing patterns of nucleotide substitutions within coding regions (Lynch and Conery, 2000). By contrast, relatively little work has been performed to assess the functional consequences of gene duplication and diversification. Here, we explore the degree to which coding versus regulatory changes have contributed to differences in gene function in duplicate genes belonging to the floral homeotic APETALA3 (AP3) lineage.
The AP3 lineage genes are a subfamily of the large MADS box family of transcription factors, which have been implicated in the regulation of a number of plant developmental processes (Irish and Kramer, 1998; Theissen et al., 2000). A major gene duplication event in the AP3 gene lineage has led to two paralogous lineages, the euAP3 and the Tomato MADS box gene 6 (TM6) gene lineages, in the core eudicot clade of the angiosperms (Kramer et al., 1998; Kramer and Irish, 1999), which suggests that this gene duplication occurred ∼125 million years ago (Magallon et al., 1999). The core eudicot TM6 lineage contains sequence motifs similar to those of the ancestral paleoAP3 lineage genes, whereas the euAP3 lineage genes possess a set of different sequence motifs and so represent a divergent paralogous lineage (Kramer et al., 1998). In particular, the TM6 and euAP3 lineage gene products are distinguished by different C-terminal domains, which likely arose by an ancestral frameshift mutation (Kramer et al., 1998, 2006; Vandenbussche et al., 2003).
Within the core eudicots, all functionally characterized AP3 genes belong to the divergent euAP3 lineage. These include the well-characterized Arabidopsis thaliana AP3 and Antirrhinum majus DEFICIENS (DEF) genes, both of which have been shown to play key roles in the specification of petal and stamen identities (Bowman et al., 1989; Carpenter and Coen, 1990; Sommer et al., 1990; Jack et al., 1994). Consistent with this role, both AP3 and DEF are expressed in petal and stamen primordia, and their expression in these organs persists through later stages of differentiation. However, there are some differences in the patterns of expression of these genes in their respective species. For instance, Antirrhinum DEF is also transiently expressed in the carpel primordia, whereas Arabidopsis AP3 is additionally expressed in a small adaxial patch of sepal cells (Jack et al., 1992; Schwarz-Sommer et al., 1992).
Members of the PISTILLATA (PI) subfamily of MADS box genes have also been shown to be required for petal and stamen specification (Bowman et al., 1989; Trobner et al., 1992; Goto and Meyerowitz, 1994). In Arabidopsis, the PI and AP3 proteins heterodimerize, which appears to be necessary for DNA binding and stable localization of this transcription factor complex to the nucleus (McGonigle et al., 1996; Riechmann et al., 1996a, 1996b). Furthermore, the AP3/PI heterodimeric complex has been shown to be responsible for the continued expression of both AP3 and PI through a positive feedback loop (Jack et al., 1992, 1994; Goto and Meyerowitz, 1994; Honma and Goto, 2000). This transcriptional complex likely includes other proteins, because AP3 and PI also physically interact with the MADS box proteins AP1 and SEP3 (Honma and Goto, 2001). The formation of distinct MADS box protein complexes in different floral organ whorls has been postulated to be responsible for organ-type specific differentiation (Honma and Goto, 2001; Pelaz et al., 2001; Theissen and Saedler, 2001). Similarly, the Antirrhinum PI lineage gene product, GLOBOSA (GLO), must heterodimerize with DEF to bind to DNA (Schwarz-Sommer et al., 1992; Trobner et al., 1992) and also forms larger MADS box protein complexes that are thought to specify organ-type differentiation (Davies et al., 1996b). However, functional analyses of the two Petunia hybrida PI ortholog genes, FBP1 (also known as Ph GLO1) and pMADS2 (Ph GLO2), as well as protein interaction data suggest that in Petunia, petal and stamen identities are specified by protein complexes that are qualitatively distinct from their Arabidopsis counterparts and whose abundance is believed to be critical for proper organ development (Vandenbussche et al., 2004). These results suggest that there is some plasticity to the types of MADS box protein complexes formed and their functions in planta.
In contrast with the euAP3 lineage, functional characterization of TM6 lineage genes has not yet been performed in any species. Several lines of evidence have suggested that TM6 lineage genes may play a role in stamen specification and/or differentiation. In Petunia, loss-of-function mutations in the euAP3 gene Ph DEF (also known as GP and pMADS1) only affect the specification of petals (van der Krol et al., 1993; Halfter et al., 1994; Tsuchimoto et al., 2000; Vandenbussche et al., 2004). Stamen identity is thought to be dependent on the action of another MADS box gene (Tsuchimoto et al., 2000), a likely candidate being the TM6 representative Ph TM6, because it is highly expressed in the third and fourth whorls (Vandenbussche et al., 2004). It has also been shown that a chimeric cDNA, containing a C-terminal paleoAP3 motif characteristic of paleoAP3 or TM6 lineage genes, fused in frame to an Arabidopsis AP3 cDNA, is sufficient to largely rescue stamen but not petal development of Arabidopsis ap3 mutant flowers (Lamb and Irish, 2003). Together, these data suggest that the core eudicot TM6 lineage genes function predominantly in stamen development, although this has not yet been critically tested. Based on these observations, we previously suggested that the advent of the divergent euAP3 lineage genes may be correlated with the de novo evolution of petals in the core eudicots (Kramer and Irish, 1999, 2000; Irish, 2003).
Here, we characterize the functions of the tomato (Solanum lycopersicum) TM6 (TM6) and euAP3 (Tomato APETALA3 [TAP3]) genes to parse the relative contributions of these gene duplicates to specifying petal and stamen development in this species. We describe loss-of-function phenotypes for TM6 and TAP3 that indicate that these tomato genes play distinct roles in flower development, with TAP3 required to specify both petal and stamen identity, whereas TM6 appears to play a role predominantly in stamen differentiation. The data we present indicate that both changes in expression and changes in protein coding functions have likely contributed to the differences in the roles of TM6 and TAP3. These results also support the idea that the divergent euAP3 lineage genes have been redeployed to a role in petal specification in the core eudicots. Surprisingly, similar analyses of the TM6 and euAP3 lineage genes in the closely related species P. hybrida indicate that these paralogous genes have subfunctionalized their roles in a different way (van der Krol et al., 1993; Tsuchimoto et al., 2000; Rijpkema et al., 2006). Together, these data demonstrate that paralogous gene duplicates can take on different roles in different lineages, illustrating the dynamic and fluid nature of subfunctionalization.
RESULTS
Identification and Phylogenetic Analyses of AP3 Lineage Genes in Tomato
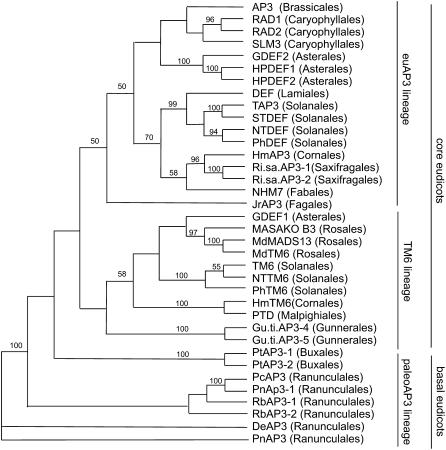
Using rapid amplification of cDNA ends (RACE), we identified the full-length coding sequence of TAP3; a partial TAP3 sequence and the coding sequence of TM6 have been published previously (Pnueli et al., 1991; Kramer et al., 1998). To place these sequences in a phylogenetic context, we performed phylogenetic analyses using neighbor joining of eudicot AP3 lineage genes and the predicted amino acid sequences of the MIK domains (Figure 1). These results confirm the placement of the tomato TM6 gene in the TM6 clade and tomato TAP3 in the euAP3 clade (Kramer et al., 1998; Kramer and Irish, 2000; Vandenbussche et al., 2003). TM6 is most closely related to TM6 representatives from other Solanales species, including P. hybrida TM6 and Nicotiana tabacum TM6, within a larger clade of core eudicot TM6 genes. Similarly, TAP3 groups together with euAP3 representatives from other Solanales species within the core eudicot euAP3 clade. These observations support the hypothesis that the duplication resulting in the TM6 and euAP3 lineages occurred coincidentally with the diversification of the core eudicots (Kramer et al., 1998; Kramer and Irish, 2000; Vandenbussche et al., 2003; Kim et al., 2004; Stellari et al., 2004). Furthermore, examination of the extant EST collection for tomato (http://www.sgn.cornell.edu/index.pl) does not reveal any other AP3 lineage genes, nor have we identified any other AP3-related genes in tomato via degenerate RT-PCR or by DNA gel blot hybridization analyses (data not shown). As such, it appears that tomato possesses a single euAP3 lineage gene, TAP3, and a single TM6 lineage gene, TM6.
Figure 1.
Neighbor-Joining Analysis of euAP3 and TM6 Lineage Genes.
Representative AP3 lineage genes from core eudicots and basal eudicots were included in the analysis; the basal eudicot Pn AP3 gene was used as the outgroup. Bootstrap values of ≥50% are shown.
