Abstract
Repression of photomorphogenesis in Arabidopsis thaliana requires activity of the COP9 signalosome (CSN), CDD, and COP1 complexes, but how these three complexes work in concert to accomplish this important developmental switch has remained unknown. Here, we demonstrate that Arabidopsis CULLIN4 (CUL4) associates with the CDD complex and a common catalytic subunit to form an active E3 ubiquitin ligase both in vivo and in vitro. The partial loss of function of CUL4 resulted in a constitutive photomorphogenic phenotype with respect to morphogenesis and light-regulated gene expression. Furthermore, CUL4 exhibits a synergistic genetic interaction with COP10 and DET1. Therefore, this CUL4-based E3 ligase is essential for the repression of photomorphogenesis. This CUL4-based E3 ligase appears to associate physically with COP1 E3 ligase and positively regulates the COP1-dependent degradation of photomorphogenesis-promoting transcription factors, whereas the CSN controls the biochemical modification of CUL4 essential for E3 activity. Thus, this study suggests a biochemical activity connection between CSN and CDD complexes in their cooperation with COP1 in orchestrating the repression of photomorphogenesis.
INTRODUCTION
Light provides critical cues for plant development. In Arabidopsis thaliana, seedlings follow drastically different developmental patterns in the presence or absence of light. Light-grown seedlings exhibit photomorphogenic development with short hypocotyls and open and expanded cotyledons. By contrast, dark-grown seedlings show skotomorphogenic development characterized by long hypocotyls, closed and unexpanded cotyledons, and apical hooks (von Arnim and Deng, 1996).
The ubiquitin–proteasome system, a common proteolysis mechanism for regulating protein degradation in all eukaryotes, has been shown to play a major role in the light control of development (Sullivan et al., 2003). The ubiquitin–proteasome system consists of two major components: the 26S proteasome responsible for the degradation of protein substrates tagged by a specific kind of polyubiquitin chain, and the ubiquitination machinery that specifically attaches a single ubiquitin or a polyubiquitin chain to a substrate. This ubiquitination system consists of a cascade mediated by three enzymes, a ubiquitin-activating enzyme (E1), a ubiquitin conjugase (E2), and a ubiquitin–protein ligase (E3). The specificity of the ubiquitin–proteasome system usually relies on the E3 ligase, and all eukaryotes possess a large number of E3 ubiquitin ligases that target distinct sets of substrate proteins. There are several known E3 ligase families, the largest of which comprises multiple groups of E3 ligases defined by distinct yet related forms of scaffolding proteins known as cullins. All cullin-based E3 ligases contain a common catalytic subunit, RBX1.
Genetic analysis of light-regulated Arabidopsis seedling development has revealed a group of pleiotropic Constitutive Photomorphogenic/De-etoliated/Fusca (COP/DET/FUS) proteins as central regulators of photomorphogenic development (Sullivan et al., 2003; Yi and Deng, 2005). Initially isolated by screening for repressors of photomorphogenesis in darkness (Wei and Deng, 1996), these regulatory proteins were later shown to play a crucial role in the ubiquitin–proteasome-mediated degradation of photomorphogenesis-promoting factors (Osterlund et al., 2000; Yi and Deng, 2005). This group of proteins carries out their functions as part of three biochemical entities: the COP1 complex, the COP9 signalosome (CSN), and the CDD (for COP10, DDB1a, and DET1) complex.
The first of these proteins to be defined molecularly, COP1 has been shown to localize in the nucleus in darkness, whereas light reduces its nuclear abundance (Deng et al., 1992; von Arnim and Deng, 1994). COP1 forms an ∼700-kD complex in vivo (Saijo et al., 2003), which functions as an E3 ubiquitin ligase to catalyze the ubiquitination of several photomorphogenesis-promoting factors, designating them as targets for proteasome-mediated degradation (Yi and Deng, 2005). These factors include at least four known transcription factors (HY5 [Osterlund et al., 2000; Saijo et al., 2003], HYH [Holm et al., 2002], LAF1 [Seo et al., 2003], and HFR1 [Duek et al., 2004; Jang et al., 2005; Yang et al., 2005]). COP1 functions as a key rate-limiting enzyme in the repression pathway (McNellis et al., 1996) and provides a point of convergence for cryptochrome and phytochrome-mediated specific light responses (Wang et al., 2001; Yang et al., 2005; Yi and Deng, 2005).
The CSN is a nucleus-enriched complex consisting of eight distinct subunits. Six of those subunits are genetically defined COP/DET/FUS proteins, and the other two are encoded by families of two genes each (Serino and Deng, 2003). The CSN is known to interact with CULLIN1 (CUL1)-based E3 ligases to deconjugate Nedd8/Rub from cullins (Lyapina et al., 2001; Schwechheimer et al., 2001). In csn mutants as well as in other cop/det/fus mutants, COP1 is depleted in the nucleus compared with the wild type in darkness, which suggests that the CSN and other COP/DET/FUS proteins play important roles in COP1 nuclear import and/or retention in the dark (Chamovitz et al., 1996; von Arnim et al., 1997).
The CDD complex comprises COP10, DET1, and DDB1a (Yanagawa et al., 2004). DET1 is a 62-kD nuclear protein lacking any known functional domain and has been shown to associate with DDB1a (Schroeder et al., 2002). COP10 encodes a small ubiquitin E2 variant protein, and both COP10 itself and the CDD complex are known to enhance the activity of several E2s (Yanagawa et al., 2004). Coimmunoprecipitation experiments have indicated that the CDD complex is able to associate with COP1 and the CSN in vivo, suggesting that the three complexes may act together to regulate the ubiquitin–proteasome-mediated degradation of photomorphogenesis-promoting factors (Yanagawa et al., 2004). However, how the individual biochemical activities of those three complexes work in concert has remained unknown.
CUL4 is a distinct member of the cullin family of proteins that is highly conserved across eukaryotic organisms. Orthologs of CUL4 exist in fission yeast, worms, flies, plants, and mammals. CUL4 loss-of-function mutants are characterized by cell elongation and decondensation of chromosomes in fission yeast (Osaka et al., 2000) and massive DNA replication in Caenorhabditis elegans embryos (Zhong et al., 2003). Deletion of CUL4A leads to lethality in mice at the early embryo stage (Li et al., 2002). CUL4 has been shown to bind RBX1, the common catalytic subunit associated with all cullin-based E3 ligases, to form the core for a family of CUL4-based ligases (Higa et al., 2003; Jia et al., 2005). In the case of several known CUL4-based E3 ligases, distinct DDB1-containing complexes associate with the CUL4-RBX1 core to form functional E3 ligases (Groisman et al., 2003; Hu et al., 2004; Wertz et al., 2004) and are thought to be responsible for recruiting their substrates (Hu et al., 2004). For example, two distinct DDB1-containing protein complexes have been associated with CUL4-based E3 ligases in mammalian DNA repair pathways (Groisman et al., 2003). Similarly, recent evidence has shown that DDB1, DET1, and COP1 can work together as part of a CUL4-based E3 ligase to regulate the ubiquitination and degradation of the transcription factor c-Jun in mammalian cells (Wertz et al., 2004). CUL4 is present in Arabidopsis, but it has remained functionally undefined until now. Because both DDB1 and DET1, together with COP10, are present in the same CDD complex (Yanagawa et al., 2004), it is of interest to know whether a CUL4-RBX1-CDD E3 ligase is present in Arabidopsis and whether it plays a role in photomorphogenesis.
Here, we report the presence of CUL4-containing E3 ubiquitin ligase complexes in Arabidopsis and reveal that at least one such complex contains RBX1 and the CDD complex. In vitro reconstitution experiments demonstrate that CUL4-RBX1 can bind the CDD complex, and together they possess ubiquitin ligase activity. Functional analysis suggests that this CUL4-based E3 is essential for repressing photomorphogenesis, possibly by positively regulating COP1-dependent events. Therefore, this CUL4-based E3 ligase plays a central role in repressing photomorphogenesis and links together the biochemical activities of the CSN and CDD complexes.
