Abstract
Chlamydia pneumoniae is an important respiratory pathogen recently associated with atherosclerosis and several other chronic diseases. Detection of C. pneumoniae is inconsistent, and standardized PCR assays are needed. Two real-time PCR assays specific for C. pneumoniae were developed by using the fluorescent dye-labeled TaqMan probe-based system. Oligonucleotide primers and probes were designed to target two variable domains of the ompA gene, VD2 and VD4. The limit of detection for each of the two PCR assays was 0.001 inclusion-forming unit. Thirty-nine C. pneumoniae isolates obtained from widely distributed geographical areas were amplified by the VD2 and VD4 assays, producing the expected 108- and 125-bp amplification products, respectively. None of the C. trachomatis serovars, C. psittaci strains, other organisms, or human DNAs tested were amplified. The amplification results of the newly developed assays were compared to the results of culturing and two nested PCR assays, targeting the 16S rRNA and ompA genes. The assays were compared by testing C. pneumoniae purified elementary bodies, animal tissues, 228 peripheral blood mononuclear cell (PBMC) specimens, and 179 oropharyngeal (OP) swab specimens obtained from ischemic stroke patients or matched controls. The real-time VD4 assay and one nested PCR each detected C. pneumoniae in a single, but different, PBMC specimen. Eleven of 179 OP specimens (6.1%) showed evidence of the presence of C. pneumoniae in one or more tests. The real-time VD4 assay detected the most positive results of the five assays. We believe that this real-time PCR assay offers advantages over nested PCR assays and may improve the detection of C. pneumoniae in clinical specimens.
Chlamydia pneumoniae is an intracellular bacterium implicated in upper and lower respiratory tract infections in humans. It has been reported to be responsible for ∼10% of cases of community-acquired pneumonia and to be an etiologic agent of bronchitis, sinusitis, and other respiratory tract illnesses (15, 17, 18, 23). Recently, C. pneumoniae was associated with several chronic diseases, including multiple sclerosis, Kawasaki's disease, and Alzheimer's disease (2, 34, 43), although these associations have been disputed by other studies (14, 19, 41). More importantly, data from numerous studies have suggested a possible link between C. pneumoniae infections and atherosclerotic vascular diseases. The reports of the association between C. pneumoniae and atherosclerosis are based on serologic and animal model studies, direct detection of the organism in atherosclerotic lesions, and preliminary clinical trials showing improved outcome among patients treated with antibiotics (16, 22, 33, 38, 40). The accumulating data demonstrating an association between C. pneumoniae and atherosclerosis are not entirely consistent; some studies show a significant association (9, 26, 31), but others do not (39, 48, 49). Moreover, it must be emphasized that evidence proving a causal role of C. pneumoniae in the pathogenesis of atherosclerosis is still lacking.
The isolation and propagation of C. pneumoniae from clinical specimens by using cell cultures is relatively labor-intensive and insensitive, and interpretation requires technical expertise (8). Serologic analysis, particularly microimmunofluorescence tests, has been extensively used; however, interpretation is problematic, since a large part of the population has preexisting immunoglobulin G antibodies from a previous exposure(s) (47). In addition, serologic methods are subjective, and there is considerable cross-reaction with other species of Chlamydia and with Bartonella (24, 30, 35, 47). Due to the difficulties with culturing and serologic analysis, a number of nucleic acid amplification assays for detecting C. pneumoniae have been developed (6). Current PCR methods are based on the amplification of a cloned PstI fragment (7), genes encoding 16S rRNA (3, 11, 28, 32), or the gene for the major outer membrane protein, known as omp1 or ompA (45). There is no commercially available PCR assay for C. pneumoniae, and results have varied widely, especially for detection of this microorganism in atheromatous lesions. For instance, the reported rate of C. pneumoniae DNA detection within atherosclerotic lesions by PCR varies from 0 and 80% (20, 44, 49), indicating a critical need for standardized assays. In an attempt to standardize the currently available C. pneumoniae diagnostic assays, an international meeting was convened by the U.S. Centers for Disease Control and Prevention (CDC) and the Canadian Laboratory Centre for Disease Control (LCDC) (8). Four PCR methods (all conventional gel-based assays) met the proposed criteria for a validated assay (7, 11, 28, 45).
