Abstract
Carotenoids and their oxygenated derivatives xanthophylls play essential roles in the pigmentation of flowers and fruits. Wild-type tomato (Solanum lycopersicum) flowers are intensely yellow due to accumulation of the xanthophylls neoxanthin and violaxanthin. To study the regulation of xanthophyll biosynthesis, we analyzed the mutant white-flower (wf). It was found that the recessive wf phenotype is caused by mutations in a flower-specific β-ring carotene hyroxylase gene (CrtR-b2). Two deletions and one exon-skipping mutation in different CrtR-b2 wf alleles abolish carotenoid biosynthesis in flowers but not leaves, where the homologous CrtR-b1 is constitutively expressed. A second β-carotene hydroxylase enzyme as well as flower- and fruit-specific geranylgeranyl diphosphate synthase, phytoene synthase, and lycopene β-cyclase together define a carotenoid biosynthesis pathway active in chromoplasts only, underscoring the crucial role of gene duplication in specialized plant metabolic pathways. We hypothesize that this pathway in tomato was initially selected during evolution to enhance flower coloration and only later recruited to enhance fruit pigmentation. The elimination of β-carotene hydroxylation in wf petals results in an 80% reduction in total carotenoid concentration, possibly caused by the inability of petals to store high concentrations of carotenoids other than xanthophylls and by degradation of β-carotene, which accumulates as a result of the wf mutation but is not due to altered expression of genes in the biosynthetic pathway.
INTRODUCTION
Plant carotenoids are C40 carbohydrates with a chain of conjugated double bonds, which creates a chromophore that absorbs light in the blue range of the spectrum. Flowers and fruits of many species are colored due to the accumulation in the chromoplasts of carotenoid pigments that provide distinct colors to the tissues, ranging from yellow to orange and red, to visually attract pollinators and facilitate seed dispersal by animals. Many plant species, including tomato (Solanum lycopersicum), accumulate yellow pigments in flowers because insects are preferentially attracted to this color (Kevan, 1983). Carotenoids, mainly xanthophylls, are the most prevalent yellow pigments found in flowers. Carotenoids are also synthesized in chloroplasts, where they play essential roles in photosynthesis in the light-harvesting systems and in the photosynthetic reaction centers (Frank et al., 1999; Demmig-Adams and Adams, 2002; Holt et al., 2004; Robert et al., 2004; Horton and Ruban, 2005; Standfuss et al., 2005).
In the last decade, carotenoid biosynthesis in plants has been described at the molecular level (reviewed in Cunningham and Gantt, 1998; Hirschberg, 2001; Fraser and Bramley, 2004). The carotenoid biosynthesis pathway begins with the formation of phytoene from geranylgeranyl diphosphate in the central isoprenoid pathway. Four dehydrogenation (desaturation) steps lead to the linear molecule lycopene, which is then cyclized at each end by either ɛ- or β-cyclase to yield β-carotene (β,β-carotene) or α-carotene (β,ɛ-carotene). Hydroxylation of these carotenes at C3 and C3′ gives rise to the xanthophylls zeaxanthin and lutein, respectively. Epoxidation at C5,C6 converts zeaxanthin to violaxanthin via the intermediate antheraxanthin. Subsequent opening of the cyclohexenyl 5-6-epoxide ring in violaxanthin gives rise to neoxanthin.
Tomato is an important model plant for the study of carotenoid biosynthesis in chromoplast-containing tissues, such as fruits. The fruits of the cultivated tomato are red owing to the accumulation of lycopene, and its flowers are yellow due to the xanthophylls violaxanthin and neoxanthin. The accumulation of lycopene in fruits is determined by differential expression of genes encoding biosynthetic enzymes during the breaker stage of fruit development (reviewed in Hirschberg, 2001). At this stage, transcription of the genes encoding phytoene synthase (Psy), phytoene desaturase (Pds), ζ-carotene desaturase (Zds), and carotene isomerase (CrtISO) is upregulated, whereas the genes for lycopene β-cyclase (Lcy-b) and lycopene ɛ-cyclase (Lcy-e) are not transcribed. Consequently, the enhanced flux of carotene in the pathway is arrested at lycopene. The substantial increase in carotenoid biosynthesis in tomato flowers is correlated with upregulation of expression of the carotenoid biosynthesis genes Psy1, Pds, Lcy-b, and Cyc-b (Giuliano et al., 1993; Corona et al., 1996; Ronen et al., 2000). The gene Lcy-e is expressed at a very low level in petals and anthers, resulting in the formation of only minute amounts of lutein in these organs. By contrast, Lcy-e is highly expressed in leaves, where lutein is the most prevalent carotenoid.
A number of mutations that alter carotenoid concentration or composition have been described in plants (reviewed in Hirschberg, 2001). Some of them affect carotenoid biosynthesis distinctively in flowers or fruits. For example, in tomato, white-flower (wf) abolishes xanthophyll accumulation in petals but does not change xanthophylls in leaves, and yellow-flesh (r) eliminates carotenoids in fruits only. These mutations suggest that carotenoid biosynthesis has unique characteristics in different organs.
To investigate the regulation of carotenoid synthesis in flowers, we analyzed the tomato mutant wf (Young and MacArthur, 1947). The recessive wf allele determines white to beige petals and pale anthers compared with intense yellow organs in the wild type (Figure 1). We report here on the cloning of two functional β-ring carotene hydroxylase genes (CrtR-b) in tomato that are responsible for the conversion of β-carotene to zeaxanthin. CrtR-b1 is constitutively expressed in leaves, whereas CrtR-b2 is active exclusively in flowers. We found that the wf phenotype is caused by a mutation in the gene CrtR-b2, thus confirming that CRTR-B1 does not play a primary role in β-ring hydroxylation in flowers. These findings together with previous data demonstrate a central role for gene duplication in the development of a chromoplast-specific carotenoid biosynthesis pathway.
Figure 1.
Flowers of wf Mutants.
(A) wf1-1 (F2 of a cross LA2370 × IL3-2) (left) and its nearly isogenic wild-type line (F2 of the same cross).
(B) wf1-2 (e1827) (left) and its isogenic line M82.
(C) Flower of wf1-2 (left) and the wild type (right) in developmental stages 1 to 3 analyzed in this work.
RESULTS
Phenotypic Characterization of wf
Flowers of the cultivated tomato accumulate high concentrations of the yellow xanthophylls violaxanthin and neoxanthin. By contrast, the corolla in the recessive mutant wf is whitish as a result of a decrease in total carotenoid content (Figure 1, Table 1). Two known wf lines, LA2370 (background genotype unknown) and LA3575 (background genotype: Ailsa Craig), were obtained from the Tomato Genetics Resource Center (University of California, Davis). In addition, we have isolated two novel wf alleles that were induced in the tomato cultivar M82 by ethyl methanesulfonate (line e1827) or fast neutron bombardment (line n5681; Menda et al., 2004; http://zamir.sgn.cornell.edu/mutants/). These single-gene recessive mutations were found to be allelic to wf by genetic crossing. We designated LA2370, e1827, and n5681 as wf1-1, wf1-2, and wf1-3, respectively. LA3575 turned out to carry the same allele as wf1-1. Both wf1-2 and wf1-3 are isogenic with the wild-type line M82.
Table 1.