RESULTS
Arabidopsis CUL4 Proteins Are Expressed in Most Tissues and Show Nuclear Localization
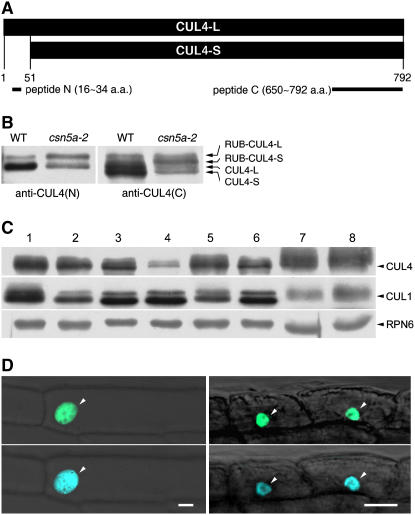
Analysis of the CUL4 mRNA sequence revealed two potential in-frame translation start sites that yield identical proteins, except that one is 50 amino acids longer than the other at the N-terminal end (designated CUL4-L and CUL4-S, respectively). The two forms of CUL4 were differentiated using polyclonal antibodies raised against two distinct antigens. Anti-CUL4(N) was generated against a 19–amino acid peptide present only in the CUL4-L N terminus, and anti-CUL4(C) was generated against the most C-terminal 143 amino acids shared by both the CUL4-L and CUL4-S forms (Figure 1A). Immunoblot analysis using anti-CUL4(N) antibodies detected two CUL4-specific bands in both the wild type and a partial loss-of-function csn5a-2 mutant, which should correspond to the rubylated and unrubylated CUL4-L proteins, based on ratio changes in the csn5a-2 mutant. As expected, anti-CUL4(C) detected four CUL4-specific bands, which should correspond to rubylated and unrubylated CUL4-L and CUL4-S (Figure 1B). This result suggests that CUL4 has two translated forms, and both of them are present in both the rubylated and unrubylated states in Arabidopsis. Although it is not clear whether there is a functional difference between the two CUL4 forms, they both contain the two structural domains of cullins as well as the CUL4-specific N-terminal motif involved in DDB1 binding, suggesting that the two forms share the same known biochemical properties (Li et al., 2006). In most of the subsequent studies, we used anti-CUL4(C) to detect all CUL4 protein forms in Arabidopsis, with CUL4-L and CUL4-S collectively designated CUL4. The CUL4-L version of the open reading frame (ORF) was used in all chimeric fusion protein constructs.
Figure 1.
Arabidopsis CUL4 Protein Forms, Expression Pattern, and Subcellular Localization.
(A) Schemes depicting the two putative CUL4 forms in Arabidopsis and the locations of the two antigens for antibody preparation. CUL4-L and CUL4-S refer to the putative large and small CUL4 proteins, respectively, with translation beginning from different start codons in the same ORF. Peptides N and C show the positions of the two antigens used for antibody preparation. a.a., amino acids.
(B) Unrubylated and rubylated CUL4 in Arabidopsis. Total soluble protein extracts from wild-type Arabidopsis and the csn5a-2 mutant were examined by protein gel blot analysis using anti-CUL4(N) and anti-CUL4(C) antibodies.
(C) CUL4 protein is expressed extensively in Arabidopsis tissues. Total soluble protein extracts from inflorescences (lane 1), stems (lane 2), roots (lane 3), siliques (lane 4), light-grown seedlings (lane 5), dark-grown seedlings (lane 6), cauline leaves (lane 7), and rosette leaves (lane 8) were examined by protein gel blot analysis using anti-CUL4 and anti-CUL1 antibodies. An anti-RPN6 antibody was used as a sample loading control.
(D) CUL4 is likely a nuclear protein. The left two panels show the localization of CUL4 in onion cells in a transient assay. The top left panel shows sGFP-CUL4 localization in epidermal cells, and the bottom left panel shows 4′,6-diamidino-2-phenylindole (DAPI) staining of the same cell to visualize the position of the nucleus (arrowheads). The right two panels show the localization of CUL4 in stable transgenic plants. The top right panel shows EGFP-CUL4 localization in light-grown seedling roots, and the bottom right panel shows DAPI staining of the same field to visualize the position of the nucleus (arrowheads). All of the images were taken using the GFP or DAPI channel of a confocal microscope. Bars = 20 μm.
As a first step to investigate the role of the CUL4 protein in plant development, the expression pattern of CUL4 in different tissues was surveyed by immunoblot analysis and compared with the known expression pattern of CUL1 (Figure 1C). We found that the CUL4 protein is present in relatively high abundance in all tissues examined except for the silique. Both onion (Allium cepa) transient and stable Arabidopsis transgenic assays with a green fluorescent protein (GFP)–labeled CUL4 fusion protein revealed that GFP-CUL4 was always localized in the nucleus (Figure 1D).
Initial Evidence for Arabidopsis CUL4-Containing E3 Ubiquitin Ligase(s)
To test whether Arabidopsis CUL4 is capable of forming E3 ubiquitin ligases as reported in other organisms, we cloned CUL4 into a yeast two-hybrid assay vector and observed its interactions with predicted components using a known CUL1-specific binding protein, ASK1, as a control. As shown in Figure 2A, CUL4 interacts with RBX1, DDB1, and COP10 but not with the CUL1-specific adaptor protein ASK1 in yeast, supporting the possible formation of a CUL4-RBX1-CDD E3 ligase complex.
Figure 2.
Evidence for a CUL4-RBX1-CDD E3 Ligase in Arabidopsis.
(A) CUL4 interacts with RBX1, DDB1a, and COP10, as shown in a yeast two-hybrid assay. The previously known interaction of DDB1a and COP10 was used as a positive control. The β-galactosidase activity resulting from the interaction is shown. Error bars represent sd (n = 4).
(B) CUL4 associates with TAP-RBX1 but not TAP-ASK1 in vivo. Total flower protein extracts prepared from wild-type Arabidopsis, 35S:TAP-ASK1, and 35S:TAP-RBX1 transgenic Arabidopsis plants were incubated with IgG-coupled Sepharose. The precipitates and total extracts were subjected to immunoblot analysis with antibodies against CUL4, CUL1, Myc, and RPN6. Arrowheads indicate protein positions, and T indicates total protein extract. Anti-RPN6 antibody was used as a pull-down control.
(C) CUL4 protein level is different among independent 35S:flag-CUL4 transgenic plants. Total protein was extracted from light-grown seedlings of the wild type and six independent transgenic lines. Protein gel blot analysis was subsequently performed using anti-flag, anti-CUL4, and anti-CUL1 antibodies. An anti-RPN6 antibody was used as a sample loading control.
(D) Flag-CUL4 associates with COP10 in vivo. Total flower protein extracts prepared from wild-type and 35S:flag-CUL4 transgenic Arabidopsis (line 10) were incubated with anti-flag antibody–conjugated agarose (α-flag). The precipitates and total extracts were subjected to immunoblot analysis with antibodies against flag and COP10. An unspecific band was used as a pull-down control. COP10(F) and COP10(P) indicate the full-length and partially degraded COP10 protein forms, respectively.
(E) CUL4 associates with flag-COP10 in vivo. Total flower protein extracts prepared from wild-type and 35S:flag-COP10 transgenic Arabidopsis were incubated with anti-flag antibody–conjugated agarose (α-flag). The precipitates and total extracts were subjected to immunoblot analysis with antibodies against flag and CUL4. An unspecific band was used as a pull-down control.
Specific in vivo association of CUL4 and RBX1 was then investigated by immunoprecipitation in two stable transgenic lines, one expressing tandem affinity purification (TAP)–tagged RBX1 (TAP-RBX1) and the other expressing tagged ASK1 (TAP-ASK1) as a control. When TAP-RBX1 and TAP-ASK1 were immunoprecipitated using IgG beads from transgenic plant extracts, CUL4 coimmunoprecipitated with TAP-RBX1 but not with TAP-ASK1 (Figure 2B). This result suggests that CUL4 associates with RBX1 but not ASK1 in Arabidopsis.
We also investigated CUL4 association with COP10 by coimmunoprecipitation using anti-flag antibodies to flag-CUL4, as our anti-CUL4 antibodies were not suitable for immunoprecipitation assays. We constructed a chimeric gene encoding the CUL4 protein with a triple FLAG epitope tag fused at the N terminus (flag-CUL4) driven by a constitutive cauliflower mosaic virus 35S promoter. After introducing the construct into Arabidopsis in a wild-type background, a transgenic line expressing a high level of the flag-CUL4 fusion protein (Figure 2C, line 10) was selected for immunoprecipitation experiments. The flag-CUL4 showed an association with full-length 21-kD COP10 from the transgenic plant extracts, whereas no interaction with the partially degraded 16-kD COP10 form was observed (Figure 2D). This finding suggests that only intact COP10 interacts with CUL4 in Arabidopsis, although the truncated form retains the ability to form the CDD complex (Yanagawa et al., 2004). Reverse coimmunoprecipitation using flag-COP10 (full-length) transgenic plant extracts (Yanagawa et al., 2004) also confirmed that CUL4 associates with flag-COP10 (Figure 2E). The reciprocity between CUL4 and COP10 coimmunoprecipitation suggests that CUL4 associates with the CDD complex, because endogenous COP10 is largely associated with DDB1 and DET1 within the CDD complex in vivo. Because CUL4 is capable of interacting directly with DDB1 and COP10 in our yeast two-hybrid assay, the CUL4 association with the CDD complex in vivo may be mediated by the direct interaction of CUL4 with COP10 and the DDB1 subunits (Figure 3H).
Figure 3.
CUL4-RBX1-CDD E3 Ligase Can Be Reconstituted in Vitro.
(A) Glutathione affinity purification of the recombinant CDD complex overexpressed in the baculovirus–insect cell system. The purified complex was analyzed by SDS-PAGE.
