Abstract
A novel real-time PCR-hybridization assay was developed for the rapid (<1 h) detection of penicillin susceptibility in Streptococcus pneumoniae. When applied to 24 pneumococcal DNA-positive cerebrospinal fluid extracts, penicillin-sensitive S. pneumoniae was detected in all instances. Real-time PCR proved more sensitive than culture, microscopy, or antigen detection and provided susceptibility data even in culture-negative cases.
Streptococcus pneumoniae is the second most frequently reported cause of pyogenic meningitis in the United Kingdom (14), with death occurring in 20 to 25% of patients (15). The rate of penicillin resistance among pneumococci is increasing (11, 12), and resistance occurs in a significant proportion (36%) of pneumococci from patients with meningitis (15). Effective management of pneumococcal meningitis depends on determination of the penicillin susceptibility of the infecting strain as swiftly as possible.
Susceptibility testing of cultures is slow, taking up to 48 h (17). Preadmission antibiotic treatment of patients in whom bacterial meningitis is suspected decreases the likelihood of successful culture. Molecular biology-based techniques applied directly to clinical material have the potential to detect the infecting organism and determine its antimicrobial susceptibility. We have previously demonstrated the value of real-time PCR in culture- and/or antigen-negative pneumococcal meningitis cases using an assay which targets the pneumolysin gene, ply (9). We now report on a complementary PCR assay for the detection of penicillin-susceptible S. pneumoniae (PSSP) and have evaluated the assay with cerebrospinal fluid (CSF) samples from patients with pneumococcal meningitis.
According to NCCLS criteria (13), the penicillin MICs for penicillin-sensitive, penicillin-intermediate-resistant, and penicillin-resistant S. pneumoniae (PRSP) are ≤0.06, 0.1 to 1.0, and ≥2 μg/ml, respectively. Penicillin resistance is due to the production of altered forms of penicillin-binding proteins (PBPs), with the production of altered PBP 2B being particularly important (3, 17). Sequence polymorphisms in the pbp2b gene result in decreased susceptibility to penicillin (MICs, >0.06 μg/ml), which has been associated with clinically relevant resistance (16) and pneumococcal meningitis treatment failures (7, 18). A seminested PCR based on this principle for the detection of PSSP and PRSP in culture-positive CSF specimens has been reported, but it is time-consuming and requires two separate reactions (4). A rapid assay that targets PBP 2B, is applicable to culture-negative specimens, and is capable of differentiating PSSP from all other nonsusceptible strains (MICs, >0.06 μg/ml) would be valuable.
Using the same platform used for the ply assay, we have developed a real-time PCR assay for the detection of PSSP using a LightCycler (LC; Idaho Technology Inc., Idaho Falls, Idaho). In the absence of a single specific determinant of penicillin resistance (8, 17), we targeted a conserved sequence of pbp2b associated with penicillin susceptibility. Successful amplification would therefore imply penicillin susceptibility, while amplification failure would infer resistance. Other investigators have used this approach (8, 17).