Carotenoid Accumulation in Petals, Anthers, Fruits, and Leaves of the Wild Type and wf Mutants
| Neoxanthin | Violaxanthin | Anteraxanthin | Lutein | Zeaxanthin | β-Carotene | Lycopene | Phytoene | Total | |
|---|---|---|---|---|---|---|---|---|---|
| Petals | |||||||||
| Wild type (M82) | 427.4 ± 32 | 192.6 ± 24 | 2.4 ± 1 | 622.4 | |||||
| wf1-1 (LA2370)a | 24.3 ± 9 | 5.6 ± 3 | 9.6 ± 3 | 65.7 ± 18 | 3.9 ± 1 | 109.1 | |||
| wf1-2 (e1827)b | 33.1 ± 6 | 7.9 ± 1 | 3.3 ± 1 | 73.7 ± 11 | 5.5 ± 2 | 123.5 | |||
| wf1-3 (n5681)b | 57.3 | 7.0 | 4.5 | 31.6 | 100.4 | ||||
| Anthers | |||||||||
| Wild type (M82) | 197.4 ± 11 | 93.6 ± 8 | 12.0 ± 2 | 3.9 ± 3 | 306.9 | ||||
| wf1-1 (LA2370)a | 42.5 ± 3 | 14.4 ± 3 | 13.8 ± 5 | 22.3 ± 7 | 93.0 | ||||
| wf1-2 (e1827)b | 51.1 ± 5 | 16.0 ± 1 | 8.9 ± 1 | 32.2 ± 4 | 108.2 | ||||
| wf1-3 (n5681)b | 56.6 | 4.2 | 2.7 | 29.2 | 92.7 | ||||
| Fruit | |||||||||
| Wild type (M82) | 0.9 ± 0.6 | 1.2 ± 0.4 | 40.6 ± 3.0 | 4.7 ± 1.0c | 47.4 | ||||
| wf1-2 (e1827)b | 0.7 ± 0.1 | 1.7 ± 1.5 | 35.0 ± 5.2 | 4.8 ± 2.0c | 42.2 | ||||
| wf1-1 (LA2370)a | 0.6 ± 0.5 | 8.1 ± 5.3 | 30.3 ± 5.1 | 8.1 ± 2.1c | 47.1 | ||||
| wf1-3 (n5681)b | 2.7 | 3.2 | 18.7 | 5.8c | 30.4 | ||||
| Leaves | |||||||||
| Wild type (M82) | 12.5 ± 3 | 28.4 ± 10 | 3.2 ± 1 | 50.6 ± 13 | 2.7 | 19.1 ± 5 | 116.6 | ||
| wf1-1 (LA2370)a | 14.7 ± 1 | 24.3 ± 3 | 1.5 ± 1 | 55.4 ± 6 | 0.8 ± 1 | 19.5 ± 2 | 116.2 | ||
| wf1-2 (e1827)b | 16.8 ± 2 | 32.1 ± 11 | 3.0 ± 1 | 59.9 ± 10 | 2.0 ± 1 | 25.8 ± 3 | 139.6 | ||
| wf1-3 (n5681)b | 15.3 | 31.0 | <0.1 | 52.6 | <0.1 | 22.3 | 121.2 |
Carotenoids were measured in fully developed (stage 3) flowers.
Background genotype unknown or hybrid.
Isogenic with M82.
Including phytofluene.
Carotenoid analysis indicated a reduction of 80 to 84% in total carotenoids in petals of the various wf mutant alleles (Table 1). The sharp decrease in the xanthophylls neoxanthin and violaxanthin was accompanied by an increase of β-carotene, a carotenoid species that in the wild type appears in minute amounts only. No difference between wf and M82 was found in lutein, which occurs in flowers in small amounts (Table 1). The phenotype of the anthers of wf was similar to that of the petals (Table 1). By contrast, no differences in carotenoid content were detected in leaves or fruits of wf compared with M82 (Table 1), and no other phenotypic changes were observed in wf plants. The limited buildup of β-carotene in wf flowers suggested the possibility of a flower-specific inhibition of β-carotene hydroxylation.
Cloning and Sequence Analysis of CrtR-b Genes
To find out whether a mutation in β-carotene hydroxylase could account for the wf phenotype, we cloned the β-carotene hydroxylase gene, CrtR-b, from tomato. A cDNA library from tomato was screened with the CrtR-b cDNA from Arabidopsis thaliana. Restriction endonuclease analysis revealed 18 positive phages with a similar digestion pattern and one positive phage that was different. Two different CrtR-b genes were subcloned into the plasmid vector pBluescript KS−. The new plasmids were designated pCrtR-b1 and pCrtR-b2, respectively. Nucleotide sequence analysis of CrtR-b1 and CrtR-b2 cDNA revealed open reading frames of 309 and 314 codons, respectively, that encode polypeptides of calculated molecular masses of 34.5 and 35.1 kD. The predicted β-carotene hydroxylase polypeptides share overall similarity of 80% and identity of 73 and 62% with the β-carotene hydroxylases from Arabidopsis. Both putative polypeptides begin with a MetAlaAla sequence, which suggests a plastid transit sequence (Gavel and von Heijne, 1990). Indeed, chloroplast transit peptide prediction software, ChloroP v1.1 (http://www.cbs.dtu.dk/services/ChloroP/; Emanuelsson and Nielsen, 1999), identified the existence of a plastid transit peptide with a proposed cleavage site between Val-59 and Cys-60 in CRTR-B1 and Val-57 and Cys-58 in CRTR-B2. The sequences of the predicted mature polypeptides after cleavage of the transit sequences are 94% similar and 86% identical.
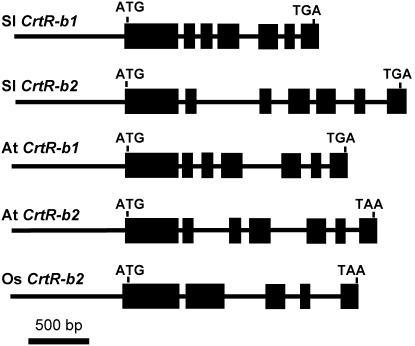
To analyze the structure of the β-carotene hydroxylase genes, we cloned the genomic DNA of both CrtR-b genes by screening a BAC library of tomato (cv Heinz 1706) (Budiman et al., 2000). Using the inserts of pCrtR-b1 and pCrtR-b2 as probes, we selected a single positive BAC clone from each probe. DNA of these BACs was digested with EcoRI (CrtR-b1) or XbaI (CrtR-b2), and the resulting fragments were cloned using the plasmid pBluescript SK−. The DNA inserts of these plasmids were sequenced, resulting in 3076 bp of CrtR-b1 and 3570 bp for the CrtR-b2 genomic sequences. Comparison with the cDNA sequence revealed seven exons in each gene, interrupted by six introns (Figure 2). Complete conservation in exon organization and splicing sites was found between tomato and Arabidopsis (Figure 2); however, there was low conservation in the length and sequence of the introns.
Figure 2.
Gene Structure of β-Carotene Hydroxylase Genes (CrtR-b).
Tomato (Sl) CrtR-b1 and CrtR-b2, Arabidopsis (At) CrtR-b1 and CrtR-2, and rice (Os) CrtR-b2. Exons are depicted as boxes.
CrtR-b2 Maps to wf
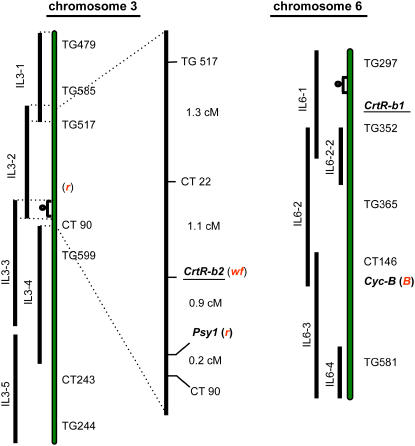
To test for possible linkage between the β-carotene hydroxylase gene and the wf phenotype, we mapped both hydroxylase genes to the tomato introgression lines (ILs; Eshed and Zamir, 1995). Restriction fragment length polymorphism (RFLP) analysis using either CrtR-b1 or CrtR-b2 as probes revealed that the first mapped to IL6-1, and CrtR-b2 was mapped exclusively to IL3-2 on the short arm of chromosome 3 but not to IL3-1 (Figure 3; Liu et al., 2003). The mutation wf was previously mapped to a similar position on chromosome 3 as CrtR-b2 (van der Biezen et al., 1994). To verify and refine previous mapping, we crossed the wf line LA2370 with IL3-2 and used 540 F2 plants for RFLP mapping and determination of flower phenotypes (Figure 3); complete cosegregation was established between wf and CrtR-b2.
Figure 3.
Genetic Mapping of CrtR-b1, CrtR-b2, and wf on the Tomato Linkage Map.
The linkage map was adapted from Eshed and Zamir (1995). The relevant chromosomal segments from Solanum pennellii that were introgressed into S. lycopersicum are represented by black bars with the names of the ILs that carry them. The high-resolution map of IL3-2, which includes the wf locus, is displayed next to chromosome 3. RFLP markers used in the fine mapping of wf and genetic distances between each pair are presented. r and B represents the loci of yellow-flesh (Psy1) and Beta (Cyc-B) mutations, respectively. cM, centimorgans.