(B) and (C) Further purification of the recombinant CDD complex by size exclusion chromatography. The TEV-cleaved GST tag and the excess COP10 protein were completely separated from the CDD complex, which migrated as a single peak species on the Superdex 200 gel filtration column (B). The fractions from the CDD complex peak were examined by SDS-PAGE (C) and appear to be approximately equimolar for the three components.
(D) and (E) CUL4-RBX1 can interact with the CDD complex to form a holo E3 ligase complex in vitro. The five-protein complex CUL4-RBX1-COP10-DDB1a-DET1 migrated as a large single-peak species on a Superdex 200 gel filtration column (D). The star indicates where COP10-DDB1a-DET1 migrates (as see in [B]). The fractions from the holo E3 ligase complex peak appear to have equimolar ratios of the five proteins, as revealed by SDS-PAGE analysis (E).
(F) and (G) In vitro ubiquitination assays with recombinant GST-RBX1-CUL4 and CDD complexes. GST-RBX1-CUL4 mediates ubiquitin chain formation, and the CDD complex can significantly enhance this process. The identical blots were probed with antibodies against flag epitope (F) or GST (G).
(H) A proposed Arabidopsis CUL4-RBX1-CDD E3 ligase supported by our data.
Reconstituted CDD Complex Forms a Stable Holo Complex with CUL4-RBX1 and Enhances Recombinant CUL4-RBX1–Mediated Polyubiquitin Chain Assembly in Vitro
Several assays were performed to probe the ubiquitination activity of this putative CUL4-containing E3 ligase. The component complexes were reconstituted by expressing and purifying the glutathione-S-transferase (GST)–fused RBX1 and CUL4 core complex and the CDD complex separately using the baculovirus/insect cell coexpression system. In the case of the CDD complex, we have made baculovirus samples that overexpress Arabidopsis GST-COP10, DDB1, and DET1 in insect cells. The expression of COP10 as a GST fusion protein allowed for a one-step glutathione affinity purification of the reconstituted complex. As shown in Figure 3A, coexpression of GST-COP10 together with both DDB1 and DET1 produced a soluble CDD complex, which could be affinity-purified from insect cell lysate. By contrast, when either DDB1 or DET1 was individually coexpressed with GST-COP10, neither associated with the GST-COP10 to form a subcomplex using the same one-step purification, suggesting that DDB1 and DET1 are both required for the formation of a soluble complex with COP10. After GST was cleaved off from COP10 within the reconstituted CDD complex, we were able to isolate an essentially native-like CDD complex by size exclusion chromatography (Figures 3B and 3C). The three proteins in the CDD complex appear to be in a 1:1:1 ratio.
Furthermore, to test whether CUL4, RBX1, COP10, DDB1, and DET1 can form an E3 complex, CUL4 and RBX1 were coexpressed in insect cells with RBX1 fused to GST. After glutathione affinity purification and thrombin cleavage of GST, the RBX1-CUL4 complex was mixed with purified recombinant CDD complex. This mixture was subsequently applied to a Superdex 200 size exclusion column. The five proteins were all eluted together as a monodispersive peak larger than either the CDD or CUL4-RBX1 complex. The five proteins in the reconstituted complex appear to be in a 1:1:1:1:1 molar ratio (Figures 3D and 3E). However, when COP1 was coexpressed with DDB1, DET1, and COP10, no association of COP1 with the soluble reconstituted CDD complex was detected (data not shown), indicating that COP1 is not an integrated part of this CUL4-RBX1-CDD E3 ligase complex.
To investigate whether the CDD complex can enhance the CUL4-RBX1 core E3 function, we performed an in vitro ubiquitination assay using recombinant CDD and CUL4-RBX1 complexes purified from insect cells and bacterially expressed Arabidopsis Ubc8, a protein closely related to Sc Ubc4/5 and Hs UbcH5A/B/C that has E2 function. Because the potential physiological substrate(s) of an Arabidopsis CUL4-RBX1 E3 remains unknown, we monitored polyubiquitin chain assembly in the reaction mixture. As shown in Figure 3F, the GST-RBX1-CUL4 complex was able to promote polyubiquitin chain formation in the presence of Arabidopsis Ubc8, detected as a ladder of ubiquitin-containing bands on an SDS-PAGE gel. Parallel immunoblot analysis using anti-GST antibody further confirmed that most of the ubiquitin chains were assembled onto GST-RBX1 (Figure 3G). Strikingly, addition of the purified recombinant CDD complex markedly enhanced the autoubiquitination of the GST-RBX1-CUL4 complex, indicating that the CDD complex can effectively increase the ubiquitination activity of the CUL4-RBX1-E2 system. This assay also confirmed that CUL4-RBX1 can form a rudimentary E3 ligase (Figure 3H).
CUL4-Containing E3 Ligases Are Modulated by CAND1 and the CSN
We then investigated whether or not the known cullin regulators CSN and CAND1 (for Cullin-Associated and Neddylation-Dissociated1) could interact with Arabidopsis CUL4 using the same yeast two-hybrid assay. As shown in Figure 4A, CUL4 interacts with CAND1 and the CSN5, CSN3, CSN8, and CSN4 subunits of the CSN in yeast.
Figure 4.
CUL4-Containing E3 Ligases Are Regulated by CSN and CAND1 through Physical Association.
(A) Evidence for direct CUL4 interaction with CSN and CAND1 by yeast two-hybrid assay. The previously known interaction of DDB1a and COP10 was used as a positive control. The β-galactosidase activity resulting from the interaction is shown. Error bars represent sd (n = 4).
(B) The flag-CUL4 associates with three representative CSN subunits (CSN3, CSN4, and CSN5) in vivo. Total flower protein extracts prepared from wild-type and 35S:flag-CUL4 transgenic Arabidopsis were incubated with anti-flag antibody–conjugated agarose (α-flag). The precipitates and total extracts were subjected to immunoblot analysis with antibodies against flag, CSN3, CSN4, and CSN5. An unspecific band was used as a pull-down control.
(C) The flag-CAND1 associates with CUL4 but not CSN in vivo. Total flower protein extracts prepared from wild-type and 35S:flag-CAND1 transgenic Arabidopsis were incubated with anti-flag antibody–conjugated agarose (α-flag). The precipitates and total extracts were subjected to immunoblot analysis with antibodies against flag, CUL4, CSN3, and CSN5. An unspecific band was used as a pull-down control.
(D) The CUL4 protein is subjected to modification by RUB in vivo. Total protein was extracted from light-grown seedlings of the wild type, csn5a-2, csn1-1 (fus6-1), csn3-1 (fus11-1), cand1-1, cop10-1, cop1-6, and det1-1, and protein gel blot analysis was subsequently performed using anti-CUL4 and anti-CUL1 antibodies. Anti-RPN6 was used as a sample loading control.
The implied association of the CSN with the CUL4-containing E3 ligase in Arabidopsis was then tested by coimmunoprecipitation assays containing flag-CUL4 and representative subunits of the CSN. Using transgenic plant extracts, we were able to detect the presence of these CSN subunits in the flag-CUL4 precipitate (Figure 4B). This result confirmed CUL4's association with the CSN in Arabidopsis.
To test for a possible association of CAND1 and CUL4 in vivo, coimmunoprecipitation experiments were performed using a flag-CAND1 transgenic line (Feng et al., 2004). As shown in Figure 4C, immunoprecipitation of flag-CAND1 using anti-flag antibody can specifically pull down CUL4 in the precipitate. This result suggests that, as in mammals, CAND1 can associate with CUL4 in Arabidopsis. At the same time, the CSN could not be pulled down by flag-CAND1. This finding indicates that CSN and CAND1 may bind CUL4 in a mutually exclusive manner, consistent with previous evidence given for CUL1, CSN, and CAND1 interactions in mammalian cells (Min et al., 2005).
It has been proposed that the RUB/NEDD8 modification of CUL1 (rubylation) acts as a positive regulator of E3 ligase activity, enhancing E2 recruitment and ubiquitination of the substrate. In the case of CUL4, rubylation and derubylation have also been shown to be required for CUL4's normal function in mammalian cells (Higa et al., 2003). To test whether Arabidopsis CUL4 is covalently modified by RUB and to investigate a possible role for the CSN as well as the CDD and COP1 complexes and CAND1 in this modification, we examined the RUB modification status of CUL4 in protein extracts from the representative csn, cand1, cop10, cop1, and det1 mutants (Figure 4D). In the weak mutant csn5a-2, some of the small bands shifted to the position of the big bands, whereas in the csn null mutants csn1-1 (fus6-1) and csn3-1 (fus11-1), all of the cellular CUL4 showed up as a broad larger band. It is likely that the upper broad band contains a mixture of the rubylated CUL4-L and CUL4-S as a result of the low gel resolution. This demonstrates conclusively that CUL4 is subject to reversible modification by RUB similar to the other cullins. In cand1-1, cop10-1, det1-1, and cop1-6 mutants, CUL4's rubylation status is similar to that in wild-type Arabidopsis, implying that CAND1 and the CDD and COP1 complexes are not directly involved in CUL4 RUB modification.