Identification of Loss-of-Function Lines for TAP3 and TM6
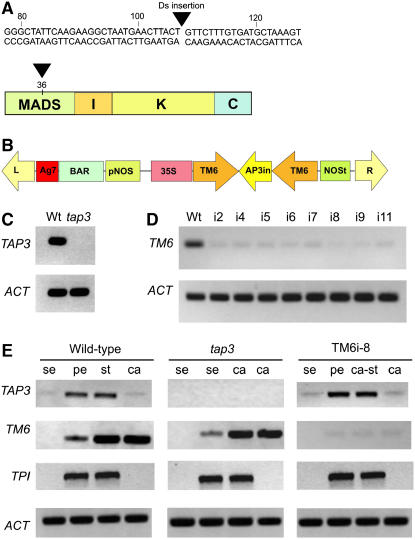
To characterize the function of AP3 lineage genes in tomato, we screened Ds insertion lines (Meissner et al., 1997, 2000) for phenotypes likely to reflect a mutation in an AP3 lineage gene. One such mutation was identified and shown to correspond to an insertion in the first exon of the TAP3 gene (Figure 2A). As expected, the expression of the TAP3 gene in the insertional mutant was completely abolished (Figure 2C). Because we did not recover any insertional mutations in the TM6 gene, we resorted to using an RNA interference (RNAi) approach to generate a loss-of-function phenotype. An RNAi construct was designed to target the last 200 nucleotides of the TM6 transcript (Figure 2B). Eight loss-of-function TM6i transgenic lines were generated, and TM6 expression was strongly reduced in all eight lines (Figure 2D). The RNAi-induced gene silencing of TM6 was gene-specific, in that transcript levels of the closest paralog, TAP3, were unaffected (Figure 2E).
Figure 2.
Characterization of Loss-of-Function Lines for TAP3 and TM6.
(A) Location of the insertion of a Ds element in the first exon of TAP3 at position +108 within the MADS domain.
(B) pTCSH1-TM6 RNAi construct used for RNAi-induced silencing of the TM6 gene.
(C) RT-PCR using TAP3-specific primers on wild-type and tap3 inflorescence tissue (stages 9 to 18); no amplification could be detected in the tap3 tissue. Amplification of the tomato ACTIN gene (ACT) was used as a control.
(D) RT-PCR using TM6-specific primers on wild-type and TM6i lines. Flowers from stages 15 to 20 showing a strong phenotype were analyzed for all transgenic lines. As expected, the level of the TM6 transcript was highly reduced in the transgenic lines.
(E) RT-PCR analyses to determine the organ-specific expression of TAP3, TM6, and TPI genes in wild-type, tap3 mutant, and TM6i8 transgenic lines. Floral organs were dissected from stage 9 to 18 flowers and pooled, and the resulting RNA was used for RT-PCR. se, sepals; pe, petals; st, stamens; ca, carpels; ca-st, carpelloid stamens.
tap3 and TM6i Loss-of-Function Lines Affect Different Aspects of Flower Development
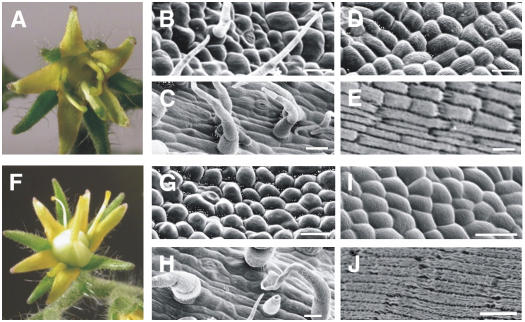
Normal tomato flowers (cv Micro-Tom) contain five or six sepals, alternating with five to six yellow petals; the reproductive organs consist of five yellow stamens forming a cone, which enclose two fused carpels that develop a multilocular ovary and a protruding style and stigma (Figure 3A). Each organ type displays characteristic epidermal cell types (Figures 3B to 3I). Wild-type sepals contain stomata and trichomes on the adaxial surface (Figure 3B). In the second whorl, the adaxial epidermis of the petals contains rounded cells, whereas the abaxial surface contains epidermal cells that are more elongate as well as sparse trichomes (Figures 3C and 3D). In the third whorl, a row of lateral and adaxial trichome hairs present on adjacent stamens interweave to form the staminal cone (Sekhar and Sawhney, 1984) (Figure 3E). The proximal regions of the anthers contain multilobed epidermal cells, whereas the epidermal cells are more elongated in the distal regions (Figures 3F and 3G). In the carpels, the epidermal cells are rounded in the proximal region and elongated in the distal region (Figures 3H and 3I).
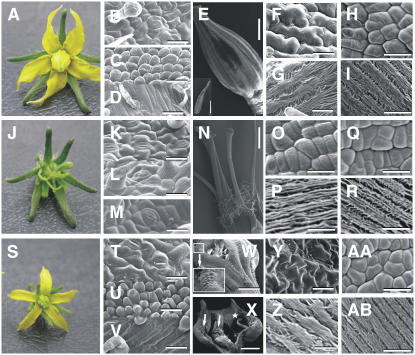
Figure 3.
Phenotypic Analyses of the tap3 Mutant and the TM6i-8 Line.
Light and scanning electron microscopy of wild-type (Micro-Tom) flowers ([A] to [I]), tap3 flowers ([J] to [R]), and TM6i-8 flowers ([S] to [AB]).
(B), (K), and (T) Adaxial epidermal cells of the first whorl organs.
(C), (L), and (U) Adaxial epidermal cells of the second whorl organs.
(D), (M), and (V) Abaxial epidermal cells of the second whorl organs.
(E) Wild-type staminal cone. The inset shows the adaxial surface of the anther.
(N) tap3 third whorl organs showing carpel-like morphology.
(W) Abaxial view of a TM6i-8 staminal cone showing partial lateral fusion (inset).
(X) Adaxial view of a TM6i-8 carpelloid stamen showing naked ovules (arrows) and stigmatic tissue (star).
(F), (O), and (Y) Abaxial epidermal cells of the proximal region of the third whorl organs.
(G), (P), and (Z) Abaxial epidermal cells of the distal region of the third whorl organs.
(H), (Q), and (AA) Epidermal cells of the proximal region of the fourth whorl organs.
(I), (R), and (AB) Epidermal cells of the distal region of the fourth whorl organs.
Bars = 40 μm ([B], [C], [G] to [I], [K] to [M], [O] to [R], [T], [U], [Z], [AA], and [AB]), 90 μm ([D], [F], and [V]), 20 μm (Y), 2 mm (inset in [E] and [N]), 1 mm ([W] and [X]), and 1.5 mm (E).
In contrast with the wild type, tap3 homozygous mutant plants develop flowers showing a classic B-class gene loss-of-function phenotype consisting of a complete transformation of the petals into sepalloid structures and stamens into carpel-like organs (Figure 3J). Scanning electron microscopy confirmed the homeotic conversion of the epidermal cells of these organs (Figures 3L, 3M, 3O, and 3P). In the second whorl, the epidermal cells had a sepal-like epidermal morphology, and stomata were observed on both the abaxial and adaxial surfaces (Figures 3L and 3M). The third whorl organs, instead of forming a cone as in the wild type, were splayed out and appeared carpelloid (Figure 3N); the third whorl carpel-like epidermal cells were indistinguishable from those of normal fourth whorl wild-type carpels (Figures 3O and 3P). The third whorl carpelloid structures produced ectopic fruit lobes that did not contain normal locules and lacked seeds (data not shown). The first and fourth whorl organs of tap3 mutant flowers appeared to be normal (Figures 3K, 3Q, and 3R). Dissection of the tap3 fourth whorl carpels revealed the presence of normal ovules that were able to set seeds when manually pollinated with wild-type pollen (data not shown).
The phenotypes produced by the TM6i5, TM6i6, TM6i8, and TM6i9 lines were characterized in detail (Tables 1 and 2, Figures 3S to 3AB). The TM6i8 and TM6i9 lines showed a more extreme phenotype in which 91 to 96% of flowers developed carpelloid stamens (Table 1, Figure 3S). The epidermal cells of these transgenic third whorl organs were smaller and arranged differently from the interlocking arrangement of comparable wild-type cells (cf. Figures 3F and 3Y). Furthermore, cells with an aberrant morphology developed in the epidermis of the proximal region of these third whorl organs (Figure 3W, inset). Third whorl organ fusion was often incomplete, as a result of the absence of interweaving lateral hairs in the proximal region (Figure 3W). Often, naked ovules developed on the adaxial face of these third whorl organs (Figure 3X). Transgenic TM6i flowers with an extreme transformation of stamens to carpelloid tissue were male-sterile, but seeds developed normally when such flowers were manually crossed with wild-type pollen. This observation indicates that the ovules of such transgenic plants were functional. When not pollinated, sterile parthenocarpic fruits developed.
Table 1.