The recently introduced real-time PCR-based fluorescence technologies have many advantages: (i) high sensitivity; (ii) high specificity due to binding of two primers and one probe; (iii) usefulness as quantitative assays; (iv) operation in a closed system, avoiding contamination; and (v) ability to provide results faster than gel-based PCR assays, allowing rapid intervention (25, 46). We have developed two real-time PCR assays for C. pneumoniae by using a fluorescent dye-labeled TaqMan probe-based system (Applied Biosystems, Foster City, Calif.) (25). Two pairs of primers and two fluorescent probes were designed based on the nucleotide sequences of two regions of the ompA gene corresponding to variable domains VD2 and VD4. In contrast to the situation for C. trachomatis and C. psittaci, the ompA VD4 of C. pneumoniae is highly conserved and is therefore a good target for a species-specific PCR (12). Here we describe the development and validation of VD2 and VD4 PCR assays.
MATERIALS AND METHODS
Bacterial isolates.
C. pneumoniae isolates and other bacterial species are described in Tables 1 and 2, respectively.
TABLE 1.
C. pneumoniae isolates tested by real-time PCR based fluorescence assays designed to detect VD2 and VD4 of the ompA gene
| Isolate | Site of isolation | Geographic location | Mycoplasma contaminationa | Sourceb |
|---|---|---|---|---|
| TW-183 | Conjunctiva | Taiwan | No | ATCC (VR2282) |
| CM-1 | Sputum | Ga. | No | ATCC (VR1360) |
| CWL-029 | Pharynx | Ga. | No | ATCC (VR1310) |
| TW-2043 | Nasopharynx | N.Y. | Yes | ATCC (VR1355) |
| TW-2023 | Nasopharynx | N.Y. | Yes | ATCC (VR1356) |
| AR-39 | Pharynx | Wash. | Yes | ATCC (535920) |
| CWL-011 | Pharynx | Ga. | No | C. Black |
| CWL-050 | Pharynx | Ga. | No | C. Black |
| A-03 | Coronary atheroma | Ky. | Yes | J. Summersgill |
| BAL-15 | BALc | N.Y. | Yes | M. Hammerschlag |
| BAL-16 | BAL | N.Y. | Yes | M. Hammerschlag |
| UL-029 | Pharynx | Ky. | No | J. Summersgill |
| AR-388 | Pharynx | Wash. | Yes | T. Grayston |
| BR-393 | Pharynx | Unknown | Yes | C. Gaydos |
| W2 | Pharynx | Wis. | No | B. MacDonald |
| W3 | Pharynx | Wis. | Yes | B. MacDonald |
| W4 | Pharynx | Wis. | No | B. MacDonald |
| W5 | Pharynx | Wis. | No | B. MacDonald |
| W6 | Pharynx | Wis. | No | B. MacDonald |
| IOL-207 | Conjunctiva | Iran | No | P. Nicolini |
| FML-7 | Nasopharynx | Norway | No | B. P. Berdal |
| FML-12 | Nasopharynx | Norway | No | B. P. Berdal |
| FML-16 | Nasopharynx | Norway | No | B. P. Berdal |
| FML-19 | Nasopharynx | Norway | No | B. P. Berdal |
| H-12 | Pharynx | Finland | Yes | P. Saiku |
| K-6 | Pharynx | Finland | Yes | P. Saiku |
| K-66 | Pharynx | Finland | Yes | P. Saiku |
| P1 (parola) | Pharynx | Finland | Yes | P. Saiku |
| UZG1 | Pharynx | Belgium | Yes | J. Ossewaarde |
| 12N | Pharynx | Finland | No | B. P. Berdal |
| 19N | Pharynx | Finland | No | B. P. Berdal |
| YK-41 | Pharynx | Japan | Yes | Y. Kanamoto |
| U172 | Unknown | Sweden | Yes | J. Boman |
| U1271 | Unknown | Sweden | Yes | J. Boman |
| U1272 | Unknown | Sweden | Yes | J. Boman |
| U1273 | Unknown | Sweden | Yes | J. Boman |
| T45953 | Unknown | Sweden | Yes | J. Boman |
| AL-1 | Unknown | Sweden | Yes | J. Boman |
| IOL-1515 | Unknown | Sweden | Yes | J. Boman |
Mycoplasma contamination was tested by an in-house PCR designed to detect the 16S rRNA gene.
ATCC, American Type Culture Collection, Manassas, Va.
BAL, bronchoalveolar lavage.
TABLE 2.