The Molecular Basis of wf Alleles
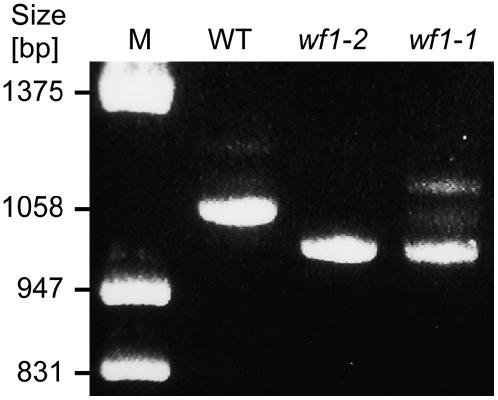
Quantitative RT-PCR analysis of CrtR-b2 mRNA in flowers indicated a similar level of expression in wf1-1, wf1-2, and the wild-type M82 (data not shown). However, the CrtR-b2 transcripts in wf1-1 and wf1-2 were shorter than that of M82 by ∼60 nucleotides (Figure 4). Sequence analysis of the CrtR-b2 cDNA from M82 (wild type) and mutant wf1-2 confirmed that the mRNA of the latter had a deletion of 60 nucleotides, which encompassed the entire length of the predicted exon 2. A point mutation from G to A was identified in the genomic sequence of CrtR-b2 from wf1-2 (Figure 5). The mutation occurred in the intron-exon junction consensus sequence at the donor site of the second intron and is expected to lead to exon skipping during RNA processing. Sequence analysis of genomic DNA and cDNA sequences of CrtR-b2 from wf1-1 revealed a 59 bp deletion at the very beginning of exon 1 (nucleotides 303 to 362; Figure 5), resulting in a short open reading frame of 29 codons due to a premature stop codon. Allele wf1-3, which was generated by fast neutron bombardment (Menda et al., 2004), carries a DNA deletion encompassing the entire CrtR-b2, as indicated by the failure to amplify this gene by PCR using a combination of specific primers.
Figure 4.
Deletions in CrtR-b2 mRNA of wf1-2 (e1827) and wf1-1 (LA2370).
Total RNA was extracted from stage 3 flowers and used as template for RT-PCR amplification. The resulting DNA products were separated by electrophoresis on 1.0% agarose gel and stained with ethidium bromide. A full-length DNA fragment of 1058 bp was generated in the wild type. Small deletions of ∼60 bp were observed in the mutants. M, DNA size markers.
Figure 5.
Mutations in the CrtR-b2 Gene from wf1-1 and wf1-2.
Genomic sequence of the first three exons of CrtR-b2 from tomato is presented. Exons are in boldface. The sequence deleted in allele wf1-1 is underlined. A transition mutation of G to A that causes skipping of the second exon in allele wf1-2 is shown at nucleotide 804.
Functional Expression of CrtR-b cDNAs in Escherichia coli
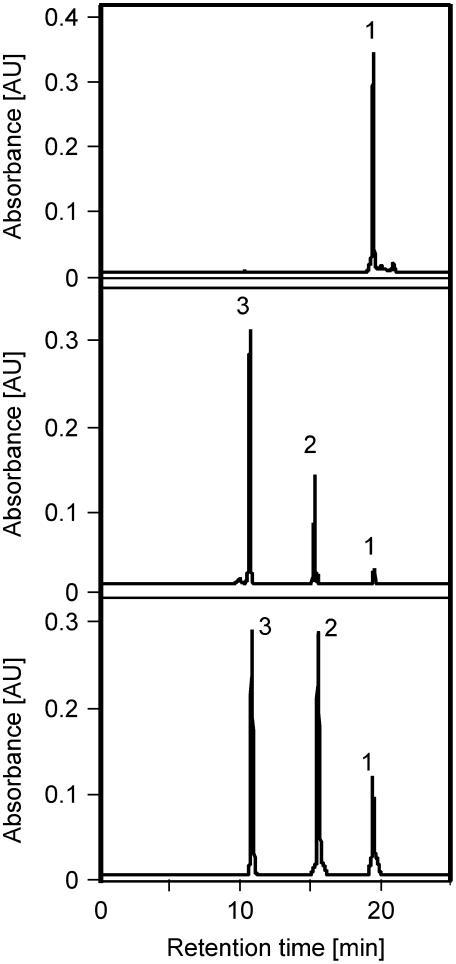
Although tomato has two CrtR-b sequences, all the wf mutations were identified in the CrtR-b2 gene. To ascertain that both CrtR-b genes code for an active β-carotene hydroxylase, we tested their enzymatic activity in E. coli cells carrying plasmid pBCAR-T (Ronen et al., 1999). These bacteria that produce β-carotene were cotransfected with plasmids pCrtR-b1 or pCrtR-b2 or with the empty vector pBluescript SK−. Carotenoids were extracted from the bacteria and analyzed by HPLC. Cells carrying pBCAR-T and pBluescript SK− yielded β-carotene, whereas cells carrying both pBCAR-T and pCrtR-b1 or pCrtR-b2 produced, in addition, zeaxanthin (β,β-carotene-3,3′-diol) and the intermediate β-cryptoxanthin (β,β-carotene-3-diol) (Figure 6, Table 2). These results indicate that both CrtR-b1 and CrtR-b2 encode active isoforms of β-carotene hydroxylase that catalyzes the hydroxylation of carbons 3 and 3′ on the β-rings of β-carotene.
Figure 6.
HPLC Analysis of Carotenoids in E. coli Cells Expressing CrtR-b1 and CrtR-b2 from Tomato.
Carotenoids were extracted from E. coli cells carrying plasmid pBETA (top); pBETA + pCrtR-b1 expressing cDNA of CrtR-b1 from tomato (middle); or pBETA + pCrtR-b2 expressing cDNA of CrtR-b2 from tomato (bottom). Peak identification: 1, β-carotene; 2, β-cryptoxanthin; 3, zeaxanthin.
Table 2.
Functional Expression of Tomato β-Carotene Hydroxylases CrtR-b1 and CrtR-b2 in E. coli Cells
| Plasmids | β-Carotene | β-Cryptoxanthin | Zeaxanthin | Othersa |
|---|---|---|---|---|
| pBETA + KS− | 98 | 2 | ||
| pBETA + pTCrtR-b1 | 4 | 25 | 71 | |
| pBETA + pTCrtR-b2 | 10 | 45 | 45 | |
| Plasmids | Lycopene | δ-Carotene | ɛ-Carotene | |
| pDELTA + KS− | 2 | 94 | 4 | |
| pDELTA + pTCrtR-b1 | 2 | 94 | 4 | |
| pDELTA + pTCrtR-b2 | 2 | 94 | 4 |
Carotenoid composition (%) was determined in E. coli cells carrying various combinations of plasmids: pBETA, carrying CrtE, CrtB, and CrtI from Erwinia uredovora and Lcy-b from tomato; pTCrtR-b1, expressing the tomato cDNA of CrtR-b1; pTCrtR-b1 and pTCrtR-b1, expressing truncated cDNA clones of CrtR-b1; pTCrtR-b2, expressing the cDNA of CrtR-b2. KS−, pBluescript KS−.
A mixture of unidentified carotenes upstream to lycopene.
To determine whether the tomato CRTR-B enzymes could add hydroxyl groups also to ɛ-ring carotenoids, we cotransfected pCrtR-b1 or pCrtR-b2 to a δ-carotene accumulating E. coli cells that carried the plasmid pDCAR (Ronen et al., 1999). No alteration in carotenoid composition was observed in these cells, indicating that both CRTR-B enzymes in tomato were incapable of hydroxylation of ɛ-ring in δ-carotene in E. coli cells (Table 2).
Expression of CrtR-b Genes in Tomato
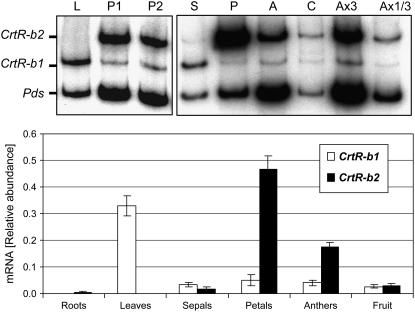
The steady state mRNA level of the two CrtR-b genes was measured in various tissues by RT-PCR and quantitative real-time RT-PCR. Total RNA was extracted from roots, leaves, flowers, and fruits of tomato (cv VF-36 and M82), and the relative amount of mRNA of CrtR-b1 and CrtR-b2 was determined with gene-specific primers. Results presented in Figure 7 indicate that the two CrtR-b genes are expressed in a tissue-specific manner. CrtR-b1 is expressed in leaves and sepals, but in flower tissues, it is expressed at low levels. By contrast, CrtR-b2 is highly expressed in petals and anthers, where yellow xanthophylls accumulate at high concentrations, and is relatively low in carpels and sepals (Figure 7). We could not detect CrtR-b2 mRNA in tomato leaves. In roots, CrtR-b2 is expressed at low levels and CrtR-b1 is hardly detected.
Figure 7.
Expression of CrtR-b1 and CrtR-b2 in Various Tomato Organs.