Quantification of Third Whorl Floral Phenotypes of Four TM6i Lines
| Lines | Class 0 | Class 1 | Class 2 | Class 3 |
|---|---|---|---|---|
| Wild type (n = 200) | 99.9% | 0.1% | 0% | 0% |
| TM6i5 (n = 372) | 24.0% | 47.0% | 25% | 4% |
| TM6i6 (n = 418) | 21.0% | 28.0% | 38% | 13% |
| TM6i8 (n = 474) | 9.0% | 5.0% | 10% | 76% |
| TM6i9 (n = 450) | 4.0% | 7.0% | 22% | 67% |
Phenotypes were grouped in four classes: class 0 consists of flowers with normal phenotype; class 1 consists of flowers with only one stamen displaying carpelloidy; class 2 consists of flowers with two to four carpelloid stamens; and class 3 consists of flowers with all carpelloid stamens.
Table 2.
Petal Size Quantification of the TM6 RNAi Line Floral Classes
| pl = 1.07 ± 0.09 | pl = 0.85 ± 0.07 | pl = 0.52 ± 0.04 | |
|---|---|---|---|
| Class | pw = 0.40 ± 0.05 | pw = 0.36 ± 0.02 | pw = 0.21 ± 0.02 |
| Class 0 | 97% | 3% | 0% |
| (n = 43) | |||
| Class 1 | 95% | 5% | 0% |
| (n = 38) | |||
| Class 2 | 15% | 58% | 27% |
| (n = 45) | |||
| Class 3 | 0% | 13% | 87% |
| (n = 45) |
Correlation of petal size with the degree of stamen transformation in the TM6i flowers. pl, mean petal length ± se; pw, mean petal width ± se. Petal length and width (in centimeters) were measured for each class; the floral classes are defined in Table 1.
Petals of the transgenic TM6i plants were also affected in terms of overall size (Table 2). The reduction in petal size was probably caused by a decrease in cell proliferation, because similar cell sizes were observed in these lines compared with the wild type (cf. Figures 3C and 3U). Furthermore, the second whorl organs of TM6i lines did not display any apparent homeotic conversions, in that they displayed normal epidermal cell types across the petal (Figures 3U and 3V).
Despite the high levels of expression of the TM6 gene in ovules and during fruit development in wild-type flowers (see below), reduction in its expression did not seem to interfere with the normal differentiation of fourth whorl organs. Transgenic TM6i carpel tissue appeared normal, as did the sepals (Figures 3T, 3AA, and 3AB).
TAP3 and TM6 Are Expressed in Distinct but Overlapping Domains in the Flower
To determine whether the differences in TM6 and TAP3 function could be ascribed to different domains of expression, we examined their spatial and temporal patterns of expression. Floral organs were dissected from flowers at stages 9 to 18 of development (tomato flower stages according to Brukhin et al. [2003]), and the individual organ types were pooled and used in RT-PCR experiments (Figure 2E). TAP3 expression could be detected at high levels in petals and stamens. TM6, by contrast, was expressed predominantly in stamens and carpels, with some expression detected in petals. This pattern of TM6 expression is similar to what has been reported previously (Pnueli et al., 1991; Lozano et al., 1998; Busi et al., 2003). We also compared these patterns with the expression pattern of the Tomato PISTILLATA gene (TPI), which we identified using an RT-PCR–based approach (see Methods). TPI transcripts were detected almost exclusively in the petals and stamens.
To further characterize these patterns of expression at early stages of floral development (before stage 9), we performed in situ hybridizations for TAP3, TM6, and TPI (Figure 4). TAP3 expression was first seen in presumptive petal primordia (Figure 4A), and by stage 3 it could be detected throughout the petal and in the subepidermal cells of stamen primordia (Figure 4B). Expression of TAP3 in petal and stamen primordia persisted until later stages of floral development (Figure 4C), and by stage 9 (Figure 4D) it was restricted to particular tissues in the differentiating petals and stamens. In stage 9 stamens, TAP3 expression was largely confined to the vascular bundle and tapetal cells of the stamen and to the lateral edges of the petals. The low levels of TAP3 expression in sepals and carpels detected by RT-PCR (Figure 2E) were not observed by in situ hybridization, presumably because of the low levels of expression in these tissues or the different stages of flower development used in the two analyses. The pattern of TAP3 expression contrasts with what we observed for TM6 expression. At stage 2, TM6 expression was more ubiquitous, throughout the petal, stamen, and carpel primordia (Figure 4E). This pattern of expression persisted through stage 3 (Figure 4F) and stage 5 (Figure 4G). By stage 9, TM6 expression was most prominent in the inner integuments of the developing ovules and also could be detected in the lateral margins of the developing petals (Figure 4H). As such, the patterns of expression of TAP3 and TM6 overlap considerably at the earliest stages of floral development but become restricted to largely distinct spatial domains within the developing flower bud by stage 9. Later in floral development (in stage 9 to 18 flowers), TM6 expression appears to become more prominent in the stamens and carpels (Figure 2E).
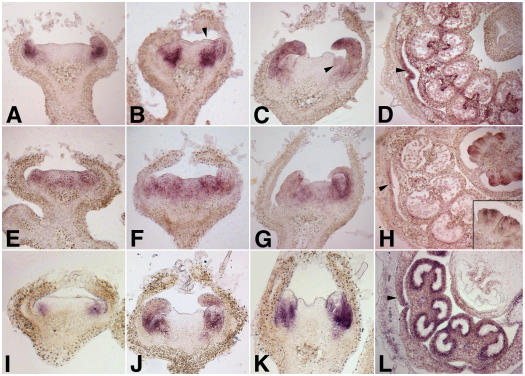
Figure 4.
In Situ Expression Analyses of TAP3, TM6, and TPI.
Expression in wild type (Micro-Tom) flower buds of TAP3 ([A] to [D]), TM6 ([E] to [H]), and TPI ([I] to [L]).
(A) TAP3 expression is apparent by stage 2, with expression seen predominantly in the petal primordia.
(B) By stage 3, expression can be seen throughout the petal primordia as well as in the subepidermal cell layers of the incipient stamen primordia (arrowhead).
(C) TAP3 expression in the petal and stamen primordia (arrowhead) at stage 5.
(D) Cross section of a flower at stage 9. TAP3 expression becomes restricted to the vascular bundle and tapetal cells of the stamens, whereas expression in the petals is seen in the lateral edges (arrowhead).
(E) Relatively low levels of TM6 expression can be seen at stage 2 throughout the presumptive petal, stamen, and carpel primordia.
(F) and (G) This pattern of expression persists through stage 3 (F) and stage 5 (G).
(H) By stage 9, TM6 expression is most prominent in the developing ovules, localized predominantly to the inner integuments (inset). Low levels of expression can also be detected in the lateral margins of the petals (arrowhead).
(I) TPI expression can be detected by early stage 2 in the presumptive petal primordia.
(J) and (K) TPI expression is observed in developing petal and stamen primordia at stage 4 (J) and stage 5 (K).
(L) By stage 9, TPI expression is seen mainly in the stamens, with high levels of expression in the tapetal cells. TPI expression can also be detected in the lateral margins of the petals (arrowhead).
By contrast, the pattern of expression of TPI was more similar to that of TAP3. TPI expression was similar to that of TAP3 at stage 2, in the presumptive petal primordia (Figure 4I). TPI expression could be detected in developing petal and stamen primordia at stages 4 (Figure 4J) and 5 (Figure 4K). By stage 9, TPI expression was restricted to particular tissues of the developing petals and stamens, with expression seen in the lateral edges of the petals and at high levels in the tapetal cells of the stamen (Figure 4L).
TAP3, Le TM6, and TPI Regulatory Interactions
To determine whether the tap3 mutation resulted in coordinate downregulation of either the TM6 or TPI gene, we examined the expression of these genes in the tap3 mutant background (Figure 2E). The levels and patterns of expression of both TM6 and TPI transcripts in the tap3 mutant flowers were similar to those of wild-type flowers, indicating that TAP3 is not required for the expression of these genes. This is in contrast with the situation in Arabidopsis, in which AP3 is required for the maintenance of PI gene expression (Goto and Meyerowitz, 1994; Honma and Goto, 2000). Similarly, downregulation of TM6 expression in the TM6i8 line had no effect on the expression levels or patterns of TAP3 or TPI (Figure 2E). These results demonstrate that the dramatic loss-of-function phenotype produced by the tap3 mutant is not attributable to coordinate downregulation of TM6 and that expression of the endogenous TM6 gene is not able to compensate for the loss of TAP3 function in petal and stamen development. Similarly, the phenotypes we observed for RNAi-induced loss of TM6 function are not the result of the coordinate loss of either TAP3 or TPI expression. Thus, TAP3 and TM6 have distinct functions in flower development.