Microorganisms (n = 84) used to test the cross-reactivity of the real-time PCR assays designed to detect VD2 and VD4 of the ompA gene of C. pneumoniae
| Genus | Species, serovar, serotype, or serogroup | No. tested |
|---|---|---|
| Chlamydia | C. trachomatis serovars A, Ba, C, D, E, F, G, H, I, J, K, and L2; C. psittaci serovars A, B, D, and E | 17 |
| Mycoplasma | M. pneumoniae, M. salivarium, M. fermentans, M. hominis, M. orale, M. faucium, M. buccale, and M. penetrans | 16 |
| Legionella | L. pneumophila, L. jordanis, L. micdadei, and L. longbeachae | 4 |
| Streptococcus | S. pneumoniae, S. pyogenes, S. agalactiae, group C, S. oralis, S. mitis, S. crista, S. gordonii, S. sanguis, S. parasanguis, and S. vestibularis | 22 |
| Haemophilus | H. influenzae serotypes a, b, c, d, e, and f; nontypeable H. influenzae; H. parainfluenzae | 8 |
| Neisseria | N. meningitidis serogroups A, B, C, Y, and W135; nongroupable N. meningitidis, N. sicca; N. flavescens | 8 |
| Bordetella | B. pertussis | 2 |
| Branhamella | B. catarrhalis | 1 |
| Staphylococcus | S. aureus | 1 |
| Corynebacterium | C. diphtheriae | 1 |
| Mycobacterium | M. tuberculosis | 1 |
| Eikenella | E. corrodens | 1 |
| Pseudomonas | P. aeruginosa | 1 |
| Proteus | P. mirabilis | 1 |
Animal tissues.
Two 8-week-old C57BL/6J female mice were inoculated intranasally with 40 μl of either sterile saline (control mouse) or sterile saline containing 2 × 106 inclusion-forming units (IFU) of C. pneumoniae strain BR-393 (infected mouse). The animals were sacrificed 3 days postinoculation. Liver, lung, and spleen tissues from each animal were tested by PCR to determine if the real-time PCR assays could detect C. pneumoniae DNA in animals following acute-phase infection.
Clinical specimens.
Specimens were collected from 317 subjects (age, ≥60 years) enrolled in an ongoing study designed to determine if C. pneumoniae infection increases the risk of first ischemic stroke. Informed written consent was obtained from all subjects, and procedures were carried out in accordance with the institutional review boards of the CDC and Columbia-Presbyterian Medical Center of New York. Specimens were collected from October 1999 to January 2001. These included 228 samples of peripheral blood mononuclear cells (PBMC) and 179 oropharyngeal (OP) swab samples. Because the specimens are part of an ongoing study, laboratory personnel remained blinded to the identities of cases and controls. Three atheromatous plaques and one lung tissue specimen obtained from a person free of C. pneumoniae and submitted to the CDC for reference diagnostic testing were also included in order to check for PCR inhibition. The PBMC specimens were analyzed by PCR, and the OP specimens were analyzed by PCR and culturing.
For PBMC fractionation, approximately 8 ml of whole blood was drawn from subjects at the Columbia-Presbyterian Medical Center of New York and transferred to Vacutainer CPT tubes (Becton Dickinson, Franklin Lakes, N.J.) within 2 h after collection. The blood samples were centrifuged at 1,500 × g for 1 h, and the mononuclear cell layer was aspirated, divided into two aliquots of approximately 200 μl each, and frozen at −70°C. PBMC preparations were shipped to the CDC in batches on dry ice. The samples collected with the OP swabs, with Dracon tips and plastic shafts (Remel Inc., Lenexa, Kans.), were placed in 3 ml of Multi-Microbe Medium M4-3 (Remel) and transported at 4°C, generally within 24 h. Although unknown for C. pneumoniae, the percentages of C. trachomatis recovery in M4-3 transport medium are reported by the manufacturer to be 82% after 8 h, 43% after 24 h, and 33% after 48 h at 2 to 8°C. Upon arrival, the swabs were vortexed in the transport medium for 1 min and pressed against the side of the tube to extract all of the liquid. One milliliter of this fluid was either stored at −70°C or immediately centrifuged at 20,000 × g for 20 min, and the pellet was used for DNA extraction. Frozen samples were thawed later for testing, centrifuged, and processed in the same manner as the fresh samples. A total of 350 μl of the original fluid was used for the cell culture inoculum.
Cell culturing.
C. pneumoniae isolates (Table 1) were propagated in HEp-2 cells as previously described (50). Briefly, the cells were maintained in Iscove's modified Dulbecco's medium (IMDM) (Gibco BRL, Grand Island, N.Y.) supplemented with 2 mM l-glutamine, 25 mM HEPES buffer, and 10% fetal calf serum. Monolayers were grown in 150-cm2 culture flasks at 36°C in a 5% CO2 atmosphere. Prior to infection, HEp-2 cells were used to seed eight 25-cm2 culture flasks and were incubated under the same conditions for 48 to 72 h, depending on monolayer confluence. The C. pneumoniae-infected HEp-2 cells were centrifuged at 1,000 × g for 1 h at 25°C, and the inoculum was removed and replaced with IMDM supplemented with cycloheximide (1 μg/ml). The cells were incubated at 36°C in a 5% CO2 atmosphere for 72 h. The monolayers were harvested and sonicated at 60 Hz for 30 s, and cellular debris was removed by centrifugation (500 × g, 10 min, 25°C). Elementary bodies were pelleted at 30,000 × g for 45 min at 4°C, resuspended in 0.5 ml of sucrose-phosphate-glutamate medium supplemented with 10% fetal calf serum, and stored in aliquots at −70°C until used.