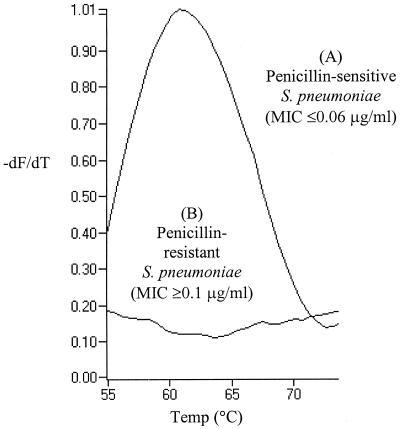
Using previously published sequence information (16), we designed primers and a bi-probe (6, 10) that targeted an 88-bp fragment of the pbp2b gene (GenBank accession no. X16022; positions 1494 to 1581). The forward and reverse primers were 5"-ATTCTTGGTATACTCAGGCT-3" (positions 1494 to 1513) and 5"-GTTTGGACCATATAGGTATTT-3" (positions 1561 to 1581), respectively. The bi-probe (5"-Cy5-CACAGCGGTCCAAGCTCT-biotin-3" [where Cy5 is cyanine 5]; positions 1531 to 1548) was designed to anneal to an internal sequence adjacent to the forward primer to confirm sequence specificity. The 10-μl PCR mixture comprised 3 mM MgCl2, a Taq-based master mixture (BioGene Ltd., Kimbolton, United Kingdom), 0.4 U of platinum anti-Taq antibody (Life Technologies Ltd., Paisley, United Kingdom), each primer at a concentration of 0.5 μM, 0.1 μM bi-probe, SYBR Green 1 (BioGene Ltd.) diluted 1:20,000, and 2 μl of DNA template. PCR cycling comprised an initial denaturation at 96°C for 1 min and 55 amplification cycles of 95°C for 0 s, 60°C for 1 s, 73°C for 3 s, with temperature transition rates of 20, 20, and 3°C/s, respectively. Fluorescence was monitored once each cycle after annealing. This was followed by a melting program of 50 to 95°C at a rate of 0.2°C/s with continuous fluorescence acquisition. Detection of the PCR product occurs as the bi-probe hybridizes to the amplicon, leading to an increase in fluorescence resonance energy transfer between the fluorophores, SYBR Green 1 and Cy5 (6, 10). Melting curve analysis was used to identify the determinant of penicillin sensitivity, which gave a characteristic melting temperature of 61 ± 2°C (Fig. 1) (2, 6, 10).
FIG. 1.
Illustrative melting curve traces obtained with a penicillin-sensitive and a penicillin-resistant strain of Streptococcus pneumoniae. (A) Representative results for all strains for which penicillin MICs are ≤0.06 μg/ml; (B) representative results for all strains for which penicillin MICs are ≥0.1 μg/ml. Melting peaks were derived by plotting the negative derivative of fluorescence with respect to temperature (−dF/dT).
The assay was evaluated with 66 isolates of S. pneumoniae, comprising 1 reference strain (NCTC 7466; National Collection of Type Cultures, London, United Kingdom) and 65 distinct clinical isolates, all of which were confirmed to be ply PCR positive (9). Penicillin MICs were determined according to NCCLS guidelines (13) and ranged from ≤0.015 to 4 μg/ml. The LC-PCR assay for penicillin susceptibility was specific, detecting all 27 PSSP isolates (MICs, ≤0.06 μg/ml). Thirty-nine strains with either resistance phenotype (20 strains with intermediate resistance [MICs, 0.1, 0.5, and 1.0 μg/ml; n = 3, 4, and 13, respectively]; 19 resistant strains [MICs, ≥2.0 μg/ml]) were LC-PCR negative, inferring alterations in the pbp2b gene sequence. Rare strains with reduced susceptibility but minimal pbp2b sequence variations have been reported (1), but none were encountered in the present study. The pbp2b PCR assay therefore successfully differentiated pneumococci for which penicillin MICs were relevant for the management of pneumococcal meningitis (i.e., MICs of ≤0.06 μg/ml versus MICs of ≥0.1 μg/ml). Furthermore, the pbp2b assay showed no evidence of cross-reaction with the taxonomically related organism S. mitis (three penicillin-sensitive strains and one resistant strain) (3, 8).