TM6 and TAP3 Play Similar Roles When Ectopically Expressed
The complete loss of petals and stamens in the tap3 mutant suggests that the endogenous level of TM6 expression is not sufficient to compensate for the loss of TAP3 function. In wild-type flowers, in fact, TM6 appears to be expressed at a lower level than TAP3, based on RNA gel blot and in situ hybridization analyses (Figure 4; data not shown). To test whether levels or domains of expression were critical for differences in TM6 and TAP3 function, we overexpressed the TAP3 and TM6 genes in the tap3 mutant. We generated transgenic lines in which the AP3 or TM6 coding region was driven by the strong constitutive 35S promoter (Benfey and Chua, 1990) and introduced these constructs into a leap3 homozygous mutant background. Two transgenic lines were obtained for each construct. Both 35S:TAP3 and 35S:TM6 were able to rescue the tap3 second whorl phenotype to somewhat different degrees (Figures 5B, 5C, 5G, and 5H). In both 35S:TM6;tap3 and 35S:TAP3;tap3 lines, the second whorl adaxial epidermis consisted of both sepal- and petal-like cells. Although stomata were still observed in the second whorl organs in the 35S:TAP3;tap3 flowers, these chimeric organs did not develop trichomes on the adaxial surface, indicating that TAP3 was able to better rescue the second whorl than TM6 (Figure 5G). The rescue of the tap3 third whorl phenotype by either 35S:TAP3 or 35S:TM6 was much more limited in that these organs largely displayed cell morphologies typical of carpel epidermal cells (Figures 5D, 5E, 5I, and 5J).
Figure 5.
Rescue of the Phenotype Conferred by tap3 by Overexpression of TM6 or TAP3.
(A) 35S:TM6;tap3 flower.
(B) and (C) Adaxial (B) and abaxial (C) epidermal cells of the 35S:TM6;tap3 second whorl organs.
(D) and (E) Epidermal cells of proximal (D) and distal (E) regions of third whorl organs of the 35S:TM6;tap3 flower.
(F) 35S:TAP3;tap3 flower.
(G) and (H) Adaxial (G) and abaxial (H) epidermal cells of the 35S:TAP3;tap3 second whorl organs.
(I) and (J) Epidermal cells of proximal (I) and distal (J) regions of third whorl organs of the 35S:TAP3;tap3 flower.
Bars in scanning electron micrographs = 20 μm.
To check whether the failure in the rescue of the third whorl was attributable to a lack of transgene expression in the third whorl organs, we performed RT-PCR with transgene-specific primers using dissected second and third whorl tissues (see Supplemental Figure 2 online). The 35S:TM6 construct was expressed at similarly high levels in both the second and third whorls, as was the 35S:TAP3 construct.
We also examined the phenotypes produced by either 35S:TAP3 or 35S:TM6 in a wild-type background. In both cases, transgenic flowers displayed a wild-type phenotype.
TM6 and TAP3 Form Qualitatively Distinct Protein Complexes
In Arabidopsis, AP3 and PI are thought to trigger stamen development by forming a quaternary complex with the AGAMOUS (AG) and SEPALLATA3 (SEP3) MADS box proteins (Honma and Goto, 2001). AP3 and PI interact with AG indirectly through the SEP3 protein, which acts to bridge the components of the complex and also appears to provide transcriptional activation functions. The overlap of expression of AP3, PI, AG, and SEP3 in the stamens can uniquely specify stamen identity. The tomato SEP ortholog TM5 is expressed in the second, third, and fourth whorls and is required for the proper differentiation of petals, stamens, and carpels (Pneuli et al., 1994). TAG1, the tomato AG ortholog, is expressed in the third and fourth whorls and specifies stamen and carpel identity (Pnueli et al., 1994).
To investigate whether TM6 and TAP3 have evolved divergent functions by forming different protein complexes, we performed yeast two-, three-, and four-hybrid assays to test TM6 and TAP3 protein interactions with TPI, TM5, and TAG1 (Table 3). Both TM6 and TAP3 strongly interact with TPI, suggesting that the PI protein interaction domain has been conserved between these AP3 lineage genes. We also observed that these dimers can bind to TM5. However, unlike TM6, TAP3 can bind to TM5 in the absence of TPI. The binding of the Arabidopsis AP3 protein to AG depends on the presence of both PI and SEP3 proteins (Honma and Goto, 2001); similarly, the tomato TAP3 protein, when expressed in conjunction with TPI and TM5, can form a quaternary complex with TAG1 (Table 3). The fact that TAG1AD-TM5+TM6BD-TPI or TAG1AD-TM5+TPIBD-TM6 does not show a positive interaction in yeast suggests that TM6 cannot replace TAP3 in the quaternary complex (Table 3). Furthermore, the positive interactions of TAG1-TM5AD+TM6BD-TPI and TAG1-TM5AD+TPIBD-TM6 can be explained by the interaction of TM5 with TM6 and TPI (Table 3). These results suggest that TM6 and TAP3 have diverged in some of their protein interaction capabilities. Furthermore, our results show that the tomato TM5 gene, unlike the Arabidopsis SEP3 gene (Honma and Goto, 2001), does not appear to have an intrinsic transcriptional activation domain in yeast, although we cannot exclude the possibility that TM5 does possess such activity in planta.
Table 3.
In Vitro Protein Interaction Assays Reveal That TAP3 Forms Qualitatively Distinct Protein Complexes Compared with TM6
| TM6BD | TAP3BD | TPIBD | TAG1BD | TM5BD | TM6BD-TPI | TPIBD-TM6 | TAP3BD-TPI | BD | |
|---|---|---|---|---|---|---|---|---|---|
| TM6AD | − | − | ++ | − | − | n | n | − | − |
| TAP3AD | − | − | ++ | − | − | − | − | n | − |
| TPIAD | ++ | ++ | − | − | − | n | n | n | − |
| TAG1AD | − | − | − | − | − | − | − | + | − |
| TM5AD | − | ++ | − | − | − | ++ | ++ | ++ | − |
| TAG1AD-TM5 | − | − | − | n | n | − | − | + | − |
| TM5AD-TAG1 | − | + | − | n | n | ++ | + | + | − |
| AD | − | − | − | − | − | − | − | − | − |
AD, Gal4 activation domain; BD, Gal4 binding domain; +, moderate interaction; ++, strong interaction; −, no interaction; n, not tested.
DISCUSSION
TAP3 and TM6 Have Divergent and Partially Redundant Functions
To define the relative contributions of AP3 lineage members to petal and stamen identity specification in tomato, we investigated the roles of the TM6 and TAP3 genes. Through the identification of a complete loss-of-function insertional mutation, we have shown that TAP3 is required to specify petal and stamen identities. In the tap3 loss-of-function mutant, we observed a dramatic conversion of petals to sepal-like structures as well as a homeotic conversion of stamens to carpel-like organs. The tap3 mutation we have identified differs in some respects from stamenless (sl), a mutation that has been postulated to be an allele of TAP3 (Gomez et al., 1999). sl is a semidominant and temperature-sensitive mutant; homozygous sl mutants grown at the restrictive temperature display a phenotype similar to the one we have described for tap3. One possibility is that sl represents a neomorphic mutation in the TAP3 locus.
In contrast with the tap3 loss-of-function mutant, the RNAi-induced loss of TM6 function results in defects predominantly in stamen development. The TM6i lines show a homeotic conversion of stamens to carpel-like organs, with concomitant formation of ectopic ovules (Figures 3W to 3Z). The TM6i lines do not display any homeotic defects in petal development; the only phenotypic effect we could observe for the downregulation of TM6 activity in petals was a reduction in overall petal size, likely caused by a reduction in cell proliferation (Table 2). Similarly, PTD, the Populus trichocarpa TM6 ortholog, has been postulated to play a role in regulating cell proliferation (Sheppard et al., 2000). Although the levels of endogenous TM6 are strongly downregulated in the TM6i lines (Figure 2), we cannot exclude the possibility that the lack of a homeotic phenotype in petals is the result of low-level residual expression in these lines. Because TAP3 and TM6 do not appear to regulate each other's expression at the transcriptional level (Figure 2E), these observations suggest that TAP3 and TM6 have acquired developmentally distinct roles in reproductive development.
It is also clear, however, that there are some similarities in the functions of the TAP3 and TM6 gene products. Our experiments (Figure 5) demonstrate that when these genes are expressed in an equivalent manner, they confer a similar, but not identical, degree of rescue to the tap3 mutant, indicating that TM6 can substitute for some aspects of TAP3 function.
Differing Expression Patterns of TAP3 and TM6 Can Partially Explain Their Divergent Functions
The tomato AP3 paralogs show quite divergent expression patterns. TAP3 is expressed strongly in the incipient and developing petal and stamen primordia until late stages, when its expression becomes restricted to various floral tissues (Figure 4). At early stages of flower development, TM6 is expressed weakly but fairly ubiquitously in petal, stamen, and carpel primordia. This expression pattern shifts such that by stage 9, strong TM6 expression is observed in developing ovules and in the transmitting tract.
The Petunia TM6 and DEF genes have also been shown to be regulated differently in that Ph TM6 is negatively regulated by the Petunia A-function BLIND gene but BLIND does not regulate the expression of Ph DEF (Tsuchimoto et al., 1993; Vandenbussche et al., 2004). An A-function gene has been identified in tomato, MADS-MC (Vrebalov et al., 2002); it is possible that this gene also differentially regulates the expression of the AP3 paralogs in tomato.