The titration of C. pneumoniae frozen stock cultures was performed with 96-well microtiter plates containing HEp-2 cells. Fifty-microliter quantities of 10-fold dilutions of C. pneumoniae stock cultures were used to inoculate triplicate wells. After incubation, the cells were fixed with methanol and stained with a Chlamydia genus-specific monoclonal antibody (Pathfinder chlamydia culture confirmation system; Bio-Rad S.A., Redmond, Wash.). The inclusions were counted by using an inverted fluorescence microscope, and the number of IFU per milliliter was calculated for each stock. Contamination with another Chlamydia species was excluded by staining the inclusions with a C. pneumoniae-specific monoclonal antibody by using a chlamydia cel Pn IF test (Cellabs, Brookvale, Australia) and PCR analysis with genus- and species-specific primers (32). Contamination with Mycoplasma was checked by a PCR assay designed to detect the 16S rRNA gene (36).
OP specimens were cultured in HEp-2 cells by using multiple centrifugations as described by Pruckler et al. (37). OP specimens (350 ml) were sonicated for 30 s, and 50 μl was used to inoculate, in triplicate, 96-well microtiter plates containing HEp-2 cells. Two plates were used for each experiment; one was stained, and the other subcultured. After centrifugation at 1,000 × g for 1 h at 25°C, the inoculum was removed and replaced with IMDM supplemented with cycloheximide (1 μg/ml), gentamicin (10 μg/ml), vancomycin (25 μg/ml), and amphotericin B (2 μg/ml). Cultures were incubated at 36°C in a 5% CO2 atmosphere. On day 3, cultures were centrifuged, and the medium was replaced. On days 4 and 5, cultures were centrifuged again, but the medium was not replaced. After the total of 7 days of incubation, the cultures were blindly passaged twice and incubated for another 3 days per passage before being considered negative. Cultures were fixed, stained, and visualized as describe before. If a potential inclusion was seen, the HEp-2 cells were scraped from the three wells, and the presence of C. pneumoniae was tested by the real-time PCR assay targeting the VD4 region. If the PCR assay was positive, the culture was considered positive, and up to five additional passages were performed in order to propagate the isolates.
DNA extraction.
DNAs from C. pneumoniae purified elementary bodies, other bacterial isolates, clinical specimens, and animal tissues were extracted with a QiaAmp DNA mini kit (Qiagen Inc., Valencia, Calif.) according to the manufacturer's instructions, eluted in 100 μl of Qiagen elution buffer, and stored at −20°C. One negative no-template control was included for every five processed specimens; in the control, sterile distilled water was added instead of specimen. The concentrations of DNAs extracted from C. pneumoniae isolates were measured with a spectrophotometer (A260).
Real-time PCR assays.
Two pairs of primers and two fluorescent probes were designed based on the nucleotide sequences of VD2 and VD4 of the ompA gene of C. pneumoniae. They were designed by using ABI Primer Express software (Applied Biosystems) and generated PCR products of 108 bp (VD2) and 125 bp (VD4). The fluorogenic probes were synthesized with a 6-carboxy-fluorescein (FAM) reporter molecule attached at the 5" end and a 6-carboxy-tetramethyl-rhodamine (TAMRA) N-hydroxysuccinimide (NHS) ester quencher dye linked to a linker arm nucleotide phosphoramidite (Glen Research, Sterling, Va.) close to the 3" end (25). The probes were synthesized with a 3"-terminal phosphate group to prevent extension during PCR. Primers and probes were synthesized in the Biotechnology Core Facility at the CDC by using a model 394-8 DNA synthesizer (Applied Biosystems) and standard phosphoramidite chemistries.
A BLAST search was performed to check the specificity of the DNA sequences of the VD4 and VD2 primer and probe sets. In addition, the targeted sequences of four isolates of C. pneumoniae (GenBank accession numbers: CWL-029, AE001652; AR-39, M69230; IOL-207, M64064; and N16, L04982) were aligned with the VD2 and VD4 sequences of C. trachomatis (GenBank accession number X77364) and C. psittaci (GenBank accession number L25436) in order to verify the species specificity of both assays (Fig. 1).
FIG. 1.