The pbp2b assay was then applied to 24 pneumococcal DNA-positive CSF samples from patients aged 1 to 76 years (median age, 41 years) with clinical and laboratory evidence suggestive of meningitis (Table 1). Gram-positive cocci were seen in 13 instances (54.2%), and samples from 12 (50%) patients (7 CSF samples and 5 blood samples) had cultural evidence of invasive pneumococcal infection; 7 of 15 CSF samples tested were found to contain pneumococcal antigen (by the Pasteurex [Sanofi Diagnostic Pasteur Ltd., Surrey, United Kingdom] and SLIDEX [BioMérieux Ltd., Hampshire, United Kingdom] assays). The majority of patients (n = 18 [75%]) had received appropriate antimicrobial therapy prior to lumbar puncture. When a sufficient volume of CSF was received (≥100 μl), the samples underwent extraction with DNAzol (Helena BioSciences, Sunderland, United Kingdom) prior to testing by PCR; the other samples were boiled for 15 min prior to testing by PCR. The 24 extracts showed no evidence of PCR inhibition (9), and the sequence of the gene for penicillin sensitivity was detected in all extracts by the pbp2b assay. The 12 S. pneumoniae isolates were identified as PSSP by traditional means of testing, confirming the accuracy of the LC assay. In view of the fact that a case of meningitis caused by PRSP was not identified in the present study, a CSF sample spiked with a PRSP strain was tested and was found to be ply positive and pbp2b negative (data not shown).
TABLE 1.
Findings for 24 CSF samples from patients with pneumococcal meningitis
| Parameter | No. (%) of samples |
|---|---|
| Gram-positive diplococci seen on microscopy | 13 (54) |
| Pneumococcal antigen detected (latex agglutination) | 7 (47)a |
| Pneumococcal culture positive | 12 (50)b |
| CSF specimen taken posttreatment | 18 (75)c |
| Real-time pneumolysin PCR positive | 24 (100)d |
| Real-time penicillin susceptibility PCR positive | 24 (100)d |
Fifteen of the 24 specimens were tested for pneumococcal antigen.
Five specimens were CSF culture negative, but blood samples from the patients were culture positive.
Eighteen patients were receiving cefotaxime; prior antimicrobial therapy was not known for six patients.
Fourteen specimens extraction underwent with DNAzol; 10 underwent extraction by boiling.
Thus, the pbp2b assay enables the direct detection of PSSP in CSF samples in which pneumococci are detected by either microscopy or antigen testing alone, as well as in culture-positive samples. These findings underscore the value of LC-PCR not only in enhancing case ascertainment (9) but also, now, in providing susceptibility data in terms of the prevalence of PSSP versus PRSP even in culture-negative cases. The assay can be completed in <1 h (this includes a 25-min cycling time), with a combined assay time of <2 h for both the ply and the pbp2b assays when samples are simply boiled prior to testing by PCR. The assay targets a region which is conserved among PSSP isolates. A PCR-positive result therefore provides a sound basis for the treatment of meningitis due to PSSP, in accordance with a locally agreed upon antibiotic treatment policy. In contrast, a PCR-negative result suggests meningitis due to PRSP and necessitates a regimen suitable for the treatment of infections caused by these strains. Despite reports of the increasing prevalence of PRSP strains, none were identified in our patient cohort. This is perhaps not surprising given that recent data suggest that only an estimated 6% of isolates in our area of the United Kingdom show resistance to penicillin, most of which is low level (5). Nevertheless, continuing surveillance remains essential for the monitoring of the changing patterns of resistance at the local, national, and international levels. Similarly, isolation of the causative organism, wherever possible, remains important for the monitoring of MIC trends, assessment of the emergence of multidrug resistance, and retrieval of serotyping data for surveillance and epidemiological purposes.
Implementation of the pbp2b PCR assay offers the prospect for patients to be taken off expensive broad-spectrum antibiotic treatment and to be treated with more specific and cheaper drugs. Moreover, the assay will aid in the assessment of the true burden of pneumococcal disease and, in the future, allow a more accurate evaluation of the impacts of new vaccines.
Acknowledgments
This work was supported in part by the PHLS Small Scientific Initiatives Fund.