Over the course of angiosperm evolution, the expression patterns of the AP3 lineage members display some variability, indicating divergence in gene regulation (Kramer and Irish, 1999; Irish, 2003; Zahn et al., 2005). Even within the Solanaceae, there are detectable differences in orthologous gene expression patterns. For instance, the Nicotiana benthamiana euAP3 gene, DEF, is expressed at high levels in the second and third whorls, but expression is detectable in all four whorls (Liu et al., 2004). This broader pattern of expression is also seen for the Nicotiana tabacum euAP3 ortholog, DEF (Davies et al., 1996a). By contrast, expression of the tomato TAP3 (Figures 2E and 4) and Petunia DEF genes is only detectable in the second and third whorls (Angenent et al., 1995). Also, the pattern of TM6 expression that we describe is quite similar to that of Petunia Ph TM6, which is initially expressed in petal, stamen, and carpel primordia, becomes predominant in stamens, and at later stages is expressed strongly in the developing placenta and ovules (Vandenbussche et al., 2004).
Furthermore, our observations suggest that TAP3, TM6, and TPI do not regulate each other's expression to a significant degree (Figure 2E). This is quite distinct from what has been observed for Arabidopsis and Antirrhinum, in which the PI and euAP3 genes cross-regulate each other's transcription to maintain continued expression (Jack et al., 1992; Trobner et al., 1992; Goto and Meyerowitz, 1994). In Petunia, the complete loss of PI lineage gene function in the phglo1 phglo2 double mutant results in the downregulation of DEF but not of TM6 (Vandenbussche et al., 2004), indicating differential regulation of the Petunia AP3 lineage genes. Together, these observations imply that the regulation of AP3 family members has diverged extensively. Furthermore, euAP3 lineage genes appear to be cross-regulated differently in tomato compared with Petunia. It is possible that, during the evolution of the Solanaceae, this mode of cross-regulation was lost from the lineage leading to modern-day tomato.
By ectopically overexpressing TAP3 and TM6, we showed that these genes could provide similar degrees of rescue to the tap3 mutant (Figure 5). These observations suggest that the differences in the patterns of expression of these paralogous genes are responsible for much of their difference in function. Although TM6 and TAP3 are both expressed in petals and stamens, they appear to be expressed at different levels and in different subpopulations of cells (Figure 4), which could explain their differences in function. We did observe some subtle differences in the degree to which rescue was affected, indicating that changes in protein-coding capabilities also are likely to play a role in specifying functional differences. Together, these observations suggest that regulatory changes play a prominent role in the diversification of TAP3 and TM6 function.
Differences in Protein–Protein Interactions: Neofunctionalization of euAP3?
In Arabidopsis, the specific amino acid residues responsible for interactions between AP3 and PI have been mapped to the K regions of these proteins; particularly important are Glu-97 and Asp-98 in PI and Asp-98 and Arg-102 in AP3 (Yang et al., 2003). These residues are conserved in the tomato TAP3 and TM6 proteins, suggesting that their interaction with TPI may be mediated by the same protein domains. However, our data show that, at least in yeast, TM6 and TAP3 have unique as well as shared protein interaction properties that likely contribute to their distinct developmental roles. These observations support the idea that the very different C-terminal domains possessed by these proteins may be important in the formation of different protein complexes, and in turn in their different biological activities. Differences in heterodimerization capabilities have also been reported for the petunia DEF and TM6 proteins; DEF interacts with both GLO1 (FBP1) and GLO2 (pMADS2), whereas TM6 interacts strongly only with GLO2 (Vandenbussche et al., 2004). In tomato, we have isolated one PI ortholog gene, TPI, whose coding sequence is more similar to that of GLO2 than GLO1. We cannot exclude the possibility that a second PI otholog, similar to GLO1, is present in tomato and may preferentially interact with TAP3.
Analyses of synonymous versus nonsynonymous substitution rates (Goldman and Yang, 1994) of euAP3 and TM6 lineage coding sequences also support the idea that these paralogous lineages have evolved different roles (G. de Martino and V. F. Irish, unpublished data). Pair-wise comparisons of the full-length coding sequences of five euAP3 genes indicate that, in all cases, the ratio of nonsynonymous to synonymous substitutions (Ka:Ks) are much less than 1. Similarly, Ka:Ks ratios for five TM6 lineage members are also all much less than 1. These observations indicate that each of these gene lineages is experiencing strong purifying selection, suggesting that characteristic lineage-specific residues contribute to differences in function.
TAP3 may have evolved new functions by interacting with different cofactors. For instance, the fact that TAP3 can complex with TAG1 could potentially be responsible for at least some of the differences seen between TAP3 and TM6 functions in the third whorl.
It is likely that TAP3 and TM6 also have differential interactions with tomato A-function gene products; however, the only tomato A-function gene characterized to date, MADS-MC, is not expressed in petal primordia (Vrebalov et al., 2002) and so likely does not participate in biologically relevant interactions with either of the tomato AP3 paralogs. However, another tomato MADS box gene that is closely related to MADS-MC has been identified (Litt and Irish, 2003) and may play such a regulatory role.
We have also shown that overexpression of TM6 can trigger petal development in the tap3 mutant, indicating that TM6 can replace TAP3 in petal-specific protein complexes. Similarly, the maize (Zea mays) paleoAP3 homolog, SILKY1, is able to rescue both petals and stamens in the Arabidopsis ap3 mutant when highly overexpressed (Whipple et al., 2004). By contrast, a chimeric Arabidopsis AP3 gene containing a paleoAP3 C-terminal motif has been shown to rescue only the ap3 third whorl phenotype (Lamb and Irish, 2003). These discrepancies may result from differing levels of transgene expression. Together, these observations suggest that, when highly overexpressed, AP3 lineage proteins that contain the paleoAP3 motif can replace euAP3 functions. In turn, these observations imply that the differences between paleoAP3 and euAP3 motif functions are likely to be attributable to differences in affinity for similar partner proteins.
Were the euAP3 Lineage Genes Recruited for the de Novo Evolution of Petals in the Core Eudicots?
Morphological and paleontological data support the idea that core eudicot petals arose as an evolutionary innovation and are nonhomologous with petals from other angiosperm lineages (Takhtajan, 1991; Drinnan et al., 1994; Ronse de Craene, 2004). We have previously suggested that the advent of the divergent euAP3 lineage genes may be correlated with this de novo evolution of petals in the core eudicots (Kramer and Irish, 1999, 2000; Irish, 2003; Lamb and Irish, 2003). The divergence in tomato TAP3 and TM6 gene functions can be attributed to changes in both the cis-regulatory and coding sequences. Together, the functional analyses of the euAP3 and TM6 lineage members in Petunia (Rijpkema et al., 2006) and tomato (this work) support the correlation of euAP3 function with the de novo derivation of petals in the core eudicot species, in that the euAP3 lineage gene in both cases plays a homeotic role in specifying petals. The role of the TM6 lineage genes appears to be more variable, though, and may reflect the gradual dispensability of this gene lineage in the core eudicots. In fact, Arabidopsis lacks a TM6 representative (Lamb and Irish, 2003), illustrating that, at least in this species, TM6 function is not necessary.
The situation in noncore eudicot species may reflect a different mechanism whereby AP3 lineage genes have acquired new roles. For instance, in monocots such as rice (Oryza sativa) and maize, the paleoAP3 representatives are involved in specifying both lodicule and stamen identity (Kang et al., 1998; Moon et al., 1999; Ambrose et al., 2000). This would suggest that, within the grass lineages, paleoAP3 genes have been recruited for the development of the grass-specific organ, the lodicule. The observation of paleoAP3 function in grasses, combined with the analyses of euAP3 and TM6 gene function in core eudicots, supports a model in which, in different angiosperm clades, different mechanisms have been used to expand the roles of these genes. In core eudicots, the duplication resulting in the euAP3 genes allowed for the recruitment of these genes to a role in petal specification; in the grasses, changes in the domain of expression of the paleoAP3 genes appear to be responsible for their new roles in lodicule specification.
Plasticity in Evolution
Through the analyses presented here and by Rijpkema et al. (2006), we have demonstrated the functional roles of euAP3 and TM6 lineage genes in different members of the Solanaceae. Two paralogous AG-related gene lineages have also been identified in the core eudicots, resulting in the AG and PLE gene lineages (Davies et al., 1999; Nitasaka, 2003; Kramer et al., 2004; Irish and Litt, 2005). Comparisons of the functions of the AG and PLE genes in Antirrhinum and Arabidopsis suggest that these gene lineages have swapped roles (Liljegren et al., 2000; Pinyopich et al., 2003; Causier et al., 2005). However, because only two species have been functionally analyzed for AG/PLE roles, it remains unclear when the paralogous gene lineages took on complementary roles. One extreme possibility is that the duplication event resulting in the AG and PLE lineages also essentially immediately resulted in the complementary and stable subfunctionalization of these genes. By contrast, our analyses of AP3 lineage genes demonstrate that parsing of biological function can be quite plastic. It is clear that the duplication event resulting in the euAP3 and TM6 lineages occurred well before the diversification of the Solanaceae. Genome-level comparisons among solanaceaeous species, including tomato and Petunia, have shown a high level of conservation among these genomes (Rensink et al., 2005), reflecting the relatively recent divergence of these taxa ∼40 million years ago (Clegg et al., 1997). Nonetheless, the euAP3 and TM6 gene functions have been parsed quite differently in tomato and Petunia, illustrating that subfunctionalization is a dynamic and fluid process.