Reduction of CUL4 Abundance Results in Constitutive Photomorphogenesis in Arabidopsis Seedlings
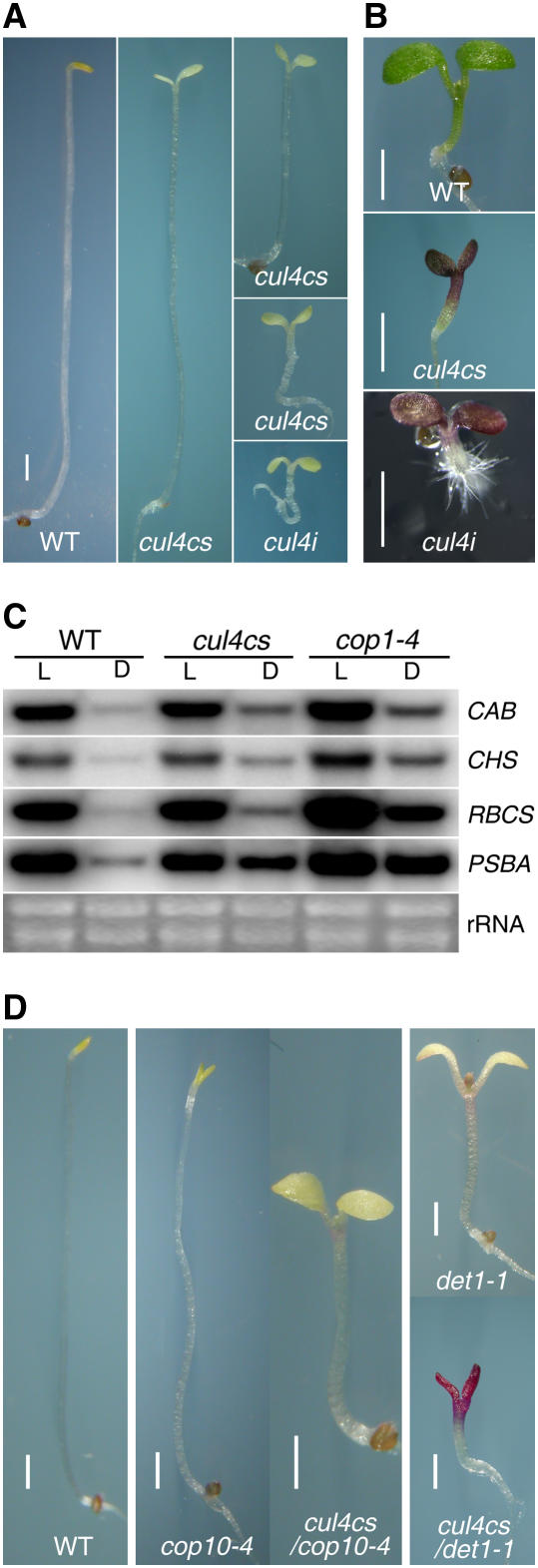
If the CDD complex acts through the CUL4-RBX1-CDD E3 ligase to achieve its repression of photomorphogenesis, it is plausible that CUL4 reduction or loss of function would result in a constitutive photomorphogenic phenotype similar to that of mutations in the CDD complex components COP10 and DET1. As a first step to test this notion, we introduced the flag-fused CUL4 ectopic expression transgene and CUL4 RNA interference (RNAi) constructs under the control of 35S promoters into wild-type Arabidopsis to generate stable transgenic plants with altered CUL4 protein levels. Interestingly, 10 of the 18 flag-CUL4 transgenic lines we obtained exhibited varying degrees of the constitutive photomorphogenic phenotype in their T2 and subsequent progeny, with opening and expansion of cotyledons as well as a variation in the length of the hypocotyls when grown in the dark (Figures 2C and 5A). When grown in the light, these same lines yielded hyperphotomorphogenic seedlings with high levels of anthocyanin, similar to the pleiotropic cop/det/fus mutants (Figure 5B). We also obtained a large number of constitutive photomorphogenic seedlings in the primary screen of constitutive CUL4 RNAi transgenic plants in the T1 generation (Figures 5A and 5B). However, these RNAi seedlings were lethal after the seedling stage and all failed to produce seeds.
Figure 5.
CUL4 Is Involved in Repressing Photomorphogenesis.
(A) CUL4 mediates a cop-like phenotype in the dark. The three CUL4 cosuppression (cul4cs) seedlings are 7-d-old dark-grown 35S:flag-CUL4 transgenic seedlings exhibiting different length hypocotyls and different degrees of opening and expansion of cotyledons (lines 1, 2, 7, and 8 in Figure 2C). The CUL4 RNAi (cul4i) yields dark-grown 35S:CUL4i transgenic seedlings exhibiting short hypocotyls and open and fully expanded cotyledons. A wild-type (Columbia-0) seedling is shown at left. Bar = 1 mm.
(B) CUL4 mediates a fusca phenotype in the light. The middle and bottom panels show 5-d-old light-grown cul4cs and cul4i seedlings, respectively, with hyperphotomorphogenic morphology and a high level of anthocyanin accumulation. The top panel shows a wild-type (Columbia-0) seedling. Bars = 1 mm.
(C) RNA gel blot analysis of steady state RNA levels of nucleus- and plastid-encoded genes. RNA levels from wide-type, cul4cs, and cop1-4 plants were analyzed. Total RNA was isolated from seedlings grown for 7 d in the light (L) or dark (D). Equal amounts of total RNA from the different plant samples were used, and four identical blots were hybridized and labeled with gene-specific probes for four different genes: CAB, CHS, RBCS, and PSBA. The rRNA band pattern was used to show equal loading.
(D) Reduction of CUL4 enhances the phenotypes of weak cop10 and det1 alleles in the dark. Different Arabidopsis lines (labeled at bottom) were grown in complete darkness for 7 d. Bars = 1 mm.
Immunoblot analysis was used to examine CUL4 protein levels in the 35S:flag-CUL4 transgenic lines with or without a cop-like phenotype. As shown in Figure 2C, all 10 lines expressing a cop-like phenotype (such as lines 1, 2, 7, and 8) have reduced levels of CUL4 (the endogenous CUL4 together with the flag-CUL4 fusion protein). All of the lines lacking a cop-like phenotype expressed the endogenous CUL4 together with a flag-CUL4 fusion protein at approximately the endogenous CUL4 levels in wild-type Arabidopsis (such as lines 10 and 13). This correlation of the cop-like phenotype and reduced CUL4 protein level was maintained in the subsequent generation, with some progeny exhibiting higher CUL4 abundance and loss of the cop-like phenotype. Thus, the cop-like phenotype seems to be a consequence of transgene-mediated cosuppression of CUL4, and lines with this phenotype were designated CUL4 cosuppression (cul4cs) lines. Changes in protein abundance in these transgenic lines appear to be limited to CUL4, because the levels of CUL1 and other tested proteins were unchanged and independent of the cop-like phenotype (Figure 2C).
Cosuppression of CUL4 Results in Increased Gene Expression of Light-Inducible Genes in Darkness
We further examined the effect of CUL4 cosuppression on light-inducible gene expression in dark-grown cul4cs seedlings. The mRNA levels of four representative light-regulated nucleus- and plastid-encoded genes were examined by RNA gel blot analysis. As shown in Figure 5C, these four representative genes are all expressed at higher levels in dark-grown cul4cs seedlings compared with their wild-type counterparts, similar to the cop1-4 mutants. This result indicates that, as in the cop/det/fus mutations, inhibition of CUL4 function also leads to constitutive expression of light-inducible genes in darkness.
Reduction of CUL4 Function Results in Synergistic Enhancement of the cop10 and det1 Weak Mutant Phenotypes
To further substantiate the functional role of CUL4 in repressing photomorphogenic development, we tested anticipated genetic interactions between CUL4 and COP10 and DET1. Therefore, we introduced the cosuppressing trait cul4cs (Figure 2C, line 2) into cop10-4 and det1-1 mutants, both of which are nonlethal and weak alleles. Remarkably, the dark-grown double mutants cul4cs cop10-4 and cul4cs det1-1 have a much more exaggerated photomorphogenic phenotype than either of their parental single mutants (Figure 5D). In the case of cul4cs det1-1, the double mutant phenotypes essentially mimic those of the det1 null mutants. These data strongly suggest that CUL4 works together with COP10 and DET1 in mediating the repression of photomorphogenesis in darkness.
The CUL4-RBX1-CDD E3 Ligase Plays a Critical Role in COP1-Dependent Degradation of the Photomorphogenesis-Promoting Transcription Factor HY5 in Darkness
Previous reports have shown that the CDD complex associates with the COP1 E3 ligase complex and that the two complexes act together in repressing photomorphogenesis (Yanagawa et al., 2004). In light of findings in this work, the reported CDD and COP1 complex association may actually correspond to the association of the CUL4-RBX1-CDD E3 ligase and the COP1 E3 ligase complexes. To test this hypothesis, coimmunoprecipitation of flag-CUL4 and COP1 was used to detect the in vivo association of CUL4 and COP1. As shown in Figure 6A, COP1 can be readily coimmunoprecipitated with flag-CUL4 from flag-CUL4 transgenic plant extracts, supporting an in vivo association of the CUL4-RBX1-CDD E3 ligase and the COP1 E3 ligase complexes in Arabidopsis. This complex-to-complex association may be mediated by direct COP10–COP1 protein contacts, as suggested by a yeast two-hybrid assay (Figure 6B) (Suzuki et al., 2002).