Top panel: Steady state levels of mRNA of CrtR-b1, CrtR-b2, and Pds were measured concomitantly by RT-PCR amplification from the same samples of total RNA. PCR was performed with 32P-labeled dCTP. Autoradiograms of PCR-amplified DNA fragments that were separated by polyacrylamide gel electrophoresis are exhibited. RNA from leaves (L) and petals from flower stages 1 (P1) and 3 (P3) are presented in the left panel. RNA from different tissues of stage 3 flowers is presented in the right panel: S, sepals; P, petals; A, anthers; and C, carpels. Samples Ax3 and Ax1/3 were amplified from anther RNA, 3-fold and one-third of the original concentration, respectively. Bottom panel: Relative levels of CrtR-b1 and CrtR-b2 mRNA from roots, leaves, flowers, and mature fruit were determined by real-time RT-PCR using gene-specific primers. Expression data were normalized to the expression of actin. Data shown are means + sd (n = 4).
Expression of Isoprenoid and Carotenoid Biosynthesis Genes in wf
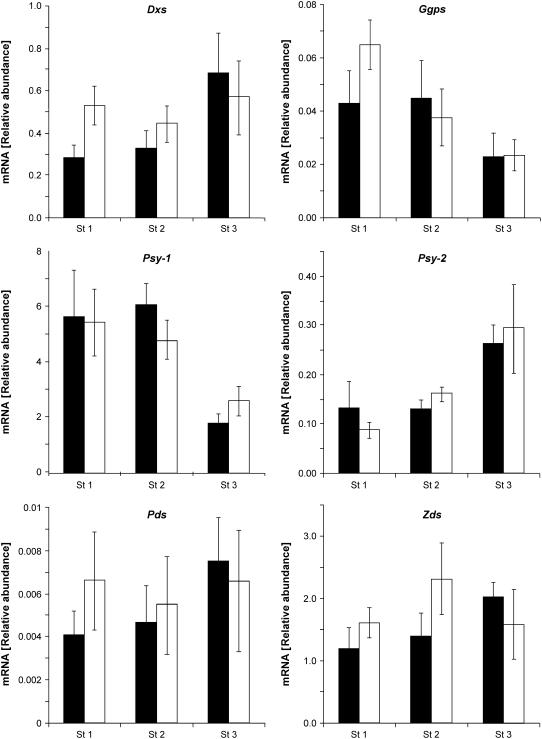
The phenotype of wf is caused by a significant reduction in total flower carotenoids, mainly due to a 90 to 95% decrease in the concentration of the yellow xanthophylls neoxanthin and violaxanthin in the petals. Since the mutations in wf occur in the β-carotene hydroxylase gene, the decrease in total carotenoids is not easily explained by a block of the biosynthetic pathway at the β-carotene hydroxylation step. To explore the possibility of alterations in transcription of genes in the pathway leading to β-carotene, mRNA levels of the genes encoding 1-deoxy-d-xylulose 5-phospate synthase (Dxs), geranylgeranyl diphosphate synthase (Ggps2), phytoene synthase (Psy1 and Psy2), phytoene desaturase (Pds), and ζ-carotene desaturase (Zds) were determined by quantitative real-time RT-PCR at three stages of flower development. No significant differences in gene expression were found between wf1-2 and the wild type (Figure 8). These results cancel out the possibility of a negative effect of the mutation on expression of key enzymes in the isoprenoid pathway.
Figure 8.
Expression of Isoprenoid and Carotenoid Biosynthesis Genes in Flowers of the Wild Type (M82) and Mutant wf1-2.
Total RNA was extracted from flowers at developmental stages 1 to 3 (Figure 1). Levels of mRNA were determined by real-time RT-PCR using gene-specific primers for the following genes: Dxs, Ggps2, Psy1, Psy2, Pds, and Zds. Expression data were normalized to the expression of actin. Data shown are means + sd (n = 5). Black bars correspond to the wild type (M82) and white bars to wf1-2.
DISCUSSION
Regulation of Carotenoid Biosynthesis in Flowers
In tomato, the concentration of total carotenoids in petals and anthers is 3- to 4-fold higher than in leaves and 8- to 10-fold higher than in mature tomato fruit. The carotenoids in flowers and fruits are considered secondary metabolites as they contribute to plant fitness but are not necessarily essential for the physiology in these tissues. On the other hand, in green leaves, carotenoids are indispensable for photosynthesis. Constitutive expression of carotenoid biosynthesis genes has been observed in all green tissues that have been examined. By contrast, accumulation of high concentrations of carotenoids in flowers and fruits is correlated with upregulation of genes that enhance the flux of the biosynthetic pathway (reviewed in Hirschberg, 2001; Bramley, 2002). Thus, the enhanced expression during tomato flower development of CrtR-b2, Psy1 (Giuliano et al., 1993; this work), Pds (Giuliano et al., 1993; Corona et al. 1996), Cyc-b, and Lcy-b (Ronen et al., 2000) results in increased carotenoid biosynthesis. Lack of lycopene ɛ-cyclase activity due to downregulation of expression in tomato flowers of Lcy-e (Ronen et al., 1999) channels the carotenoid pathway in tomato flowers to the β-xanthophylls neoxanthin and violaxanthin. In flowers of marigold (Tagetes), high level of expression of the genes of Psy1, Pds, Zds, and Lcy-e but not Lcy-b bring about the accumulation of the α-branch xanthophyll lutein (Moehs et al., 2001). Upregulation of Psy1, Pds, and Zds was reported in flowers of Gentiana (Zhu et al., 2002) and of Psy1 in flowers of Narcissus pseudonarcissus (Schledz et al., 1996) and Cucumis sativus (Vishnevetsky et al., 1997). Taken together, these data indicate that in flowers, similarly to fruits, gene expression, most probably at the level of transcription, is the major mechanism that determines the flux of carotenoid biosynthesis to the various xanthophylls.
Why Are Flowers of wf Nearly Colorless?
Here, we describe the cloning from tomato of the wf gene. Surprisingly, lack of carotenoids in flowers of this mutant is caused by a lesion in the enzyme β-carotene hydroxylase, which functions downstream from β-carotene in the pathway leading to the xanthophylls violaxanthin and neoxanthin. It is intriguing that inhibition of β-carotene hydroxylation leads to a reduction of 80% in total carotenoid concentration in the petals of wf rather than to the replacement of the xanthophylls by an equivalent amount of β-carotene and upstream carotenes. Three possible mechanisms may explain the decline in total carotenoids: (1) reduced synthesis due to downregulation of the pathway, possibly at the transcriptional level; (2) inability of petals to accumulate carotenoid species other than xanthophylls; and (3) degradation of β-carotene, which accumulates as a result of the wf mutation.
Because expression of genes encoding the rate-limiting isoprenoid and carotenoid enzymes DXS (Rodriguez-Concepcion et al., 2001) and PSY (Romer et al., 2000; Paine et al., 2005), as well as genes for other enzymes in the pathway (GGPS2, PDS, and ZDS), is not altered in wf (Figure 8), downregulation of the carotenoid pathway at the transcriptional level as a result of the mutation in CrtR-b2 can be ruled out. Although carotenoid biosynthesis is regulated mainly at the transcriptional level, one cannot exclude a mechanism of feedback inhibition, at the enzymatic or protein level, for example by β-carotene, of the rate-limiting enzymes DXS or PSY.
Posttranscriptional regulation has been described for PSY2 in tomato fruit (Fraser et al., 1999) and in the MEP pathway enzymes in Arabidopsis (Sauret-Gueto et al., 2006), specifically of DXS (Guevara-García et al., 2005).
An alternative explanation that links the white phenotype to a failure in accumulation of carotene species is supported by the mechanisms of sequestration in chromoplasts that enable storage of high concentrations of xanthophylls (Vishnevetsky et al., 1999). Most of the xanthophylls in tomato flowers are esterified with fatty acids. These xanthophyll-esters are necessary for both stabilization and accumulation by association with specific polypeptides. Since β-carotene cannot be esterified, its accumulation in chromoplasts is very limited. This suggestion for limited storage capability is in agreement with the fact that during early stages of flower development, when carotenoids still exist in low concentrations, the difference in carotenoid content between wf and the wild type is relatively small. For example, in flowers at stage 1 (2 d before anthesis; Figure 1), total carotenoid concentration was 345 ± 48 μg·g−1 fresh weight (FW) in the wild type (M82) and 226 ± 34 μg·g−1 FW in wf1-2, whereas in fully developed flowers (stage 3), it was 676 ± 72 μg·g−1 FW in the wild type and 147 ± 14 μg·g−1 FW in wf1-2.