METHODS
Phylogenetic Analyses
Sequences of AP3 lineage genes from selected basal and core eudicots were obtained from the National Center for Biotechnology Information GenBank. The MIK domains were aligned using MacVector (Accelrys) and adjusted manually (see Supplemental Figure 1 online). The neighbor-joining analysis was performed and bootstrapped 2000 times using PAUP*b10 (Swofford, 2000).
In Situ Hybridizations
In situ hybridizations were performed according to published protocols (Jackson, 1991) with minor modifications. Tomato (Solanum lycopersicum cv Micro-Tom) wild-type floral buds were fixed in 4% paraformaldehyde in PBS and embedded in Paraplast Plus tissue-embedding medium (Tyco Healthcare). For the TAP3 antisense probe, a 563-bp probe was made using the TAP3F primer (5′-TTGTTCGATCTGTACCAGAAG-3′) and the TAP3RT7 primer containing the T7 promoter (5′-CATAATACGACTCACTATAGGGTTCTTGTGAACAAACATTGC-3′). For the TM6 antisense probe, a 909-bp probe was made using the TM6F primer (5′-AAGAAGATTGAAAACTTGAC-3′) and the TM6RT7 primer containing the T7 promoter (5′-CATAATACGACTCACTATAGGGTAAGTCGAAAAGGAAAATTG-3′). For the TPI antisense probe, a 400-bp probe was made using the TPIF primer (5′-CAATCAACTTACCCATAAAGAGC-3′) and the TPIRT7 primer containing the T7 promoter (5′-CATAATACGACTCACTATAGGGCCAACATGAAACAGAGTCTTAGC-3′). Hybridizations were performed at 48°C, and subsequent washes were done at 52°C.
Identification of TPI and TAP3 Sequences
Poly(A) mRNA was extracted from total RNA using Magnetight oligo(dT) beads (Novagen). Single-stranded cDNA was synthesized by priming with oligo(dT)25 from 500 ng of poly(A+) RNA. Two degenerate primers complementary to the K domain and the PI motif of PI-like genes were used to amplify the tomato PI ortholog, TPI, cDNA (TPIF, 5′-GGATGC(T/A)AA(G/A)CATGA(G/A)(A/C)AI(T/C)GA(A/G)ATA-3′; TPIR, 5′-TGIA(A/G)(A/G)TTIGGITGIA(A/T)(T/G)GGITG-3′).
PCR was performed using the following regime: 10 cycles of 20 s of denaturing at 94°C, 30 s of annealing at 38°C, and a 1-min extension at 72°C. The program was completed by 30 cycles of 20 s of denaturing at 94°C, 30 s of annealing at 42°C, and an extension time of 1 min at 72°C. The TPI cDNA full-length sequence was obtained by RACE reactions according to the manufacturer's protocol (Gibco BRL). A partial sequence of the TAP3 gene has already been isolated (accession number AF052868). To obtain the 5′ terminus of the TAP3 cDNA, we performed 5′ RACE reactions (primer sequences for RACE are available upon request).
Isolation of the tap3 Loss-of-Function Mutation
Inverse PCR was performed with Ds-specific primers (Ds5216, 5′-TTGTATATCCCGTTTCCGTTCCGTT-3′; Ds7665, 5′-TTTCGTTTCCGTCCCGCAAGTTAAATA-3′) using genomic DNA from line EE1153, leading to the identification of an insertion in the tomato TAP3 gene. PCR was performed using the Expand Long Template PCR system (Boehringer) with the following program: 10 cycles of 10 s of denaturing at 94°C, 30 s of annealing at 60°C, and 5 min at 68°C; 20 cycles of 10 s at 94°C, 30 s at 60°C, and 5 s + 20 s at 68°C; and a final step of 7 min at 68°C. Further screening of the individual plants by PCR using Ds- and TAP3-specific primers showed that these plants were heterozygous for the insertion (primer sequences are available upon request). Cosegregation of the Ds element and the mutant phenotype was confirmed by both DNA gel blot analyses and PCR.
Transformation Constructs and Plant Transformation
Plant transformations were all performed using cv Micro-Tom. We generated a TM6 RNAi construct to target part of the C terminus and the 3′ untranslated region of the endogenous TM6 transcript. A 220-bp fragment was amplified by PCR with TM6-specific primers (TM6-617F, 5′-CATTGCACCCCAATCTTCAAAACG-3′; TM6-837R, 5′-CAAGATCTGCTTAACACAGAACCAAATCCAGACTCG-3′). The second intron of Arabidopsis thaliana AP3 was used for the hairpin of the double-stranded RNA.
The TM6+/AP3int/TM6− construct was inserted into the binary vector pTCSH1 (Hardtke et al., 2000) in the XhoI/XbaI sites. For the TM6 and TAP3 overexpression constructs, the complete open reading frame of TM6 or TAP3 was amplified by PCR and inserted in XhoI/XbaI sites of the pTCSH1 binary vector. pTCSH1 binary vectors containing the sequence of interest were electroporated into the Agrobacterium tumefaciens LBA 4404 strain and used in independent tomato transformations.
Micro-Tom was transformed using a protocol derived from a previously published method (Fillatti et al., 1987) with some modifications. Seeds were germinated on Murashige and Skoog medium at 25°C under constant light. Explants of 7-d-old cotyledons were incubated for 40 min with the Agrobacterium LBA 4404 strain containing the construct of interest (grown to log phase). After incubation, the explants were quickly dried on sterile paper and transferred to a cocultivation medium containing 2% glucose and 100 μM acetosyringone. After 2 d of cocultivation in the dark, the explants were transferred to D1 medium containing 1 mg/L zeatin, 200 mg/L timentin, 0.05 mg/L indole-3-acetic acid, and 25 mg/L glufosinate ammonium (Sigma-Aldrich) under constant light. After 2 to 3 weeks, the regenerated calli were transferred to the same medium except that zeatin was reduced to 0.1 mg/L for shoot formation. Shoots of 1- to 1.5-cm plantlets were cut from the calli, dipped into a rooting powder solution, and transferred into soil. The T1 transgenic plants were checked for the presence of the transgene by PCR amplification of the BAR gene (BAR-F, 5′-AGACAAGCACGGTCAACTTCCGTA-3′; BAR-R, 5′-CGATGACAGCGACCACGCTCTT-3′). The T2 generation lines were selected by screening the seed progeny on Murashige and Skoog medium containing 25 mg/L glufosinate ammonium and 0.5% sucrose.
For the tap3 rescue experiments, cotyledons from progeny of TAP3/tap3 heterozygous plants were used in the transformation; homozygous mutant tap3 transgenic plants were identified by PCR.
Scanning Electron Microscopy
Plant samples were fixed overnight in a 3:1 ethanol:glacial acetic acid solution, gradually dehydrated to 100% ethanol, and dried in a critical point drier. Samples were dissected with a stereomicroscope by removing some parts to reveal the organs to be examined. The resulting samples were analyzed using previously published protocols (Irish and Sussex, 1990).
RT-PCR
Total RNA was isolated using Trizol reagent (Life Technologies) according to the manufacturer's protocol. Five micrograms of total RNA was used as a template for first-strand cDNA synthesis in a volume of 20 μL with 1 μL of Superscript II RT (Gibco BRL). The RT reaction was diluted 1:5, and 2 μL was used for PCR. The primers used were as follows: TM61F (5′-ATGGGACGGGGAAAAATTGAGATCAAGAAGATT-3′) and TM6-570R (5′-GCAAATGCCACAGCAGAGAGTGG-3′) for TM6; TAP3-1F (5′-AATGGGCTATTCAAGAAGGCTAATG-3′) and TAP3-580R (5′-CACCTCCACTGTGAAGATGATTATG-3′) for TAP3; TPI-1F (5′-CATCATGGGGAGAGGTAAAATAG-3′) and TPI-760R (5′-GGTAATAATTCCAACATGAAACAG-3′) for TPI; and LEACTF (5′-CAGGATTTGCTGGTGATGCTCCTC-3′) and LEACTR (5′-GAGGTACGACCGCTAGCATACAG-3′) for ACTIN (accession number AB199316). The PCR regime was 25 s of denaturing at 94°C, 25 s of annealing at 56°C, and an extension time of 1 min at 72°C, with 32 cycles for the TM6 gene and 28 cycles for the ACTIN, TPI, and TAP3 genes.
Yeast Two-, Three-, and Four-Hybrid Analysis
TM6, TAP3, TPI, TM5, and TAG1 IKC domains were used to test protein interactions in yeast assays. For the two-hybrid analysis, single proteins were fused with the GAL4 DNA binding domain in the pGBT9 vector (Clontech) and with the GAL4 activation domain in the pGAD424 vector (Clontech). For the three- and four-hybrid analyses, two genes on the same vector were expressed under the control of two independent ADH1 promoters. Protein interactions were confirmed by testing different vector combinations: (X − AD; Y) + (W − BD; Z), (W; Z − AD) + (X; Y − BD), etc. Two independent transformations for each vector combination were performed, and five colonies per transformation were used for the assay. The β-galactosidase liquid assay was performed using a protocol available at http://www.fhcrc.org/science/labs/gottschling/yeast/Bgal.html.