Figure 6.
The CUL4-RBX1-CDD E3 Ligase Can Enhance COP1-Mediated HY5 Degradation.
(A) The flag-CUL4 associates with COP1 in vivo. Total flower protein extracts prepared from wild-type and 35S:flag-CUL4 transgenic Arabidopsis were incubated with anti-flag antibody–conjugated agarose (α-flag). The precipitates and total extracts were subjected to immunoblot analysis with antibodies against flag and COP1. A nonspecific band was used as a pull-down control.
(B) A possible direct COP10 and COP1 interaction is supported by a yeast two-hybrid assay. The previously known interaction of DDB1a and COP10 was used as a positive control. The β-galactosidase activity resulting from the interaction is shown. Error bars represent sd (n = 4).
(C) HY5 is degraded less efficiently in the cul4cs mutants than in wild-type Arabidopsis. Four-day-old light-grown seedlings of wild-type Arabidopsis and cul4cs plants were transferred to complete darkness. Samples were collected at different time points starting from the transfer (indicated at top) and blotted with anti-HY5 and anti-RPN6 antibodies.
To further test whether CUL4 directly affects COP1 activity, we examined COP1-mediated degradation of the photomorphogenesis-promoting transcription factor HY5 in cul4cs seedlings. It has been shown that the extent of photomorphogenic development is correlated directly with the abundance of HY5 (Osterlund et al., 2000). The COP1 ubiquitin E3 ligase specifically targets HY5 for ubiquitination and subsequent degradation by the 26S proteasome in darkness (Saijo et al., 2003). A kinetic analysis was performed of HY5's degradation rate after transfer of the light-grown cul4cs and wild-type seedlings to darkness (Figure 6C). HY5 protein degradation in darkness was markedly slower in cul4cs transgenic plants compared with wild-type plants, indicating that CUL4 is important for COP1-mediated HY5 degradation in darkness.
Arabidopsis CUL4 Is Also Involved in Many Other Developmental Processes
As in other organisms, the CUL4-RBX1 E3 core may interact with adaptors other than the CDD complex to form distinct E3 ligases in Arabidopsis. Indeed, an Arabidopsis DDB1 mutant resulted in an embryo-lethal phenotype, different from the cop-like phenotype of the cop10 and det1 mutants (Schroeder et al., 2002), implying that DDB1 participates in other developmental pathways, potentially by forming other distinct functional complexes, as has been shown in mammals (Groisman et al., 2003; Hu et al., 2004). Thus, CUL4 cosuppression lines should display other phenotypes in addition to the defect in repression of photomorphogenesis conferred by the CUL4-RBX1-CDD E3 ligase. Indeed, examination of CUL4 cosuppression plants at different stages of Arabidopsis growth and development reveals multiple developmental defects.
At the seedling stage, all of the cul4cs plants were smaller than their wild-type counterparts. Frequently, the cul4cs plants had seedlings with triple cotyledons (Figure 7A). In the vegetative phase after the seedling stage, cul4cs plants had rosettes smaller than the wild type, with various irregular shapes and patterns (Figure 7B). In the reproductive phase, the phenotypic differences between wild-type and cul4cs plants became even more drastic. The cul4cs plants exhibited a severe dwarfism, accompanied by an increase in the number of secondary inflorescences and leaves. When the photoperiod was changed from the normal 16 h of light/8 h of darkness to 12 h of light/12 h of darkness, the number of secondary inflorescences of cul4cs plants was increased and that of the wild type remained unchanged (Figure 7C). Notably, the cul4cs plants invariably produced lobed, asymmetrical, or curled rosette leaves (Figure 7D). Different cul4cs lines displayed different degrees of severity in the abnormal leaf shape phenotype. Moreover, cul4cs flowers and siliques were smaller than their wild-type counterparts (Figures 7E and 7F).
Figure 7.
Multifaceted Developmental Defects of cul4cs Mutant Plants.
(A) Seven-day-old light- and dark-grown wild-type and cul4cs seedlings. L and D indicate light and dark, respectively.
(B) Three-week-old wild-type and cul4cs plants under the 16L/8D (16 h of light/8 h of dark each day) condition.
(C) Eight-week-old wild-type plants under the 16L/8D condition and cul4cs plants under both the 16L/8D and 12L/12D conditions. For (A) to (C), photographs in the same panel were taken at the same magnification.
(D) Rosette leaves from 5-week-old cul4cs plants. The bottom right image was taken with a twofold magnification compared with the others.
(E) Comparison of wild-type and cul4cs flowers.
(F) Comparison of wild-type and cul4cs siliques.
DISCUSSION
This work provides the biochemical and functional analysis of Arabidopsis CUL4 and describes a biochemical mechanism for the coordinated regulation of photomorphogenesis. Since the first COP/DET/FUS protein was identified in 1989 (Chory et al., 1989), many proteins and their associated complexes have been identified and characterized, but how all are choreographed to accomplish the light-regulated developmental switch has remained a matter of speculation. Our data show that an E3 ligase, which comprises Arabidopsis CUL4, RBX1, and the CDD complex, plays an essential and central role in repressing photomorphogenesis through biochemical interactions with CSN and COP1. We further show that Arabidopsis CUL4 may be involved in many other developmental processes in addition to the repression of photomorphogenesis.
The CUL4-RBX1-CDD E3 Ligase in Arabidopsis and Its Interactions with the COP9 Signalosome
Our studies confirmed that CUL4 interacts with RBX1 and the CDD complex to form an E3 ligase in plants. The in vitro reconstitution assay suggested that the presence of the CDD complex enhanced the polyubiquitin chain formation catalyzed by the CUL4-RBX1 core. Our data are consistent with previous findings that CUL4 always associates with RBX1 and a DDB1-containing complex in forming ubiquitin E3 ligase complexes in mammalian systems (Groisman et al., 2003; Hu et al., 2004). The Arabidopsis CDD complex fills the role of the DDB1-containing complex and also includes DET1 and COP10, possibly a plant-specific ubiquitin-conjugating enzyme variant (Yanagawa et al., 2004). When reconstituting this complex in insect cells, we further observed that DDB1 and DET1 are both required for the formation of a soluble complex with COP10. This is consistent with our previous observation that the CDD complex was undetectable in the det1-1 mutant (Yanagawa et al., 2004). The ability to reconstitute the five-protein complex (CUL4-RBX1-COP10-DDB1a-DET1) in vitro and the presence of recombinant CDD complex markedly enhanced CUL4-RBX1–mediated polyubiquitin chain assembly, indicating that CUL4-RBX1 may work through association with the CDD complex to form a functional E3 ligase in Arabidopsis that targets a specific set of substrates involved in photomorphogenesis.
A study in mammalian cells suggested that DET1 and COP1 could form an adaptor module to accomplish the observed CUL4-RBX1-DDB1–based E3 ligase function (Wertz et al., 2004). Although our data support an association between the CUL4-RBX1-CDD E3 ligase and COP1 E3 ligase complexes, Arabidopsis COP1 is unlikely to be a direct component of a CUL4-RBX1-CDD E3 ligase complex for several reasons. First, when Arabidopsis COP10, DDB1, DET1, and COP1 were coexpressed in insect cells, only COP10, DDB1, and DET1 could form a stable ternary complex, whereas COP1 was excluded (Figure 3A) (X. Tang and N., Zheng, unpublished data). Second, in Arabidopsis, the size of the CDD complex is ∼300 kD, whereas the majority of COP1 is present in an ∼700-kD protein complex (Saijo et al., 2003; Yanagawa et al., 2004). Third, Arabidopsis β-glucuronidase (GUS)-COP1 and GFP-COP1 were enriched in the nuclear speckles in darkness, and some of them moved into the cytoplasm when cells were exposed to light in a cell type–specific manner (von Arnim and Deng, 1994; Ang et al., 1998). On the other hand, Arabidopsis GUS-DET1 and GFP-DET1 were found to localize exclusively in the nucleus (Pepper et al., 1994; Schroeder et al., 2002).
As in the case of all other cullins, Arabidopsis CUL4's derubylation was also mediated by the CSN (Figure 4). As in the case of Arabidopsis CUL1 (Schwechheimer et al., 2001), a rubylation and derubylation cycle for CUL4 may also be important for the CUL4-based E3 ligase function in vivo. A possible mechanism for the required CSN role in CUL-based E3 ligase activity was suggested in a recent study in both yeast and mammals that showed that the CSN plays a role in maintaining the stability of the adaptor in cullin-RING ubiquitin ligases (Wee et al., 2005). In the case of Arabidopsis, the CDD complex seems to fit the role of the adaptor for a CUL4-RBX1–based E3 ligase. Therefore, we might infer that the CSN is capable of stabilizing the CDD complex, a notion consistent with our prior finding that the CDD complex was found to be unstable in csn8-1 (cop9-1) and csn1-1 (fus6-1), which abolished CSN function (Suzuki et al., 2002).