Enzymatic cleavage of carotenoids may reduce the carotenoid level, as demonstrated in the mutant ccd1-1 of Arabidopsis (Auldridge et al., 2006). Analysis by gas chromatography–mass spectrometry of volatiles produced by tomato flowers revealed that in wf, the levels of β-ionone and β-cyclocytral, both produced by an oxidative cleavage of β-carotene, was 24- and 7-fold, respectively, higher in wf1-2 than in M82 (E. Lewinsohn, unpublished data). This result indicates that cleavage of β-carotene does take place in the petals; therefore, reduced total carotenoids in wf could be due to this degradation.
Two Non-Heme β-Carotene Hydroxylase Enzymes in Tomato
Two CrtR-b genes that encode β-carotene hydroxylase exist in tomato. The products of both are capable of proper hydroxylation of the β-rings of β-carotene in E. coli cells to give β-cryptoxanthin and zeaxanthin, but they fail to hydroxylate the ɛ-ring in δ-carotene. Comparison of the deduced amino acid sequence of CRTR-B from different plants identified a conserved His-rich domain that is found in other non-heme di-iron monooxygenases (Bouvier et al., 1998). This SUR2-type (syringomycin response protein 2) domain occurs in enzymes such as sterol oxidase, sterol desaturase, and fatty acid hydroxylase that use iron-activated oxygen for catalyzing oxygenation or desaturation of hydrophobic molecules. Biochemical characterization in bell pepper (Capsicum annum) has determined that iron, ferredoxin, and ferredoxin oxidoreductase are needed for the enzymatic activity of β-hydroxylation (Bouvier et al., 1998).
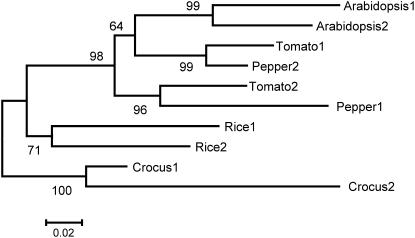
Two bona fide β-carotene hydroxylase enzymes exist also in Arabidopsis and pepper. A phylogenetic analysis based on amino acid sequences of β-carotene hydroxylases that was conducted using MEGA version 3.1 (Kumar et al., 2004) shows that tomato CRTR-B1 is more similar to one of the CrtR-b genes of bell pepper (CRTR-B2) than to its homolog in tomato CrtR-b2 (Figure 9). Likewise, CRTR-B2 of tomato is more similar to pepper CRTR-B1 than to the tomato CRTR-B1. The pattern of expression of the CrtR-b genes described here indicates that in tomato, one enzyme, CRTR-B1, functions in carotenoid biosynthesis in photosynthetic tissues and the second enzyme, CRTR-B2, operates in flowers in the accumulation of carotenoids as secondary metabolites for pigmentation.
Figure 9.
Phylogenetic Tree Based on Amino Acid Sequences of β-Carotene Hydroxylases in Higher Plants.
The following sequences have been analyzed: Arabidopsis 1, Arabidopsis 2, Crocus 1, Crocus 2, Pepper 1 (CRTR-B2), Pepper 2 (CRTR-B1), Rice 1, Rice 2, Tomato 1, and Tomato 2. Alignment of the amino acid sequences of these polypeptides is presented in Supplemental Figure 1 online.
In pepper, expression of CrtR-b1 is upregulated in fruit chromoplasts where massive accumulation of xanthophylls takes place (Bouvier et al., 1998). Thus, both tomato CrtR-b2 and pepper CrtR-b1 are upregulated in xanthophyll-accumulating tissues in flowers and fruits, respectively. The similarity between the tomato CrtR-b2 and pepper CrtR-b1 genes extends also to their chromosomal location in the genome as they are mapped to chromosome 6 in both pepper and tomato. Similarly, tomato CrtR-b2 and pepper CrtR-b1 are both mapped to chromosome 3 (Figure 3; Thorup et al., 2000). These similarities in CrtR-b between pepper and tomato suggest that the CrtR-b gene duplication preceded the separation of tomato and pepper during evolution. Moreover, the gene structure is identical in all hydroxylases of tomato, pepper, and Arabidopsis, and the splicing sites of the six introns are completely conserved. Altogether, these data can be explained if all CrtR-b genes in these species evolved from the same ancestral gene.
In Arabidopsis, which does not produce carotenoids in the petals, both genes are expressed in photosynthetic tissues under normal growth conditions. However, the mRNA level of β-carotene hydroxylase 1 is 10- to 50-fold higher than β-carotene hydroxylase 2 (Tian and DellaPenna, 2001). There is no colored chromoplast-containing tissue in Arabidopsis, and the reason for the existence of three β-hydroxylase genes, two non-heme β-carotene hydroxylases (Tian and DellaPenna, 2001) and a p450 oxygenase (Kim and DellaPenna, 2006), has remained unexplained.
Interestingly, we have observed that CrtR-b2 is expressed in tomato roots, although at a level that is <1% of that in flowers (Figure 7). Expression of CrtR-b1 in roots is at least 15-fold lower than CrtRb-2. Nevertheless, root architecture in wf, as well as overall plant morphology, do not show any phenotype (data not shown), implying that abscisic acid is not significantly altered. Since abscisic acid is produced from β-xanthophylls, it is likely that a third β-carotene hydroxylase, possibly a P450-type enzyme (Kim and DellaPenna, 2006), is active in tomato roots.
The formation of a limited amount of xanthophylls in flowers of wf indicates the existence of low-level β-ring hydroxylation activity that can be provided by CRTR-B1, which is expressed at a low level, or by a third β-carotene hydroxylase. Another possible source of β-carotene hydroxylation in wf flowers is a residual β-carotene hydroxylation activity by the ɛ-ring hydroxylase that has been suggested (Tian et al., 2004).
Gene Duplication and the Evolution of a Chromoplast-Specific Carotenoid Biosynthesis Pathway
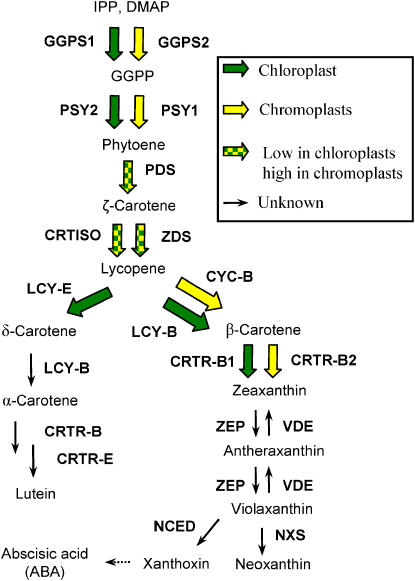
In tomato, four carotenoid biosynthesis enzymes, geranylgeranyl diphosphate synthase (Ggps; TC120732 and TC127610) (Tomato Expression Database, Cornell; http://ted.bti.cornell.edu/), phytoene synthase (Psy1 and Psy2), lycopene β-cyclase (Lcy-b and Cyc-B), and β-carotene hydroxylase (CrtR-b1 and Crtr-b2), are each encoded by at least two genes. In each of these enzymes, one isoform is constitutively expressed in leaves, whereas the other is specific for chromoplasts in flowers and/or fruits (Figure 10). The phytoene synthase encoded by Psy2 is constitutively expressed in photosynthetic tissues (Bartley and Scolnik, 1993; Fraser et al., 1999), and the one encoded by Psy1 is expressed in flowers and fruits only (Bartley et al., 1992; Fraser et al., 1999). A null mutation in Psy1 in the r mutant eliminates most of the carotenoids in fruits (Fray and Grierson, 1993) but not in leaves. The lycopene β-cyclase encoded by Lcy-b is expressed in leaves and at a relatively low level in flowers, and the β-cyclase encoded by Cyc-B is expressed exclusively in flowers and also at low levels in fruits (Pecker et al., 1996; Ronen et al., 2000). A null mutation in the Cyc-B gene, named old-gold, causes accumulation of low levels of the red pigment lycopene in the flowers and eliminates β-carotene in fruits but has no effect on carotenoids in leaves (Ronen et al., 2000). The mutation wf that revealed a flower-specific CrtR-b emphasizes gene duplication as a major mechanism underlying the establishment of a parallel carotenoid biosynthesis pathway that is activated in chromoplasts for high accumulation of pigments.
Figure 10.
Carotenoid Biosynthesis Pathway in Tomato.
Cyc-B, chromoplast-specific lycopene β-cyclase; Ggps, geranylgeranyl diphosphate synthase; Lcy-b, lycopene β-cyclase; Lcy-e, lycopene ɛ-cyclase; CrtR-b, β-ring hydroxylase; CrtR-e, ɛ-ring hydroxylase; Nxs, neoxanthin synthase; Pds, phytoene desaturase; Psy, phytoene synthase; Vde, violaxanthin de-epoxidase; NCED, 9-cis-epoxycarotenoid dioxygenase; Zds, ζ-carotene desaturase; Zep, zeaxanthin epoxidase.