Accession Numbers
Sequences of AP3 lineage genes from selected basal and core eudicots were obtained from GenBank: AP3 (AF115814), RAD1 (X89113), RAD2 (X89108), SLM3 (X80490), GDEF1 (AJ009724), GDEF2 (AJ009725), HPDEF1 (AF180364), HPDEF2 (AF180365), Ph DEF (X69946), Ph TM6 (AF230704), Nt DEF (X96428), Nt TM6 (AY577817), Le TM6 (X60759), Le AP3 (AF052868 and this study), DEF (X52023), Hm AP3 (AF230702), Hm TM6 (AF230703), MASAKO B3 (AB055966), Md TM6 (AB081093), Md MADS13 (Aj251116), PTD (AF057708), Jr AP3 (AJ313089), NMH7 (L41727), Ri.sa. AP3-1 (AY337758), Ri.sa. AP3-2 (AY337759), Gu.ti. AP3-4 (AY337756), Gu.ti. AP3-5 (AY337757), Pn AP3-1 (AF052873), Pn AP3-2 (AF052874), De AP3 (AF052875), Rb AP3-1 (AF052876), Rb AP3-2 (AF130869), Pc AP3 (AF052872), Pt AP3-1 (AF052870), Pt AP3-2 (AF052871), STDEF (X67511). Sequence data from this article can be found in the GenBank data library under the following accession numbers: TPI (DQ674531) and TAP3 (DQ674532).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment Used for Phylogenetic Analyses.
Supplemental Figure 2. RT-PCR Analyses of TAP3 and TM6 Expression Levels.
Supplementary Material
Acknowledgments
We thank Queenie Tan for her help in generating the RNAi constructs. We appreciate the kindness of Christian Hardtke and Tamara Western for providing laboratory facilities for part of this work. We thank members of the Irish laboratory and David Weiss for constructive comments on the manuscript and Tom Gerats and Michiel Vandenbussche for communicating results before publication. This work was supported by Grant 0110731 from the National Science Foundation to V.F.I.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Vivian F. Irish (vivian.irish@yale.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.106.042978.
References
- Ambrose, B.A., Lerner, D.R., Ciceri, P., Padilla, C.M., Yanofsky, M.F., and Schmidt, R.J. (2000). Molecular and genetic analyses of the Silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5 569–579. [DOI] [PubMed] [Google Scholar]
- Angenent, G.C., Busscher, M., Franken, J., Dons, H.J., and van Tunen, A.J. (1995). Functional interaction between the homeotic genes fbp1 and pMADS1 during petunia floral organogenesis. Plant Cell 7 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey, P.N., and Chua, N.-H. (1990). The cauliflower mosaic virus 35S promoter: Combinatorial regulation of transcription in plants. Science 250 959–966. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brukhin, V., Hernould, M., Gonzalez, N., Chevalier, C., and Mouras, A. (2003). Flower development schedule in tomato Lycopersicon esculentum cv. Sweet Cherry. Sex. Plant Reprod. 15 311–320. [Google Scholar]
- Busi, M.V., Bustamante, C., D'Angelo, C., Hidalgo-Cuevas, M., Boggio, S.B., Valle, E.M., and Zabaleta, E. (2003). MADS-box genes expressed during tomato seed and fruit development. Plant Mol. Biol. 52 801–815. [DOI] [PubMed] [Google Scholar]
- Carpenter, R., and Coen, E.S. (1990). Floral homeotic mutations produced by transposon-mutagenesis in Antirrhinum majus. Genes Dev. 4 1483–1493. [DOI] [PubMed] [Google Scholar]
- Causier, B., Castillo, R., Zhou, J., Ingram, R., Xue, Y., Schwarz-Sommer, Z., and Davies, B. (2005). Evolution in action: Following function in duplicated floral homeotic genes. Curr. Biol. 15 1508–1512. [DOI] [PubMed] [Google Scholar]
- Clegg, M.T., Cummings, M.P., and Durbin, M.L. (1997). The evolution of plant nuclear genes. Proc. Natl. Acad. Sci. USA 94 7791–7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, B., Di Rosa, A., Eneva, T., Saedler, H., and Sommer, H. (1996. a). Alteration of tobacco floral organ identity by expression of combinations of Antirrhinum MADS-box genes. Plant J. 10 663–677. [DOI] [PubMed] [Google Scholar]
- Davies, B., Egea-Cortines, M., de Andrade Silva, E., Saedler, H., and Sommer, H. (1996. b). Multiple interactions among floral homeotic MADS box proteins. EMBO J. 16 4330–4343. [PMC free article] [PubMed] [Google Scholar]
- Davies, B., Motte, P., Keck, E., Saedler, H., Sommer, H., and Schwarz-Sommer, Z. (1999). PLENA and FARINELLI: Redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development. EMBO J. 18 4023–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnan, A.N., Crane, P.R., and Hoot, S.B. (1994). Patterns of floral evolution in the early diversification of non-magnoliid dicotyledons (eudicots). In Early Evolution of Flowers, P.K. Endress and E.M. Friis, eds (New York: Springer-Verlag), pp. 93–122.
- Fillatti, J., Kiser, J., Rose, B., and Comai, L. (1987). Efficient transformation of tomato and the introduction and expression of a gene for herbicide tolerance. In Tomato Biotechnology, D. Nevins and R. Jones, eds (New York: Alan R. Liss), pp. 199–210.
- Force, A., Lynch, M., Pickett, F.B., Amores, A., Yan, Y.L., and Postlethwait, J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman, N., and Yang, Z. (1994). A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11 725–736. [DOI] [PubMed] [Google Scholar]
- Gomez, P., Jamilena, M., Capel, J., Zurita, S., Angosto, T., and Lozano, R. (1999). Stamenless, a tomato mutant with homeotic conversions in petals and stamens. Planta 209 172–179. [DOI] [PubMed] [Google Scholar]
- Goto, K., and Meyerowitz, E.M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8 1548–1560. [DOI] [PubMed] [Google Scholar]
- Halfter, U., Ali, N., Stockhaus, J., Ren, L., and Chua, N.-H. (1994). Ectopic expression of a single homeotic gene, the Petunia gene green petal, is sufficient to convert sepals to petaloid organs. EMBO J. 13 1443–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S., Gohda, K., Osterlund, M.T., Oyama, T., Okada, K., and Deng, X.W. (2000). HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19 4997–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2000). The Arabidopsis floral homeotic gene PISTILLATA is regulated by discrete cis-elements responsive to induction and maintenance signals. Development 127 2021–2030. [DOI] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409 469–471. [DOI] [PubMed] [Google Scholar]
- Irish, V.F. (2003). The evolution of floral homeotic gene function. Bioessays 25 637–646. [DOI] [PubMed] [Google Scholar]
- Irish, V.F., and Kramer, E.M. (1998). Genetic and molecular analysis of angiosperm flower development. Adv. Bot. Res. 28 197–230. [Google Scholar]
- Irish, V.F., and Litt, A. (2005). Flower development and evolution: Gene duplication, diversification and redeployment. Curr. Opin. Genet. Dev. 15 454–460. [DOI] [PubMed] [Google Scholar]
- Irish, V.F., and Sussex, I.M. (1990). Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2 741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack, T., Brockman, L.L., and Meyerowitz, E.M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68 683–697. [DOI] [PubMed] [Google Scholar]
- Jack, T., Fox, G.L., and Meyerowitz, E.M. (1994). Arabidopsis homeotic gene APETALA3 ectopic expression: Transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76 703–716. [DOI] [PubMed] [Google Scholar]
- Jackson, D. (1991). In situ hybridisation in plants. In Molecular Plant Pathology: A Practical Approach, D.J. Bowles, S.J. Gurr, and P. McPherson, eds (Oxford, UK: Oxford University Press), pp. 163–174.