The CDD Complex of CUL4-RBX1-CDD E3 Ligase Is a Unique Adaptor with Novel Biochemical Activity
Interestingly, the CDD complex is a unique kind of adaptor, unlike any reported to date for cullin-based E3 ligases. This is because in addition to its assumed role in substrate recognition, the CDD complex also has intrinsic E2 enhancement activity (Yanagawa et al., 2004). This E2 enhancement function contributes to the activity of an E3 ligase in addition to its known function of conferring a favorable spatial configuration of the E3 substrate and E2, which is typical of a normal E3 adaptor. Thus, the CUL4-RBX1-CDD E3 ligase in Arabidopsis has an additional novel catalytic function not found in any of the other known cullin-based E3 ligases.
Role of CUL4-RBX1-CDD E3 Ligase in the Repression of Photomorphogenesis
In this study, we provide strong evidence for a role of the CUL4-RBX1-CDD E3 ligase in repressing photomorphogenesis in Arabidopsis. Given that a reduction of the CUL4 protein level results in a similar cop phenotype in darkness, we can now add CUL4 to the list of known repressors of photomorphogenesis, which includes the COP10 and DET1 subunits of the CDD complex. The cop-like phenotype of the CUL4 reduction-of-function seedlings is evident in both plant morphology and physiology and in the light regulation of gene expression. Thus, in many respects, CUL4 represents a new COP/DET/FUS-like protein. We also provide genetic evidence that CUL4 acts together in the same pathway with COP10 and DET1 (Figure 5D). This is evident in the strong synergistic enhancement of phenotype in double mutant lines, which results from the combined effects of a CUL4 reduction-of-function mutation and the weak alleles of the COP10 and DET1 genes. The similar phenotype and observed genetic interaction between CUL4 and COP10 or DET1 strongly support the conclusion that the CUL4-RBX1-CDD E3 ligase functions in the repression of photomorphogenesis.
The association between Arabidopsis CUL4 and COP1 suggests that the CUL4-RBX1-CDD and COP1 E3 ligases likely act together in the repression of photomorphogenesis (Figure 6A). This conclusion is further supported by the fact that CUL4 is important for the COP1 E3 ligase–targeted degradation of the photomorphogenesis-promoting transcription factor HY5 (Figures 6B and 6C) (Osterlund et al., 2000; Saijo et al., 2003).
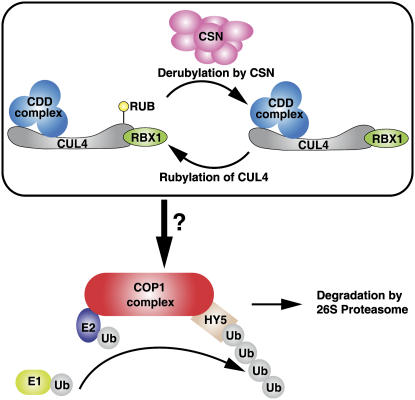
Together, our data suggest a general framework for how all three known COP/DET/FUS protein complexes work in concert to repress photomorphogenesis (Figure 8). CUL4-RBX1 associates with the CDD complex to form an E3 ligase, whose function depends on the derubylation activity of the CSN. The CUL4-RBX1-CDD E3 ligase may repress photomorphogenesis through enhancing COP1 activity as an E3 ligase. Our working model suggests that the CUL4-RBX1-CDD E3 ligase positively regulates COP1 activity, although the precise biochemical mechanism remains unclear. The fact that COP10, one of the CUL4-RBX1-CDD E3 ligase components, was observed to interact directly with COP1 in a yeast two-hybrid assay suggests that these two E3 ligase complexes are capable of associating in vivo. This view is supported by the results of coimmunoprecipitation assays as well (Figure 6A) (Yanagawa et al., 2004). It is plausible that the CUL4-RBX1-CDD E3 ligase may mediate some forms of ubiquitination (such as monoubiquitination or Lys-63 linkage polyubiquitination) on COP1 or its complex component(s) to enhance its E3 ligase activity. Alternatively, the CUL4-RBX1-CDD E3 ligase may mediate the degradation of a repressor closely associated with the COP1 E3 ligase complex. The latter hypothesis may help to explain why, after the loss of COP10, the peak of the COP1 complex(es) was shifted to a larger fraction (Saijo et al., 2003).
Figure 8.
A Working Model of CUL4 and the Three-Protein Complex in Mediating the Repression of Photomorphogenesis.
In the dark, the CUL4-RBX1-CDD complex positively regulates COP1-mediated degradation of light-regulated transcriptional factors such as HY5. CUL4-RBX1-CDD E3 ligase activity is regulated through the rubylation and derubylation cycle, with CSN mediating derubylation. It remains unknown how this CUL4-based E3 ligase positively regulates COP1 E3 ligase activity.
CUL4 Plays Other Roles in Arabidopsis Development
The importance of CUL4-based E3 ligases has only recently been recognized. These regulatory complexes have since been implicated in several cellular and development pathways, from cell cycle control to genomic stability, in organisms as diverse as fission yeast and mammals. Although our major focus in this study is one particular CUL4-based E3 ligase, the CUL4-RBX1-CDD E3 ligase, and its role in repressing photomorphogenesis, the observed range of defects in cul4cs plants at different developmental stages (Figure 7) clearly indicates that other CUL4-based E3 ligases may serve important functions. It is of interest that several phenotypes of the cul4cs plants are very similar to phenotypes of the CSN weak mutants (Peng et al., 2001a, 2001b; Schwechheimer et al., 2001; Gusmaroli et al., 2004). Various genetic studies suggest that the CSN is necessary for the optimal activity of multiple cullin-containing E3 ligases (Schwechheimer et al., 2001; Feng et al., 2003; Groisman et al., 2003; Wang et al., 2003). It is most likely that Arabidopsis CSN and CUL4 also work together to mediate many other development processes besides photomorphogenesis.
Some phenotypes observed in cul4cs mutant plants resemble defects resulting from other types of E3 ligase mutations. For example, the outgrowth of the secondary inflorescences in wild-type plants is inhibited by the apical dominance of the primary shoot apex through the production of the plant hormone auxin. It has previously been shown that plants with reduced CSN levels also exhibit a decreased auxin response (Schwechheimer et al., 2001). The phenotype of lost apical dominance was thought to result from defects in groups of CUL1-based E3 ubiquitin ligases. In our study, we also observed similar apical dominance defects in cul4cs mutants even though CUL1 protein level was not affected (Figure 2C). Therefore, CUL4 may also participate in the auxin response. Elucidating the roles of other CUL4-based E3 ligases will be an important aim of future research.
METHODS
Plant Material and Growth Conditions
The wild-type Arabidopsis thaliana plants used in this study were of the Columbia-0 ecotype. All Arabidopsis growth conditions were essentially as described (Feng et al., 2004) unless specified otherwise. Mutants and transgenic lines are as follows: csn5a-2 (csn5aT) (Gusmaroli et al., 2004), csn1-1 (fus6-1) (Castle and Meinke, 1994), csn3-1 (fus11-1) (Peng et al., 2001b), cand1-1 (Feng et al., 2004), cop10-1 (Wei et al., 1994), cop10-4 (Suzuki et al., 2002), det1-1 (Pepper et al., 1994), cop1-4 and cop1-6 (McNellis et al., 1994), 35S:TAP-RBX1 and 35S:TAP-ASK1 (Rubio et al., 2005), 35S:flag-CAND1 (Feng et al., 2004), and 35S:flag-COP10 (Yanagawa et al., 2004).
Arabidopsis CUL4 and DDB1 cDNA Clones
The full-length ORF of CUL4-L was amplified by RT-PCR from wild-type Arabidopsis seedlings using the forward primer 5′-CCGGAATTCGGTACCATGTCTCTTCCTACCAAACG-3′ and the reverse primer 5′-ATAAGAATGCGGCCGCCTAAGCAAGATAATTGTATA-3′. The EcoRI/NotI fragment of the PCR product was cloned into pEG202 (Origene Technologies) and then sequenced. This construct is designated pEG-CUL4 and served as the PCR template for all subsequent cloning of CUL4 into other vectors. Arabidopsis has two highly identical (likely functionally redundant) DDB1 genes (Schroeder et al., 2002), and the full-length cDNA clone corresponding to DDB1a was used for all of our analyses described in this work.
Generation of Transgenic Arabidopsis Plants
A KpnI/SalI fragment containing the full-length CUL4 ORF was inserted into pF3PZPY122 (Feng et al., 2003). Then, an XbaI/SalI fragment containing the inserted DNA was subcloned into pJIM19(Kan), a plant binary vector that includes a kanamycin resistance marker and the 35S promoter of the Cauliflower mosaic virus. Wild-type Arabidopsis plants were used in the transformation of the 35S:flag-CUL4 transgene. Transgenic plants were selected with kanamycin (100 μg/mL; Sigma-Aldrich).