It is interesting to note that other enzymes in the carotenoid biosynthesis pathway, phytoene desaturase (Pds), ζ-carotene desaturase (Zds), CrtISO, zeaxanthin epoxidase (Zep), and violaxanthin de-epoxidase (Vde), are encoded by single genes in the tomato genome (Liu et al., 2003). The reason why these genes have not been duplicated in evolution is unknown. The duplicated carotenoid genes encode rate-controlling biosynthetic enzymes. None of the enzymes Pds, Zds, and CrtISO is a bottleneck in the biosynthetic flux of carotenoids in fruits and flowers of tomato, since almost no carotenoid intermediates accumulate in these tissues. Therefore, their duplication for the enhancement of carotenoid synthesis in chromoplasts may not be necessary. Alternatively, duplication of genes for these enzymes is less likely to evolve if their mode of action is in the form of a large heteromeric complex. In such a case, parallel duplication of all three genes would be required to duplicate a whole function to be selected in evolution.
Plants of wf show no evidence of any phenotypic alteration other than in the color of the flowers. The carotenoid composition in their leaves and fruits is similar to the wild type. This is in compliance with the finding that CrtR-b2 is expressed exclusively in flowers. Among the known eight wild species of tomato only two, Solanum pimpinellifolium and Solanum cheesmanii, have red- or orange-colored fruits. The rest have green to pale yellow fruits. However, the color of flowers in all wild species of tomato is a strong yellow as a result of accumulation of xanthophylls, mainly neoxanthin (data not shown). We therefore conclude that the duplication and preservation of a second carotene hydroxylase gene in tomato that is dedicated to the generation of yellow pigments in flowers has been under strong selection pressure. Genetic evidence derived from ILs indicates the existence of CrtR-b2 that is also responsible for flower coloration in the green-fruited species Solanum pennellii and Solanum habrochaites. We suggest that the generation by gene duplication of a chromoplast pathway of carotenoid biosynthesis had occurred initially to enhance carotenogenesis in flowers, and only later in evolution were these genes recruited to increase pigmentation in fruit chromoplasts in the colored-fruited species S. cheesmanii, S. pimpinellifium, and S. lycopersicum.
METHODS
Plants and Bacteria
Tomato (Solanum lycopersicum) varieties VF-36 and M82 were used as the wild type for pigment analysis and RNA measurement. For genetic mapping IL3-1 and IL3-2, derived from crosses between S. lycopersicum cv M82 and the wild species Solanum pennellii (LA716) (Eshed and Zamir, 1995) were used. Seeds for wf mutants LA2370 and LA3575 and for the wild species LA1245 (Solanum pimpinellifolium), LA1305 (Solanum peruvianum), LA0361 (Solanum habrochaites; formerly Solanum hirsutum), LA1306 (Solanum chmielewskii), LA1340 (S. pennellii), LA1406 (Solanum cheesmanii), and LA1329 (Solanum parviflorum) were obtained from the Tomato Genetics Resource Center (University of California, Davis). Mutant lines e1827 (wf1-2) and n5681 (wf1-1) carrying wf mutations were isolated from M82 following mutagenesis and screening (Menda et al., 2004; http://zamir.sgn.cornell.edu/mutants/). Escherichia coli strain XL1-Blue grown on Luria-Bertani (LB) medium was used in all experiments that are described in this work.
Extraction of DNA from Tomato and RFLP Mapping
Genomic DNA was prepared from 15 mg of leaf tissue as described (Eshed and Zamir, 1995). RFLP on the tomato genomic DNA was done using the DNA markers TG479, TG585, TG517, CT22, and CT90 (Tanksley et al., 1992) and cDNA clones pSlCrtR-b1, pSlCrtR-b2 (this work), and Psy1.
Tomato Genomic and cDNA Libraries
The BAC genomic library of S. lycopersicum cv Heinz 1706 (http:\\www.clemson.edu) (Budiman et al., 2000) was screened to identify the genomic sequences of CrtR-b1 and CrtR-b2. Following screening one-third of the library with each of these sequences as probes, five BAC clones were identified.
Tomato (cv VF-36) leaf cDNA library in λ gt10 vector was screened using CrtR-b1 cDNA from Arabidopsis thaliana as a probe. Altogether 900,000 phage plaques were screened. Nineteen positive phages were isolated from the library. For CrtR-b2 cDNA, which lacked the 5′ end, the full putative open reading frame was amplified using reverse transcription with an oligo16T primer followed by PCR with the primers 5′-ACTGAATTTGGATCCTCCACC-3′ (forward) and 5′-CTACAGCCTAAATACAATCGC-3′ (reverse). Sequence analysis was done as previously described (Ronen et al., 1999).
Expression of CrtR-b1 and CrtR-b2 in E. coli
For expression in E. coli of carotenoid biosynthesis genes, plasmid pBCAR-T, carrying CrtE, CrtB, and CrtI from Erwinia uredovora and Lcy-b from tomato, and plasmid pDCAR, carrying the same genes but Lcy-e from tomato replaced Lcy-b, were used (Ronen et al., 1999). To express CrtR-b1 in E. coli, the 5′ end of the cDNA sequences was truncated (nucleotides 1 to 205), and the remaining sequence from nucleotide 206 to the 3′ end was cloned in the EcoRV site of the plasmid vector pBluescript KS−. Similarly, the cDNA of CrtR-b2 (Y14810) from nucleotide 16 to the 3′ end was inserted in the EcoRV site of pBluescript KS−. The recombinant plasmids were designated pTCrtR-b1 and pTCrtR-b2. Inserts were sequenced to identify possible PCR-derived mutations. To enhance the expression of CrtR-b1 and CrtR-b2, isopropylthio-β-galactoside (24 mg/L) was added to the LB medium. E. coli cells were grown overnight at 37°C followed by 5 d of shaking at room temperature for pigment accumulation.
Pigment Extraction and Analysis
Extraction of carotenoids from bacteria and plants followed previously described protocols (Ronen et al., 1999). Saponification of flower pigments was done in ethanol:KOH (60% [w/v]) 9:1 overnight at 4°C. The carotenoids were separated with di-ethyl-ether following addition of NaCl to a final concentration of 1.2%. Analysis by HPLC was previously described (Ronen et al., 1999). Carotenoids were identified by their characteristic absorption spectra and typical retention time, which corresponded to standard compounds of lycopene, β-carotene, and zeaxanthin. Carotenoid quantification was performed by integrating the peak areas using Millennium chromatography software (Waters).
DNA Sequence Analysis
DNA and RNA were extracted from 200 mg of leaf and flower tissue according to previously described protocols (Ronen et al., 1999). PCR amplifications were performed with the Hot Star Taq Master kit (Qiagen). Thermocycling conditions were 95°C for 10 min followed by 36 cycles of 94°C for 15 s, 60°C for 45 s, and 72°C for 1 to 2 min. The purified PCR products were separated on 1.3% agarose gel and stained with ethidium bromide to verify that a single product in the expected length was amplified. PCR products were sequenced as described by Ronen et al. (1999).
The following gene-specific primers were used for the PCR and sequencing reactions: for CrtR-b1, 5′-CCATCCTTCCACCTTTCTCC-3′ (forward) and 5′-TTCTTATCAATGAAGAAGGGTGA-3′ (reverse); for CrtR-b2, 5′-ATTTAGAAGGGAGTGAGACT-3′ (forward) and 5′-GCGATTGTATTTAGGCTGTAG-3′ (reverse).
Sequence Data Analysis
Sequences of β-carotene hydroxylases in different plants species were examined by comparing the presumed mature polypeptides after cleavage of the transit peptides (see Supplemental Figure 1 online). The following sequences were analyzed: Arabidopsis 1 (locus NP-200070) amino acid residues 89 to 303, Arabidopsis 2 (AAC49443) residues 91 to 310, Crocus 1 (CAC95130) residues 81 to 305, Crocus 2 (AAT84408) residues 72 to 291, pepper 1 (CRTR-B2; locus CAA70427) residues 105 to 315, pepper 2 (CRTR-B1; locus CAA70888) residues 106 to 316, rice 1 (AAP54790) residues 76 to 292, rice 2 (XP-473611) residues 102 to 303, tomato 1 (CRTR-B1; locus CAB55625) residues 99 to 309, and tomato 2 (CRTR-B2; locus CAB55626) residues 103 to 314. Alignment of these amino acid sequences was done with the ClustalW program followed by manual adjustments (Vector NTI Suite 5.5). Phylogenetic analyses based on amino acid sequences were calculated with MEGA software version 3.1 (Kumar et al., 2004). A phylogeny tree was calculated by maximum parsimony with bootstrap resampling of 1000 replicates.