Alignment of primer and probe sequences with VD2 and VD4 regions of the ompA genes of C. pneumoniae, C. trachomatis serovar D (C. t.) and C. psittaci serovar C (C. p.). Dots indicate identity with C. pneumoniae isolate CWL-029.
Reactions were prepared with a 96-well MicroAmp optical plate (Applied Biosystems) by the addition of a 5-μl aliquot of extracted DNA to 20 μl of a PCR master mixture consisting of 1× TaqMan universal PCR master mix (Applied Biosystems), 200 nM each primer, and 100 nM fluorescent probe. Primers and probes had been previously titrated to check for amplification efficiency. Amplification and detection were performed with an AB Prism 7700 sequence detection system (Applied Biosystems) by using the manufacturer's standard protocols. The thermal cycling conditions were 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. Standard procedures for the operation of the model 7700 system were followed in this study, including the use of all default program settings, with the exception of reaction volume, which was changed from 50 to 25 μl. Cycle threshold (CT) values, defined as the fraction of a cycle number at which the measured fluorescence generated by the released reporter molecule during cleavage exceeds a fixed threshold value above the baseline, were automatically calculated by the instrument for each reaction. Target gene copy values were derived from a standard curve generated by plotting the CT values for 10-fold serial dilutions of 109 to 100 copies of the VD4 PCR product. Each run contained at least six no-template controls to establish the baseline emission intensity of the quenched reporter dye. Negative controls (one for every five extracted DNA samples) were included, as was a positive control consisting of 1 ng of DNA extracted from C. pneumoniae isolate ATCC VR1360. The VD4 PCR product was obtained by amplification of the DNA extracted from the same isolate, following by purification with a QIAquick PCR purification kit (Qiagen) and precipitation with 1 volume of isopropanol and 0.1 volume of 3 M sodium acetate. The pellet was washed twice with ethanol, dried, and resuspended in 50 μl of Qiagen elution buffer. The DNA concentrations were determined by using a spectrophotometer (A260), and the number of copies was calculated. The 10-fold serial dilutions were dispensed in single-use amounts to avoid freeze-thaw and were stored at −20°C until needed.
The sensitivities of both real-time PCR assays were evaluated by testing approximately 1 ng of DNA from each of 39 C. pneumoniae strains (Table 1) and by testing 10-fold serial dilutions of DNA extracted from purified elementary bodies of C. pneumoniae ATCC VR1360. The specificity was determined by testing DNA extracted from other bacterial species commonly found in the human respiratory tract (Table 2) and human placental DNA (Sigma, St. Louis, Mo.). In order to identify potential inhibitors in the samples or in the PCR itself, three pools of repeatedly negative DNA extracts from clinical specimens (five PBMC specimens, five OP specimens, and three atheromatous plaques) and one lung tissue specimen were spiked with 10-fold serial dilutions of purified C. pneumoniae DNA (ATCC VR1360) ranging from 106 to 102 copies. Amplification plots were compared to those obtained by using only the serial dilutions (10−1 to 10−5) of the C. pneumoniae isolate as DNA templates. In addition, amplification of the human β-globin gene was used as a control for PCR inhibition. A 5-μl aliquot of DNA extracted from each clinical specimen was tested in a separate reaction for the presence of amplified DNA by using the real-time PCR protocol described by Tucker et al. (46).
Comparison to nested PCR assays.
The performances of the VD2 and VD4 real-time PCR assays in detecting C. pneumoniae DNA were compared to those of two nested PCRs, one targeting the 16S rRNA gene (32) and the other targeting a different region of the ompA gene (45). The latter PCR uses the technique of “touchdown,” in which the annealing temperature is lowered 1°C every two cycles from 65 to 55°C. The four assays were compared by (i) determining the sensitivity for detecting C. pneumoniae DNA, (ii) testing tissues from animals infected with C. pneumoniae, (iii) testing OP and PBMC specimens obtained from subjects enrolled in the stroke case control study, and (iv) examining the ability of the assays to detect C. pneumoniae in OP specimens relative to that of cell culturing. The sensitivities of the four methods were compared by using DNA extracted from C. pneumoniae ATCC VR1360 (titer, 7 × 105 IFU/ml). The stock culture was 10-fold serially diluted in culture medium containing HEp-2 cells to keep the level of the DNA background constant. DNA was extracted from each dilution. Assuming 100% DNA isolation efficiency, the amount of IFU used for amplification ranged from 1.4 IFU to 0.00014 IFU per PCR. For all four PCRs, the amplification reactions were run in a volume of 50 μl containing 5 μl of DNA in triplicate and on the same day to avoid freeze-thaw of DNA extracts. The nested assays were performed according to published protocols, with a slight modification made for the inner primer sequences of the 16S rRNA nested PCR (32) in order to decrease the difference in melting temperatures between the primers (Table 3). The concentrations of the PCR components and cycling conditions were the same, except that 1.5 mM MgCl2 and a 62°C annealing temperature were used in the 16S rRNA inner PCR. For testing of clinical specimens or animal tissues, three reactions were carried out for each DNA extract (undiluted and 1:5 and 1:25 dilutions) (M. Zhang, B. P. Holloway, W. L. Thacker, S. B. Schwartz, and D. F. Talkington, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. G18, 1999), except for the nested PCR targeting the ompA gene (45), used only for undiluted clinical specimens. Positive amplification visualized by 1.5% agarose gel electrophoresis was confirmed by sequencing of the PCR product. A positive result in a single PCR determination was considered a positive test. Several measures were applied to all PCRs to avoid carryover contamination of specimens by amplified products. Separate rooms were used for preparing specimens, setting up PCRs, and analyzing products. The samples and the PCR master mixtures were prepared in two dedicated class II laminar safety cabinets. A separate set of pipettes was devoted to each step of the reaction, and aerosol barrier pipette tips as well as disposable gloves and gowns were used all the time.