To make the CUL4 RNAi constructs, a CUL4 cDNA fragment (1707 to 2176 bp) was amplified and cloned into the XhoI/HindIII and EcoRI/SpeI sites of pSK-int (Guo et al., 2003) in antisense/sense directions, respectively. The DNA fragment containing a cauliflower mosaic virus 35S promoter and antisense/sense CUL4 fragment was excised by XhoI/SpeI and cloned into the same site of the binary vector pJIM19(Kan).
To obtain 35S:EGFP-CUL4 transgenic plants, an EcoRI/SalI-digested CUL4 fragment was inserted into pCAMBIA1200/35SP/EGFP binary vector. The resulting construct was then transformed into wild-type Arabidopsis, and transgenic plants were selected with hygromycin (20 μg/mL; Roche).
RNA Gel Blot Analysis
For RNA gel blot analysis, Arabidopsis seedlings were grown in different light conditions as indicated above. Total RNA was extracted using RNeasy plant mini kits (Qiagen). Blotting was performed as described previously (Yang et al., 2005). Fragments of CAB, CHS, RBCS, and PSBA used for probe labeling were generated by PCR.
Antibodies and Immunoblot Assays
We generated two polyclonal CUL4 antibodies designated anti-CUL4(N) and anti-CUL4(C). Anti-CUL4(N), a rabbit polyclonal antibody to CUL4 (N-SDDSSYSSPPMKKAKNDLH-C), was raised against a synthetic peptide derived from the Arabidopsis CUL4 protein. For anti-CUL4(C), the C-terminal region of CUL4 (143 C-terminal residues) was expressed in the vector pET-28b in Escherichia coli strain BL21(DE3) (Novagen). The fusion protein with a 6× His tag was purified and injected into rabbits as antigen. Polyclonal anti-CUL4 antibodies were purified from rabbit serum using the purified GST-CUL4 with glutathione Sepharose 4B (Amersham Pharmacia).
Other primary antibodies used in this study include anti-CUL1 (Schwechheimer et al., 2001), anti-CSN3 (Peng et al., 2001b), anti-CSN4 (Serino et al., 1999), anti-CSN5 (Kwok et al., 1998), anti-COP10 (Yanagawa et al., 2004), anti-COP1 (Saijo et al., 2003), anti-HY5 (Osterlund et al., 2000), anti-RPN6 (Kwok et al., 1999), anti-GST (Sigma-Aldrich), anti-MYC (Sigma-Aldrich), and anti-flag (Sigma-Aldrich).
For the immunoblot analysis, Arabidopsis tissues were homogenized in an extraction buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 0.1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, and 1× complete protease inhibitor (Roche). The extracts were centrifuged twice at 13,000g for 10 min at 4°C, and the protein concentration in the supernatant was determined by Bradford assay (Bio-Rad). Protein samples were boiled in sample buffer, run on SDS-PAGE gels (8, 12, or 15%), and blotted onto polyvinylidene difluoride membranes (Millipore). The blots were probed with different primary antibodies.
Transient Expression in Onion Epidermal Cells
Full-length CUL4 ORF was subcloned into the BsrGI-NotI site of vector pUC18-sGFP (Chiu et al., 1996) to yield the construct pUC18-sGFP-CUL4. The procedure for transient expression in living onion (Allium cepa) epidermal cells using particle bombardment was performed as described previously (Ang et al., 1998). After bombardment, onion cell layers were incubated for 24 h at 22°C in the light. The cell layers were then mounted in DAPI staining buffer and examined by confocal microscopy.
Preparation of Recombinant Arabidopsis CDD and CUL4-RBX1 Complex
Arabidopsis DDB1a, DET1, and CUL4-L cDNAs were subcloned into the pFastBac vector (Invitrogen), whereas Arabidopsis COP10 and RBX1 were subcloned into a customized GST fusion pFastBac-based vector with a TEV cleavage site. Baculovirus samples expressing each individual protein were made according to the manufacturer's protocol (Invitrogen). Amplified virus was then used to transfect a monolayer culture of Hi5 cells with 80% confluence. Two days after the infection, insect cells were harvested and resuspended in a lysis buffer containing 20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 5 mM DTT, and a protease inhibitor cocktail. After brief sonication and centrifugation at 15,000 rpm for 30 min, the supernatant was applied to a gravity glutathione-affinity column, which was subsequently washed with 20 column volumes of lysis buffer. The resin-bound proteins were eluted with 5 column volumes of elution buffer containing 50 mM Tris-HCl, pH 8.0, 200 mM NaCl, and 10 mM reduced glutathione. The final purified products were analyzed by SDS-PAGE with Coomassie Brilliant Blue R 250 staining. The Arabidopsis CDD complex purified by glutathione affinity chromatography was subsequently cleaved by adding TEV protease after the eluted proteins were concentrated to >0.5 mg/mL. The protein samples (CDD itself or mixed together with CUL4-RBX1) were loaded onto a Superdex 200 column (Amersham Biosciences). Fractions corresponding to each elution peak were analyzed by SDS-PAGE followed by Coomassie Brilliant Blue R 250 staining.
In Vitro Ubiquitination Assays
Arabidopsis Ubc8 was overexpressed and purified from E. coli as a His-tagged protein. Ubiquitination reactions were performed in a total volume of 20 μL (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10 mM ATP, 400 ng of GST-E1, 100 ng of His-At Ubc8, and 4 μg of flag-ubiquitin). In addition, 1 μg of GST-RBX1-CUL4 complex and/or 1 μg of CDD complex was added to the reaction. Reactions were incubated for 1 h at room temperature and terminated by the addition of SDS loading buffer. For each reaction, 10 μL of reaction mixture was analyzed by SDS-PAGE followed by protein gel blotting with either the anti-flag or anti-GST antibody.
Yeast Two-Hybrid and in Vivo Pull-Down Analyses
The two-hybrid interaction assay in yeast was performed as described previously (Serino et al., 1999) except that the LexA fusion constructs and the activation domain fusion constructs were cotransformed into yeast strain L40 (Invitrogen). β-Galactosidase activity was assayed as described previously (Kwok et al., 1998). An EcoRI-SalI fragment containing full-length CUL4 ORF was subcloned into pJG4-5 (Origene Technologies) to make the CUL4 prey construct. DDB1a was subcloned into pEG202 (Origene Technologies) to make a bait construct. Each subunit of the COP9 signalosome (Serino et al., 1999), CAND1 (Feng et al., 2004), COP10 (Suzuki et al., 2002), COP1 (Ang et al., 1998), RBX1 (Schwechheimer et al., 2001), and ASK1 (Figueroa et al., 2005) was subcloned into pEG202 and pJG4-5 as described (Origene Technologies).
The flag antibody pull-down and IgG pull-down experiments were performed as described previously (Feng et al., 2003; Saijo et al., 2003) with minor modifications. The composition of the lysis/binding/washing buffer was 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 10 mM NaF, 2 mM Na3VO4, 25 mM β-glycerolphosphate, 10% glycerol, 0.1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, and 1× complete protease inhibitor (Roche). The acid elution and concentration steps were omitted.
Accession Numbers
The full-length cDNA sequence accession number for Arabidopsis CUL4 in GenBank is NM_123990, and the Arabidopsis Genome Initiative locus number is At5g46210.
Acknowledgments
We thank Yan Guo for the binary vector pCAMBIA1200/35SP/EGFP, Huishan Guo for the vector pSK-int, Ligeng Ma for providing helpful advice during this work, Yanfeng Liu for camera operation, and Yutao Shen for plant material preparation. We also thank Valerie J. Karplus and Ning Wei for critical reading of the manuscript. This research was supported by the National Institute of Biological Sciences at Beijing and by Grant GM-47850 to X.W.D. from the U.S. National Institutes of Health.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Xing Wang Deng (xingwang.deng@yale.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.106.043224.