Measurement of mRNA by RT-PCR
Protocols for RNA extraction and quantification by RT-PCR were previously described (Ronen et al., 1999). Total RNA was isolated from fruit, flowers (sepals, petals, anthers, and carpels), and leaves using TRI-REAGENT according to the manufacturer's protocol (Molecular Research Center). The amplification procedure by PCR consisted of 17 cycles of 1 min at 95°C, 1 min at 58°C, and 1 min at 72°C. For the purpose of quantification, 5 μCi of 32P-dCTP (specific activity 3000 Ci mmole−1) was included in the PCR reaction mixture. Various initial concentrations of mRNA, ranging over a 9-fold difference, were used to demonstrate a linear ratio between concentration of the template cDNA (corresponding to the mRNA) and the final PCR products.
To rule out the possibility that genomic DNA contaminated the RNA samples, the primers were designed so as to span intron sequences. The following primers were used for the PCR amplification: for Pds, 5′-TTGTGTTTGCCGCTCCAGTGGATAT-3′ (forward) and 5′-GCGCCTTCCATTGAAGCCAAGTAT-3′ (reverse); for CrtR-b1, 5′-GATCTTAGCTACTCGCTTGG-3′ (forward) and 5′-CACCTTTCCTCATTGATAAGA-3′ (reverse); and for CrtR-b2, 5′-AATCCGCCTCAACCGCCC-3′ (forward) and 5′-CTACAGCCTAAATACAATCGC-3′ (reverse).
Products of the PCR amplification were separated by electrophoresis in 7% polyacrylamide gels. The gels were dried and autoradiographed. The amount of DNA was determined by measuring the radioactivity using a phosphor imager (MacBAS V2.5; Fujix) and exposure to x-ray film.
Measurement of mRNA by Real-Time RT-PCR
Samples of leaves, roots, sepals, petals, anthers, and fruits from wf1-2 and its isogenic line M82 were harvested and frozen immediately in liquid nitrogen. RNA was extracted from 200 mg of tissue as previously described by Ronen et al. (1999). Amplification of the RNA samples with intron-exon junction primers confirmed that RNA samples were not contaminated by genomic DNA.
The real-time PCR reactions consisted of 6 μL cDNA, 1.5 μL 3 μM primers, 7.5 μL Failsafe real-time PCR PreMix selection kit, and 0.3 μL Failsafe real-time PCR system DNA polymerase (Epicentre), and the reaction was performed on a Rotor-Gene 2000 thermocycler (Corbett Research).
Thermocycling conditions were 95°C for 2 min followed by 40 cycles of 95°C for 15 s, 60°C for 10 s, 72°C for 15 s, and florescence acquisition at 79°C.
Melting curve analysis and sequencing verified PCR product specificity. For quantification, calibration curves were run simultaneously with experimental samples, and Ct calculations were performed by the Rotor-Gene 5.0 software (Corbett Research). The actin gene served as reference for normalization.
The following primers for the PCR amplifications were used: for Dxs, 5′-AGCTTCCGGCTGGAAACAAA -3′ (forward) and 5′-CTAGCACAATAGCAGCATCC-3′ (reverse); for Ggps2, 5′-GTACCTCGCTACCGCTACA-3′ (forward) and 5′-TAATCCCACATTAGGGTTACC-3′ (reverse); for Psy1, 5′-AACTTGTTGATGGCCCAAAC-3′ (forward) and 5′-CTGTATCGGACAAAGCACCA-3′ (reverse); for Psy2, 5′-AGTTCTGCTAGTAGATGGCC-3′ (forward) and 5′-GGGCACTAGAGATCTTGCAT-3′ (reverse); for Pds, 5′-GCAGAAGGCGTCCAGTTTAG-3′ (forward) and 5′-TGCACACAAAGTGCTCAACA-3′ (reverse); for Zds, 5′-CATGTCAAAGGCCACTCAGA-3′ (forward) and 5′-ACGGTAACAACAGGCACTCC-3′ (reverse); for CrtR-b1, 5′-TTGGTGCTGCTGTAGGAATG-3′ (forward) and 5′-GCAATGAGGCCTTTATGGAA-3′ (reverse); for CrtR-b2, 5′-ACATGTTCGTTCACGATGGA-3′ (forward) and 5′-TTGTCCGAGTGATGAAGCTG-3′ (reverse); and for actin, 5′-TTGCTGACCGTATGAGCAAG-3′ (forward) and 5′-GGACAATGGATGGACCAGAC-3′ (reverse).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers. cDNA: Arabidopsis CrtR-b, U58919; tomato actin, BT013524; tomato CrtR-b1, Y14809; tomato CrtR-b2, Y14810; tomato Dxs, AF143812; tomato Ggps2, SGN-U223568; tomato Pds, X59948; tomato Psy1, Y00521; tomato Psy2, L23424; tomato Zds, AF195507. Genomic sequences: Arabidopsis CrtR-b1, U58919; Arabidopsis CrtR-2, NM_124636; rice CrtR-b2, NM_197521; tomato CrtR-b2, DQ650804. Polypeptides: Arabidopsis CRTR-B1, NP-200070; Arabidopsis CRTR-B2, AAC49443; Crocus CRTR-B1, CAC95130; Crocus CRTR-B2, AAT84408; pepper CRTR-B2, CAA70427; pepper CRTR-B1, CAA70888; CRTR-B1, AAP54790; rice CRTR-B2, XP-473611; tomato CRTR-B1, CAB55625; tomato CRTR-B2, CAB55626.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Alignment of Amino Acid Sequences of β-Carotene Hydroxylases in Higher Plants Used for Calculating the Phylogenetic Tree.
Supplementary Material
Acknowledgments
We thank Rod Wing (Clemson University, Clemson, SC) for providing us with the tomato genomic BACs, Efraim Lewinsohn and Einat Bar (Agriculture Research Organization, Newe Yaar) for the gas chromatography–mass spectrometry analysis of volatiles and communicating of unpublished data, David Kachanovsky (Hebrew University) for his skillful advice on quantitative PCR and fruitful discussions, and Zach Lipman (Hebrew University) for valuable comments on the manuscript. This work was supported by Israel Science Foundation Grant 677/01. Work in the laboratory of J.H. was carried out under the auspices of the Avron Even-Ari Minerva Center.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Joseph Hirschberg (hirschu@vms.huji.ac.il).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.039966.