TABLE 3.
PCR assays for C. pneumoniae detection
| Assay format | Target | Primer or probe designation | Primer or probe sequence (5" → 3") | Amplicon size (bp) | Taq DNA polymerase | DNA/PCR (μl) | Source or reference |
|---|---|---|---|---|---|---|---|
| Real-time VD2 | ompA | VD2F | CGT TGG TTT ATT CGG AGT TA | 108 | Hot start | 5/25 | This study |
| VD2R | CCA AGA GAA AGA GGT GTC TGT | ||||||
| VD2 probe | FAM-TGT AAA TGC AAA TGA ACT ACC AAA CGT TTC-TAMRA | ||||||
| Real-time VD4 | ompA | VD4F | TCC GCA TTG CTC AGC C | 125 | Hot start | 5/25 | This study |
| VD4R | AAA CAA TTT GCA TGA AGT CTG AGA A | ||||||
| VD4 probe | FAM-TAA ACT TAA CTG CAT GGA ACC CTT CTT TAC TAG G-TAMRA | ||||||
| Nested touchdown | ompA | CP1 | TTA CAA GCC TTG CCT GTA GG | 333 | Regular | 10/50 | 45 |
| CP2 | GCG ATC CCA AAT GTT TAA GGC | ||||||
| CPC | TTA TTA ATT GAT GGT ACA ATA | 207 | |||||
| CPD | ATC TAC GGC AGT AGT ATA GTT | ||||||
| Nested | 16S rRNA | OF | ACG GAA TAA TGA CTT CGG | 436 | Hot start | 10/50 | 32 (Modified) |
| OR | TAC CTG GTA CGC TCA AAT | ||||||
| IF | CGG AAT AAT GAC TTC GGT TGT TAT TTA G | 224 | |||||
| IR | TCA TCG CCT TGG TGG GCT T |
Statistical analysis.
The VD2 PCR, VD4 PCR, 16S rRNA nested PCR, ompA nested PCR, and culture results for C. pneumoniae detection were compared by using Fisher's exact test. Statistical significance was defined as a P value of <0.05.
RESULTS
Analytical sensitivity and specificity.
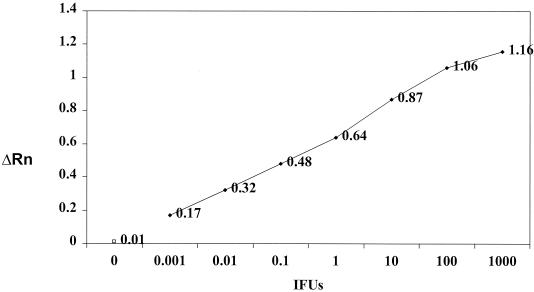
The real-time VD2 and VD4 assays successfully amplified DNAs from all C. pneumoniae isolates (Table 1). The CT values ranged from 16 to 24 cycles. The lowest level of detection of purified DNA extracted from C. pneumoniae isolate ATCC VR1360 was 0.001 IFU per PCR when either VD2 (data not shown) or VD4 (Fig. 2) was targeted, corresponding to less than 10 copies of C. pneumoniae DNA. No amplified DNA products were detected when DNAs extracted from C. psittaci, C. trachomatis, or the other bacterial species listed in Table 2 or human DNA was tested for specificity.
FIG. 2.
Sensitivity limits of the ompA VD4 real-time PCR assay. Tenfold dilutions of DNA extracted from C. pneumoniae isolate ATCC VR1360 and with a known titer (IFU per milliliter) were used in the assay. The number of IFU per dilution, fluorescence measurement, and data analysis were determined as described in Materials and Methods. The limit of detection of the real-time PCR assay was 0.001 IFU per PCR. ΔRn, fluorescence emission values. Open square, negative control.