References
- Ang, L.H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., and Deng, X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1 213–222. [DOI] [PubMed] [Google Scholar]
- Castle, L.A., and Meinke, D.W. (1994). A FUSCA gene of Arabidopsis encodes a novel protein essential for plant development. Plant Cell 6 25–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamovitz, D.A., Wei, N., Osterlund, M.T., von Arnim, A.G., Staub, J.M., Matsui, M., and Deng, X.W. (1996). The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86 115–121. [DOI] [PubMed] [Google Scholar]
- Chiu, W., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6 325–330. [DOI] [PubMed] [Google Scholar]
- Chory, J., Peto, C., Feinbaum, R., Pratt, L., and Ausubel, F. (1989). Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58 991–999. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., Matsui, M., Wei, N., Wagner, D., Chu, A.M., Feldmann, K.A., and Quail, P.H. (1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71 791–801. [DOI] [PubMed] [Google Scholar]
- Duek, P.D., Elmer, M.V., van Oosten, V.R., and Fankhauser, C. (2004). The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr. Biol. 14 2296–2301. [DOI] [PubMed] [Google Scholar]
- Feng, S., Ma, L., Wang, X., Xie, D., Dinesh-Kumar, S.P., Wei, N., and Deng, X.W. (2003). The COP9 signalosome interacts physically with SCF COI1 and modulates jasmonate responses. Plant Cell 15 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, S., Shen, Y., Sullivan, J.A., Rubio, V., Xiong, Y., Sun, T.P., and Deng, X.W. (2004). Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. Plant Cell 16 1870–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa, P., Gusmaroli, G., Serino, G., Habashi, J., Ma, L., Shen, Y., Feng, S., Bostick, M., Callis, J., Hellmann, H., and Deng, X.W. (2005). Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell 17 1180–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman, R., Polanowska, J., Kuraoka, I., Sawada, J., Saijo, M., Drapkin, R., Kisselev, A.F., Tanaka, K., and Nakatani, Y. (2003). The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113 357–367. [DOI] [PubMed] [Google Scholar]
- Guo, H.S., Fei, J.F., Xie, Q., and Chua, N.H. (2003). A chemical-regulated inducible RNAi system in plants. Plant J. 34 383–392. [DOI] [PubMed] [Google Scholar]
- Gusmaroli, G., Feng, S., and Deng, X.W. (2004). The Arabidopsis CSN5A and CSN5B subunits are present in distinct COP9 signalosome complexes, and mutations in their JAMM domains exhibit differential dominant negative effects on development. Plant Cell 16 2984–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa, L.A., Mihaylov, I.S., Banks, D.P., Zheng, J., and Zhang, H. (2003). Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 5 1008–1015. [DOI] [PubMed] [Google Scholar]
- Holm, M., Ma, L.G., Qu, L.J., and Deng, X.W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J., McCall, C.M., Ohta, T., and Xiong, Y. (2004). Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 6 1003–1009. [DOI] [PubMed] [Google Scholar]
- Jang, I.C., Yang, J.Y., Seo, H.S., and Chua, N.H. (2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 19 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, S., Kobayashi, R., and Grewal, S.I. (2005). Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat. Cell Biol. 7 1007–1013. [DOI] [PubMed] [Google Scholar]
- Kwok, S.F., Solano, R., Tsuge, T., Chamovitz, D.A., Ecker, J.R., Matsui, M., and Deng, X.W. (1998). Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell 10 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok, S.F., Staub, J.M., and Deng, X.W. (1999). Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J. Mol. Biol. 285 85–95. [DOI] [PubMed] [Google Scholar]
- Li, B., Ruiz, J.C., and Chun, K.T. (2002). CUL-4A is critical for early embryonic development. Mol. Cell. Biol. 22 4997–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T., Chen, X., Garbutt, K.C., Zhou, P., and Zheng, N. (2006). Structure of DDB1 in complex with a paramyxovirus V protein: Viral hijack of a propeller cluster in ubiquitin ligase. Cell 124 105–117. [DOI] [PubMed] [Google Scholar]
- Lyapina, S., Cope, G., Shevchenko, A., Serino, G., Tsuge, T., Zhou, C., Wolf, D.A., Wei, N., and Deshaies, R.J. (2001). Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292 1382–1385. [DOI] [PubMed] [Google Scholar]
- McNellis, T.W., Torii, K.U., and Deng, X.W. (1996). Expression of an N-terminal fragment of COP1 confers a dominant-negative effect on light-regulated seedling development in Arabidopsis. Plant Cell 8 1491–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis, T.W., von Arnim, A.G., Araki, T., Komeda, Y., Misera, S., and Deng, X.W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min, K.W., Kwon, M.J., Park, H.S., Park, Y., Yoon, S.K., and Yoon, J.B. (2005). CAND1 enhances deneddylation of CUL1 by COP9 signalosome. Biochem. Biophys. Res. Commun. 334 867–874. [DOI] [PubMed] [Google Scholar]
- Osaka, F., Saeki, M., Katayama, S., Aida, N., Toh, E.A., Kominami, K., Toda, T., Suzuki, T., Chiba, T., Tanaka, K., and Kato, S. (2000). Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 19 3475–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 462–466. [DOI] [PubMed] [Google Scholar]
- Peng, Z., Serino, G., and Deng, X.W. (2001. a). Molecular characterization of subunit 6 of the COP9 signalosome and its role in multifaceted developmental processes in Arabidopsis. Plant Cell 13 2393–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Z., Serino, G., and Deng, X.W. (2001. b). A role of Arabidopsis COP9 signalosome in multifaceted developmental processes revealed by the characterization of its subunit 3. Development 128 4277–4288. [DOI] [PubMed] [Google Scholar]
- Pepper, A., Delaney, T., Washburn, T., Poole, D., and Chory, J. (1994). DET1, a negative regulator of light-mediated development and gene expression in Arabidopsis, encodes a novel nuclear-localized protein. Cell 78 109–116. [DOI] [PubMed] [Google Scholar]
- Rubio, V., Shen, Y., Saijo, Y., Liu, Y., Gusmaroli, G., Dinesh-Kumar, S.P., and Deng, X.W. (2005). An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 41 767–778. [DOI] [PubMed] [Google Scholar]
- Saijo, Y., Sullivan, J.A., Wang, H., Yang, J., Shen, Y., Rubio, V., Ma, L., Hoecker, U., and Deng, X.W. (2003). The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17 2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, D.F., Gahrtz, M., Maxwell, B.B., Cook, R.K., Kan, J.M., Alonso, J.M., Ecker, J.R., and Chory, J. (2002). De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr. Biol. 12 1462–1472. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., Callis, J., Crosby, W.L., Lyapina, S., Deshaies, R.J., Gray, W.M., Estelle, M., and Deng, X.W. (2001). Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 292 1379–1382. [DOI] [PubMed] [Google Scholar]
- Seo, H.S., Yang, J.Y., Ishikawa, M., Bolle, C., Ballesteros, M.L., and Chua, N.H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423 995–999. [DOI] [PubMed] [Google Scholar]
- Serino, G., and Deng, X.W. (2003). The COP9 signalosome: Regulating plant development through the control of proteolysis. Annu. Rev. Plant Biol. 54 165–182. [DOI] [PubMed] [Google Scholar]
- Serino, G., Tsuge, T., Kwok, S., Matsui, M., Wei, N., and Deng, X.W. (1999). Arabidopsis cop8 and fus4 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. Plant Cell 11 1967–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, J.A., Shirasu, K., and Deng, X.W. (2003). The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nat. Rev. Genet. 4 948–958. [DOI] [PubMed] [Google Scholar]
- Suzuki, G., Yanagawa, Y., Kwok, S.F., Matsui, M., and Deng, X.W. (2002). Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev. 16 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim, A., and Deng, X.W. (1996). Light control of seedling development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 215–243. [DOI] [PubMed] [Google Scholar]
- von Arnim, A.G., and Deng, X.W. (1994). Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79 1035–1045. [DOI] [PubMed] [Google Scholar]
- von Arnim, A.G., Osterlund, M.T., Kwok, S.F., and Deng, X.W. (1997). Genetic and developmental control of nuclear accumulation of COP1, a repressor of photomorphogenesis in Arabidopsis. Plant Physiol. 114 779–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Ma, L.G., Li, J.M., Zhao, H.Y., and Deng, X.W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294 154–158. [DOI] [PubMed] [Google Scholar]
- Wang, X., Feng, S., Nakayama, N., Crosby, W.L., Irish, V., Deng, X.W., and Wei, N. (2003). The COP9 signalosome interacts with SCF UFO and participates in Arabidopsis flower development. Plant Cell 15 1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee, S., Geyer, R.K., Toda, T., and Wolf, D.A. (2005). CSN facilitates cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat. Cell Biol. 7 387–391. [DOI] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.W. (1996). The role of the COP/DET/FUS genes in light control of Arabidopsis seedling development. Plant Physiol. 112 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., Kwok, S.F., von Arnim, A.G., Lee, A., McNellis, T.W., Piekos, B., and Deng, X.W. (1994). Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. Plant Cell 6 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz, I.E., O'Rourke, K.M., Zhang, Z., Dornan, D., Arnott, D., Deshaies, R.J., and Dixit, V.M. (2004). Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303 1371–1374. [DOI] [PubMed] [Google Scholar]
- Yanagawa, Y., Sullivan, J.A., Komatsu, S., Gusmaroli, G., Suzuki, G., Yin, J., Ishibashi, T., Saijo, Y., Rubio, V., Kimura, S., Wang, J., and Deng, X.W. (2004). Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 18 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., Lin, R., Sullivan, J., Hoecker, U., Liu, B., Xu, L., Deng, X.W., and Wang, H. (2005). Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17 804–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, C., and Deng, X.W. (2005). COP1—From plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15 618–625. [DOI] [PubMed] [Google Scholar]
- Zhong, W., Feng, H., Santiago, F.E., and Kipreos, E.T. (2003). CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 423 885–889. [DOI] [PubMed] [Google Scholar]