References
- Auldridge, M.E., Block, A., Vogel, J.T., Dabney-Smith, C., Mila, I., Bouzayen, M., Magallanes-Lundback, M., DellaPenna, D., McCarty, D.R., and Klee, H.J. (2006). Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 45 982–993. [DOI] [PubMed] [Google Scholar]
- Bartley, G.E., and Scolnik, P.A. (1993). cDNA cloning, expression during development, and genome mapping of PSY2, a second tomato gene encoding phytoene synthase. J. Biol. Chem. 268 25718–25721. [PubMed] [Google Scholar]
- Bartley, G.E., Viitanen, P.V., Bacot, K.O., and Scolnik, P.A. (1992). A tomato gene expressed during fruit ripening encodes an enzyme of the carotenoid biosynthesis pathway. J. Biol. Chem. 267 5036–5039. [PubMed] [Google Scholar]
- Bouvier, F., Keller, Y., d'Harlingue, A., and Camara, B. (1998). Xanthophyll biosynthesis: Molecular and functional characterization of carotenoid hydroxylases from pepper fruits (Capsicum annuum L.). Biochim. Biophys. Acta 1391 320–328. [DOI] [PubMed] [Google Scholar]
- Bramley, P.M. (2002). Regulation of carotenoid formation during tomato fruit ripening and development. J. Exp. Bot. 53 2107–2113. [DOI] [PubMed] [Google Scholar]
- Budiman, M.A., Mao, L., Wood, T.C., and Wing, R.A. (2000). A deep-coverage tomato BAC library and prospects toward development of an STC framework for genome sequencing. Genome Res. 10 129–136. [PMC free article] [PubMed] [Google Scholar]
- Corona, V., Aracri, B., Kosturkova, G., Bartley, G.E., Pitto, L., Giorgetti, L., Scolnik, P.A., and Giuliano, G. (1996). Regulation of a carotenoid biosynthesis gene promoter during plant development. Plant J. 9 505–512. [DOI] [PubMed] [Google Scholar]
- Cunningham, F.X., Jr., and Gantt, E. (1998). Genes and enzymes of carotenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 557–583. [DOI] [PubMed] [Google Scholar]
- Demmig-Adams, B., and Adams III, W.W. (2002). Antioxidants in photosynthesis and human nutrition. Science 298 2149–2153. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., and Nielsen, H. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed, Y., and Zamir, D. (1995). An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141 1147–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, H.A., Young, A., Britton, G., and Cogdell, R.J. (1999). The Photochemistry of Carotenoids. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Fraser, P.D., and Bramley, P.M. (2004). The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 43 228–265. [DOI] [PubMed] [Google Scholar]
- Fraser, P.D., Kiano, J.W., Truesdale, M.R., Schuch, W., and Bramley, P.M. (1999). Phytoene synthase-2 enzyme activity in tomato does not contribute to carotenoid synthesis in ripening fruit. Plant Mol. Biol. 40 687–698. [DOI] [PubMed] [Google Scholar]
- Fray, R.G., and Grierson, D. (1993). Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol. Biol. 22 589–602. [DOI] [PubMed] [Google Scholar]
- Gavel, M., and von Heijne, G. (1990). A conserved cleavage site motif in chloroplast transit peptides. FEBS Lett. 261 455–458. [DOI] [PubMed] [Google Scholar]
- Giuliano, G., Bartley, G.E., and Scolnik, P.A. (1993). Regulation of carotenoind biosynthesis during tomato development. Plant Cell 5 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara-García, A., San Román, C., Arroyo, A., Cortés, M.E., Gutiérrez-Nava, M.L., and León, P. (2005). Characterization of the Arabidopsis clb6 mutant illustrates the importance of posttranscriptional regulation of the methyl-D-erythritol 4-phosphate pathway. Plant Cell 17 628–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg, J. (2001). Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant Biol. 4 210–218. [DOI] [PubMed] [Google Scholar]
- Holt, N.E., Fleming, G.R., and Niyogi, K.K. (2004). Toward an understanding of the mechanism of nonphotochemical quenching in green plants. Biochemistry 43 8281–8289. [DOI] [PubMed] [Google Scholar]
- Horton, P., and Ruban, A. (2005). Molecular design of the photosystem II light-harvesting antenna: Photosynthesis and photoprotection. J. Exp. Bot. 56 365–373. [DOI] [PubMed] [Google Scholar]
- Kevan, P.G. (1983). Floral colors through the insect eye: What they are and what they mean. In Handbook of Experimental Pollination Biology, C.E. Jones and R.J. Little, eds (New York: Van Nostrand Reinhold Company), pp. 3–30.
- Kim, J., and DellaPenna, D. (2006). Defining the primary route for lutein synthesis in plants: The role of Arabidopsis carotenoid β-ring hydroxylase CYP97A3. Proc. Natl. Acad. Sci. USA 103 3474–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- Liu, Y.S., Gur, A., Ronen, G., Causse, M., Damidaux, R., Buret, M., Hirschberg, J., and Zamir, D. (2003). There is more to tomato fruit colour than candidate carotenoid genes. Plant Biotechnol. J. 1 195–207. [DOI] [PubMed] [Google Scholar]
- Menda, N., Semel, Y., Peled, D., Eshed, Y., and Zamir, D. (2004). In silico screening of a saturated mutation library of tomato. Plant J. 38 861–872. [DOI] [PubMed] [Google Scholar]
- Moehs, C.P., Tian, L., Osteryoung, K.W., and DellaPenna, D. (2001). Analysis of carotenoids biosynthetic gene expression during marigold petal development. Plant Mol. Biol. 45 281–293. [DOI] [PubMed] [Google Scholar]
- Paine, J.A., Shipton, C.A., Chaggar, S., Howells, R.M., Kennedy, M.J., Vernon, G., Wright, S.Y., Hinchliffe, E., Adams, J.L., Silverstone, A.L., and Drake, R. (2005). Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat. Biotechnol. 23 482–487. [DOI] [PubMed] [Google Scholar]
- Pecker, I., Gabbay, R., Cunningham, F.X., Jr., and Hirschberg, J. (1996). Cloning and characterization of the cDNA for lycopene b-cyclase from tomato reveals decrease in its expression during fruit ripening. Plant Mol. Biol. 30 807–819. [DOI] [PubMed] [Google Scholar]
- Robert, B., Horton, P., Pascal, A.A., and Ruban, A.V. (2004). Insights into the molecular dynamics of plant light-harvesting proteins in vivo. Trends Plant Sci. 9 385–390. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Concepcion, M., Ahumada, I., Diez-Juez, E., Sauret-Gueto, S., Lois, L.M., Gallego, F., Carretero-Paulet, L., Campos, N., and Boronat, A. (2001). 1-Deoxy-D-xylulose 5-phosphate reductoisomerase and plastid isoprenoid biosynthesis during tomato fruit ripening. Plant J. 27 213–222. [DOI] [PubMed] [Google Scholar]
- Romer, S., Fraser, P.D., Kiano, J.W., Shipton, C.A., Misawa, N., Schuch, W., and Bramley, P.M. (2000). Elevation of the provitamin A content of transgenic tomato plants. Nat. Biotechnol. 18 666–669. [DOI] [PubMed] [Google Scholar]
- Ronen, G., Carmel-Goren, L., Zamir, D., and Hirschberg, J. (2000). An alternative pathway to b-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. Proc. Natl. Acad. Sci. USA 97 11102–11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen, G., Cohen, M., Zamir, D., and Hirschberg, J. (1999). Regulation of carotenoid biosynthesis during tomato fruit development: Expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. Plant J. 17 341–351. [DOI] [PubMed] [Google Scholar]
- Sauret-Gueto, S., Botella-Pavia, P., Flores-Perez, U., Martinez-Garcia, J.F., San Roman, C., Leon, P., Boronat, A., and Rodriguez-Concepcion, M. (2006). Plastid cues posttranscriptionally regulate the accumulation of key enzymes of the methylerythritol phosphate pathway in Arabidopsis. Plant Physiol. 141 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schledz, M., Al-Babili, S., von Lintig, J., Haubruck, H., Rabbani, S., Kleinig, H., and Beyer, P. (1996). Phytoene synthase from Narcissus pseudonarcissus: Functional expression, galactolipid requirement, topological distribution in chromoplasts and induction during flowering. Plant J. 10 781–792. [DOI] [PubMed] [Google Scholar]
- Standfuss, J., Terwisscha van Scheltinga, A.C., Lamborghini, M., and Kuhlbrandt, W. (2005). Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 A resolution. EMBO J. 24 919–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley, S.D., et al. (1992). High density molecular linkage maps of the tomato and potato genomes. Genetics 132 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorup, T.A., Tanyolac, B., Livingstone, K.D., Popovsky, S., Paran, I., and Jahn, M. (2000). Candidate gene analysis of organ pigmentation loci in the Solanaceae. Proc. Natl. Acad. Sci. USA 97 11192–11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, L., and DellaPenna, D. (2001). Characterization of a second carotenoid β-hydroxylase gene from Arabidopsis and its relationship to the LUT1 locus. Plant Mol. Biol. 47 379–388. [DOI] [PubMed] [Google Scholar]
- Tian, L., Musetti, V., Kim, J., Magallanes-Lundback, M., and DellaPenna, D. (2004). The Arabidopsis LUT1 locus encodes a member of the cytochrome p450 family that is required for carotenoid ɛ-ring hydroxylation activity. Proc. Natl. Acad. Sci. USA 101 402–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen, E.A., Overduin, B., Nijkamp, H.J., and Hille, J. (1994). Integrated map of chromosome 3. TGC Report 44 8–10. [Google Scholar]
- Vishnevetsky, M., Ovadis, M., Itzhaki, H., and Vainstein, A. (1997). CHRC, encoding a chromoplast-specific carotenoid-associated protein, is an early gibberellic acid-responsive gene. J. Biol. Chem. 272 24747–24750. [DOI] [PubMed] [Google Scholar]
- Vishnevetsky, M., Ovadis, M., and Vainstein, A. (1999). Carotenoid sequestration in plants: The role of carotenoid-associated proteins. Trends Plant Sci. 4 232–235. [DOI] [PubMed] [Google Scholar]
- Young, P.A., and MacArthur, J.W. (1947). Horticultural characters of tomatoes. Tex. Agric. Exp. Stn. Bull. 698 34. [Google Scholar]
- Zhu, C., Yamamura, S., Koiwa, H., Nishihara, M., and Sandmann, G. (2002). cDNA cloning and expression of carotenogenic genes during flower development in Gentiana lutea. Plant Mol. Biol. 48 277–285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.