Evaluation of PCR inhibition in spiked specimens.
The pools of negative clinical specimens (PBMC, OP swabs, or atheromatous plaques) and the lung tissue spiked with serial dilutions of C. pneumoniae DNA ranging from 106 to 102 copies did not show any inhibition when VD4 was targeted. In a similar manner, the VD2 assay was positive at all concentrations tested, except for 100 copies of C. pneumoniae DNA in the atheromatous plaque tissue pool. The human β-globin gene was detected in all 14 specimens tested as a control for inhibition. The C. pneumoniae DNA-spiked specimens showed CT values similar to those obtained for reactions containing only purified DNA up to the dilution 10−3, corresponding to 10,000 copies. Some variability in CT values was seen for the higher dilutions in both VD2 and VD4 assays (data not shown). It is possible that this variability occurred because of some degree of inhibition in the presence of a low copy number or some other unknown limiting factor.
Comparison to nested PCR assays.
The sensitivity limits of the VD2 and VD4 real-time assays were compared to those of two nested PCR assays by using DNA extracted from dilutions of C. pneumoniae cultures with known titers. The limit of detection for all four assays was 0.001 IFU per PCR when 10-fold serial dilutions in Tris buffer of DNA extracted from the stock culture were tested. The sensitivities of the VD4 and VD2 assays were 0.014 and 0.14 IFU of C. pneumoniae per PCR, respectively, when a C. pneumoniae culture was serially diluted in HEp-2 cells (to mimic eukaryotic DNA levels in clinical samples) and the DNA was extracted from each dilution. With a standard curve, the average minimal amounts of C. pneumoniae DNA detected in the three replicates of the VD4 and VD2 real-time PCR assays were 12 and 83 copies, respectively. The detection limit for the nested PCR targeting the 16S rRNA gene was 0.014 IFU per PCR, and that of the nested PCR targeting the ompA gene was 0.14 IFU per PCR.
The performances of the VD2 and VD4 assays were compared to those of nested PCR assays by using DNAs extracted from lung, liver, and spleen tissues of a mouse infected with C. pneumoniae. All four assays detected C. pneumoniae only in the lung tissue of the infected animal. The three DNA extracts obtained from the lung tissue (undiluted and 1:5 and 1:25 dilutions) were found positive by all PCR assays. The tissues from the sham-inoculated control mouse were PCR negative.
A total of 228 PBMC specimens were analyzed by the four PCR assays. The real-time VD4 PCR assay and the 16S rRNA nested PCR assay each detected C. pneumoniae in a single, but different, PBMC specimen. All PBMC specimens were found negative by the VD2 and ompA PCR assays. The identity of the C. pneumoniae PCR product detected by the 16S rRNA nested PCR was confirmed by DNA sequencing. The discrepancy between the VD4 and 16S rRNA nested PCR assays was not resolved because both specimens were found negative by the other assays. The CT and relative copy number values determined for the PBMC specimen found positive by the VD4 assay were 38 cycles and 1.3 copies, respectively. Two of five repeated VD4 reactions were positive with undiluted DNA extracts only, confirming the small amount of C. pneumoniae DNA in this sample. In contrast, the human β-globin gene was successfully amplified from all 228 PBMC specimens.
Among the 179 OP specimens tested by PCR, 3 (1.7%) were found positive by both the VD2 and the VD4 assays (Table 4). An additional three samples (1.7%) were found positive by the VD4 assay only. Three samples (1.7%) were found positive by the 16S rRNA nested PCR but were not reactive in any other assay (Table 4). The DNA sequences of the 16S rRNA PCR products were confirmed to be those of C. pneumoniae. The nested PCR targeting the ompA gene did not detect C. pneumoniae DNA in any of the specimens. Interestingly, two OP specimens found positive by the 16S nested PCR and one of the six specimens found positive by the VD4 PCR were found positive only when 1:5 dilutions of the DNA extract were used. This result could have been caused by the presence of PCR inhibitors in the undiluted samples. Attempts to repeat the reactions with diluted DNA extracts were not successful, possibly because of DNA degradation during storage. All OP specimens found positive by the VD2 or VD4 assay had CT values of 35 cycles or higher, suggesting a low DNA copy number in those specimens. The relative copy number calculated by the standard curve for C. pneumoniae DNA in those specimens was between 17 and 47 copies. The human β-globin gene was successfully amplified in all but two OP specimens, suggesting the presence of PCR inhibitors. Those two specimens were found negative by the four PCR assays and culturing.