REFERENCES
- 1.Beall, B., R. R. Facklam, D. M. Jackson, and H. H. Starling. 1998. Rapid screening for penicillin susceptibility of systemic pneumococcal isolates by restriction enzyme profiling of the pbp2B gene. J. Clin. Microbiol. 36:2359-2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernard, P. S., M. J. Lay, and C. T. Wittwer. 1998. Integrated amplification and detection of the C677T point mutation in the methylenetetrahydrofolate reductase gene by fluorescence resonance energy transfer and probe melting curves. Anal. Biochem. 255:101-107. [DOI] [PubMed] [Google Scholar]
- 3.Dowson, C. G., T. J. Coffey, C. Kell, and R. A. Whiley. 1993. Evolution of penicillin resistance in Streptococcus pneumoniae; the role of Streptococcus mitis in the formation of a low affinity PBP2B in S. pneumoniae. Mol. Microbiol. 9:635-643. [DOI] [PubMed] [Google Scholar]
- 4.du Plessis, M., A. M. Smith, and K. P. Klugman. 1998. Rapid detection of penicillin-resistant Streptococcus pneumoniae in cerebrospinal fluid by a seminested-PCR strategy. J. Clin. Microbiol. 36:453-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.George, R. C., and A. Melegaro. 2001. Invasive pneumococcal infection England and Wales 1999. Commun. Dis. Rep. Wkly. 11:6-12. [Google Scholar]
- 6.Gibson, J. R., N. A. Saunders, B. Burke, and R. J. Owen. 1999. Novel method for rapid determination of clarithromycin sensitivity in Helicobacter pylori. J. Clin. Microbiol. 37:3746-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heffelfinger, J. D., S. F. Dowell, J. H. Jorgensen, K. P. Klugman, L. R. Mabry, D. M. Musher, J. F. Plouffe, A. Rakowsky, A. Schuchat, C. G. Whitney, and the Drug-Resistant Streptococcus pneumoniae Therapeutic Working Group. 2000. Management of community-acquired pneumonia in the era of pneumococcal resistance. Arch. Intern. Med. 160:1399-1408. [DOI] [PubMed] [Google Scholar]
- 8.Jalal, H., S. Organji, J. Reynolds, D. Bennett, E. O'Mason, Jr., and M. R. Millar. 1997. Determination of penicillin susceptibility of Streptococcus pneumoniae using the polymerase chain reaction. J. Clin. Pathol. Mol. Pathol. 50:45-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kearns, A. M., R. Freeman, O. M. Murphy, P. R. Seiders, M. Steward, and J. Wheeler. 1999. Rapid PCR-based detection of Streptococcus pneumoniae DNA in cerebrospinal fluid. J. Clin. Microbiol. 37:3434.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kearns, A. M., A. J. L. Turner, C. E. Taylor, P. W. George, R. Freeman, and A. R. Gennery. 2001. LightCycler-based quantitative PCR for the rapid detection of human herpesvirus 6 DNA in clinical material. J. Clin. Microbiol. 39:3020-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klugman, K. P. 1990. Pneumococcal resistance to antibiotics. Clin. Microbiol. Rev. 3:171-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leggiadro, R. J. 2000. Penicillin-nonsusceptible pneumococcus. Int. J. Antimicrob. Agents 14:123-127. [DOI] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. 1998. Performance standards for antimicrobial susceptibility tests (M100-S8). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 14.Public Health Laboratory Service. 1999. Communicable disease report. Commun. Dis. Rep. Wkly. 9:337-340. [Google Scholar]
- 15.Schuchat, A., K. Robinson, J. D. Wenger, L. H. Harrison, M. Farley, A. L. Reingold, L. Lefkowitz, and B. A. Perkins. 1997. Bacterial meningitis in the United States in 1995. N. Engl. J. Med. 337:970-976. [DOI] [PubMed] [Google Scholar]
- 16.Smith, A. M., and K. P. Klugman. 1995. Alterations in penicillin-binding protein 2B from penicillin-resistant wild-type strains of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 39:859-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ubukata, K., Y. Asahi, A. Yamane, and M. Konno. 1996. Combinational detection of autolysin and penicillin-binding protein 2B genes of Streptococcus pneumoniae by PCR. J. Clin. Microbiol. 34:592-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waghorn, D. J. 1992. Meningitis due to multiply resistant pneumococcus. J. R. Soc. Med. 85:113-114. [DOI] [PMC free article] [PubMed] [Google Scholar]