- Kang, H.-G., Jeon, J.-S., Lee, S., and An, G. (1998). Identification of class B and class C floral organ identity genes from rice plants. Plant Mol. Biol. 38 1021–1029. [DOI] [PubMed] [Google Scholar]
- Kim, S., Yoo, M., Albert, V.A., Farris, J.S., Soltis, P.S., and Soltis, D.E. (2004). Phylogeny and diversification of B-function genes in angiosperms: Evolutionary and functional implications of a 260-million year old duplication. Am. J. Bot. 91 2102–2118. [DOI] [PubMed] [Google Scholar]
- Kramer, E.M., Dorit, R.L., and Irish, V.F. (1998). Molecular evolution of petal and stamen development: Gene duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149 765–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, E.M., and Irish, V.F. (1999). Evolution of genetic mechanisms controlling petal development. Nature 399 144–148. [DOI] [PubMed] [Google Scholar]
- Kramer, E.M., and Irish, V.F. (2000). Evolution of petal and stamen developmental programs: Evidence from comparative studies of the lower eudicots and basal angiosperms. Int. J. Plant Sci. 161 (suppl.), S29–S40. [Google Scholar]
- Kramer, E.M., Jaramillo, M.A., and Di Stilio, V.S. (2004). Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics 166 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, E.M., Su, H.J., Wu, C.C., and Hu, J.M. (2006). A simplified explanation for the frameshift mutation that created a novel C-terminal motif in the APETALA3 gene lineage. BMC Evol. Biol. 6 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, R.S., and Irish, V.F. (2003). Functional divergence within the APETALA3/PISTILLATA floral homeotic gene lineages. Proc. Natl. Acad. Sci. USA 100 6558–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren, S.J., Ditta, G.S., Eshed, Y., Savidge, B., Bowman, J.L., and Yanofsky, M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404 766–770. [DOI] [PubMed] [Google Scholar]
- Litt, A., and Irish, V.F. (2003). Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: Implications for the evolution of floral development. Genetics 165 821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Nakayama, N., Schiff, M., Litt, A., Irish, V.F., and Dinesh-Kumar, S.P. (2004). Virus induced gene silencing of a DEFICIENS ortholog in Nicotiana benthamiana. Plant Mol. Biol. 54 701–711. [DOI] [PubMed] [Google Scholar]
- Lozano, R., Angosto, T., Gomez, P., Payan, C., Capel, J., Huijser, P., Salinas, J., and Martinez-Zapater, J.M. (1998). Tomato flower abnormalities induced by low temperatures are associated with changes of expression of MADS-box genes. Plant Physiol. 117 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and Conery, J.S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and Force, A. (2000). The probability of duplicate gene preservation by subfunctionalization. Genetics 154 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magallon, S., Crane, P.R., and Herendeen, P.S. (1999). Phylogenetic pattern, diversity, and diversification of eudicots. Ann. Mo. Bot. Gard. 86 297–372. [Google Scholar]
- McGonigle, B., Bouhidel, K., and Irish, V.F. (1996). Nuclear localization of the Arabidopsis APETALA3 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes Dev. 10 1812–1821. [DOI] [PubMed] [Google Scholar]
- Meissner, R., Chague, V., Zhu, Q., Emmanuel, E., Elkind, Y., and Levy, A.A. (2000). A high throughput system for transposon tagging and promoter trapping in tomato. Plant J. 22 265–274. [DOI] [PubMed] [Google Scholar]
- Meissner, R., Jacobson, Y., Melamed, S., Levyatuv, S., Shalev, G., Ashri, A., Elkind, Y., and Levy, A. (1997). A new model system for tomato genetics. Plant J. 12 1465–1472. [Google Scholar]
- Moon, Y.H., Jung, J.Y., Kang, H.G., and An, G. (1999). Identification of a rice APETALA3 homologue by yeast two-hybrid screening. Plant Mol. Biol. 40 167–177. [DOI] [PubMed] [Google Scholar]
- Nitasaka, E. (2003). Insertion of an En/Spm-related transposable element into a floral homeotic gene DUPLICATED causes a double flower phenotype in the Japanese morning glory. Plant J. 36 522–531. [DOI] [PubMed] [Google Scholar]
- Ohno, S. (1970). Evolution by Gene Duplication. (New York: Springer).
- Pelaz, S., Tapia-Lopez, R., Alvarez-Buylla, E.R., and Yanofsky, M.F. (2001). Conversion of leaves into petals in Arabidopsis. Curr. Biol. 11 182–184. [DOI] [PubMed] [Google Scholar]
- Pinyopich, A., Ditta, G.S., Savidge, B., Liljegren, S.J., Baumann, E., Wisman, E., and Yanofsky, M.F. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424 85–88. [DOI] [PubMed] [Google Scholar]
- Pnueli, L., Abu-Abeid, M., Zamir, D., Nacken, W., Schwarz-Sommer, Z., and Lifschitz, E. (1991). The MADS box gene family in tomato: Temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant J. 1 255–266. [PubMed] [Google Scholar]
- Pneuli, L., Hareven, D., Broday, L., Hurwitz, C., and Lifshitz, E. (1994). The TM5 MADS box gene mediates organ differentiation in the three inner whorls of tomato flowers. Plant Cell 6 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli, L., Hareven, D., Rounsley, S.D., Yanofsky, M.F., and Lifschitz, E. (1994). Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell 6 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensink, W.A., Lee, Y., Liu, J., Iobst, S., Ouyang, S., and Buell, C.R. (2005). Comparative analyses of six solanaceous transcriptomes reveal a high degree of sequence conservation and species-specific transcripts. BMC Genomics 6 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., Krizek, B.A., and Meyerowitz, E.M. (1996. a). Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 93 4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., Wang, M., and Meyerowitz, E.M. (1996. b). DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 24 3134–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijpkema, A., Royaert, S., Zethof, J., van der Weerden, G., Gerats, T., and Vandenbussche, M. (2006). Functional divergence within the DEF/AP3 lineage: An analysis of PhTM6 in Petunia hybrida. Plant Cell 18 1819–1832. [DOI] [PMC free article] [PubMed]
- Ronse de Craene, L.P. (2004). Floral development of Berberidopsis corallina: A crucial link in the evolution of flowers in the core eudicots. Ann. Bot. (Lond.) 94 741–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer, Z., Hue, I., Huijser, P., Flor, P.J., Hansen, R., Tetens, F., Lonnig, W.-E., Saedler, H., and Sommer, H. (1992). Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: Evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 11 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhar, K.N.C., and Sawhney, V.K. (1984). A scanning electron microscope study of the development and surface features of floral organs of tomato (Lycopersicon esculentum). Can. J. Bot. 62 2403–2413. [Google Scholar]
- Sheppard, L.A., Brunner, A.M., Krutovskii, K.V., Rottmann, W.H., Skinner, J.S., Vollmer, S.S., and Strauss, S.H. (2000). A DEFICIENS homolog from the dioecious tree black cottonwood is expressed in female and male floral meristems of the two-whorled unisexual flowers. Plant Physiol. 124 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer, H., Beltran, J.-P., Huijser, P., Pape, H., Lonnig, W.-E., Saedler, H., and Schwarz-Sommer, Z. (1990). Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO J. 9 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellari, G.M., Jaramillo, M.A., and Kramer, E.M. (2004). Evolution of the APETALA3 and PISTILLATA lineages of MADS-box-containing genes in the basal angiosperms. Mol. Biol. Evol. 21 506–519. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. (2000). PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods). (Sunderland, MA: Sinauer Associates).
- Takhtajan, A. (1991). Evolutionary Trends in Flowering Plants. (New York: Columbia University Press).
- Theissen, G., Becker, A., Di Rosa, A., Kanno, A., Kim, J.T., Muenster, T., Winter, K.-W., and Saedler, H. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42 115–149. [PubMed] [Google Scholar]
- Theissen, G., and Saedler, H. (2001). Plant biology. Floral quartets. Nature 409 469–471. [DOI] [PubMed] [Google Scholar]
- Trobner, W., Ramirez, L., Motte, P., Hue, I., Huijser, P., Lonnig, W.E., Saedler, H., Sommer, H., and Schwarz-Sommer, Z. (1992). Globosa—A homeotic gene which interacts with Deficiens in the control of Antirrhinum floral organogenesis. EMBO J. 11 4693–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto, S., Mayama, T., van der Krol, A., and Ohtsubo, E. (2000). The whorl-specific action of a petunia class B floral homeotic gene. Genes Cells 5 89–99. [DOI] [PubMed] [Google Scholar]
- Tsuchimoto, S., van der Krol, A.R., and Chua, N.-H. (1993). Ectopic expression of pMADS3 in transgenic petunia phenocopies the petunia blind mutant. Plant Cell 5 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche, M., Theissen, G., Van de Peer, Y., and Gerats, T. (2003). Structural diversification and neo-functionalization during floral MADS-box gene evolution by C-terminal frameshift mutations. Nucleic Acids Res. 31 4401–4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche, M., Zethof, J., Royaert, S., Weterings, K., and Gerats, T. (2004). The duplicated B-class heterodimer model: Whorl-specific effects and complex genetic interactions in Petunia hybrida flower development. Plant Cell 16 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Krol, A.R., Brunelle, A., Tsuchimoto, S., and Chua, N.H. (1993). Functional analysis of petunia floral homeotic MADS box gene pMADS1. Genes Dev. 7 1214–1228. [DOI] [PubMed] [Google Scholar]
- Vrebalov, J., Ruezinsky, D., Padmanabhan, V., White, R., Medrano, D., Drake, R., Schuch, W., and Giovannoni, J. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296 343–346. [DOI] [PubMed] [Google Scholar]
- Whipple, C.J., Ciceri, P., Padilla, C.M., Ambrose, B.A., Bandong, S.L., and Schmidt, R.J. (2004). Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development 131 6083–6091. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Fanning, L., and Jack, T. (2003). The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 33 47–59. [DOI] [PubMed] [Google Scholar]
- Zahn, L.M., Leebens-Mack, J., DePamphilis, C.W., Ma, H., and Theissen, G. (2005). To B or not to B a flower: the role of DEFICIENS and GLOBOSA orthologs in the evolution of the angiosperms. J. Hered. 96 225–240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.