TABLE 4.
Comparison of real-time and nested PCR assays with cell culturing for detection of C. pneumoniae in 179 oropharyngeal specimens
| No. (%) of oropharyngeal specimens | Resulta of:
|
|||||
|---|---|---|---|---|---|---|
| Culturingb | Real-time TaqMan
|
16S rRNA nested PCR | ompA nested touchdown PCR | |||
| VD2 | VD4 | |||||
| 2 (1.1) | + | + | + | − | − | |
| 2 (1.1) | + | − | − | − | − | |
| 1 (0.6) | − | + | + | − | − | |
| 2 (1.1) | Overgrowthc | − | + | − | − | |
| 1 (0.6) | − | − | + | − | − | |
| 1 (0.6) | Overgrowth | − | − | + | − | |
| 2 (1.1) | − | − | − | + | − | |
| 6 (3.3) | Overgrowth | − | − | − | − | |
| 162 (90.5) | − | − | − | − | − | |
+, positive; −, negative.
One to three chlamydial inclusions were visualized in three wells. Only one of the positive cultures could be propagated after three blind passages. The other three positive cultures could not be propagated, and results were confirmed by PCR analysis of the infected monolayers.
Overgrowth by respiratory flora.
Comparison to culturing.
The PCR and culture results for C. pneumoniae detection in OP specimens were compared (Table 4). Four (2.2%) of the 179 OP specimens were culture positive. Very few chlamydial inclusions were observed in positive cultures (one to three per culture). They were confirmed as C. pneumoniae inclusions by VD4 PCR analysis of the infected HEp-2 cell monolayers. The inclusions of one culture were successfully propagated by serial passage. Two of four culture-positive samples were also found positive by the VD2 and VD4 real-time PCR assays but not by either nested PCR assay (Table 4). Successful recovery of C. pneumoniae was not possible for nine OP specimens due to overgrowth of the respiratory flora. Three of those samples were found PCR positive, two by the VD4 assay and one by the 16S rRNA nested PCR assay (Table 4). Overall, 6.1% (11 of 179) OP specimens showed evidence of the presence of C. pneumoniae in one or more of the five methods. The VD4 assay detected the most positive results (3.3% positives), followed by culturing (2.2% positives) and then VD2 and 16S nested PCR assays (1.7% positives). The nested touchdown PCR targeting the ompA gene was the least sensitive test.
We compared the proportions of OP specimens found positive by the VD4 assay with the proportions of OP specimens found positive by any of other four assays and found no statistically significant difference among the assays, except for the comparison of the VD4 PCR assay results (6 of 179 positive) with the ompA nested PCR assay results (0 of 179 positive), which was statistically significant (P = 0.01).
DISCUSSION
A large number of PCR assays have been developed to detect C. pneumoniae infection. This large number is partly due to the potential impact of demonstrating a causal role for C. pneumoniae in atherosclerosis and partly due to the difficulty in isolating the bacteria by culturing (6, 8). Current PCR assays are noncommercial assays that use various primers, reaction conditions, and methods of product visualization. In a multicenter comparison of PCR methods for the detection of C. pneumoniae in endarterectomy specimens, the positivity rates varied between 0 and 60% for the different methods (1).
Real-time PCR-based fluorescence assays have advantages over nested or single-step gel-based assays. First, fluorescent probes make the assays more specific than a non-probe-based PCR. Second, they require less manipulation, reducing the potential for amplification product carryover. This second point is an enormous advantage, especially over nested PCR technology. Finally, by use of a standard curve for the target of interest, relative copy number values can be determined for any unknown sample. Quantitative assays may be useful for determining the chlamydial load in C. pneumoniae carriers, especially for epidemiologic studies or for monitoring the effect of therapy in treatment trials.
This study describes the optimization of two real-time PCR assays by using the validation criteria established during a meeting for the standardization of C. pneumoniae diagnostic assays (8). The CDC-LCDC recommendations stated that all new PCR assays should be compared to at least one of the four selected assays that met the proposed validation criteria (three single-step PCR assays [7, 11, 28] and one nested touchdown PCR assay [45]). Nested PCR assays are usually more sensitive than single-step PCR assays for the detection of C. pneumoniae in respiratory and PBMC specimens (3, 4, 29, 42). The sensitivities of the VD2 and VD4 assays were compared to those of the nested PCR assays described by Tong and Sillis (45) and Messmer et al. (32). Using purified C. pneumoniae DNAs as templates, the limits of C. pneumoniae detection of the real-time PCRs were equivalent to those of the nested PCRs (0.001 IFU per PCR). Similar results for an ompA nested PCR were reported by Mahony et al. (29) in a comparison of five PCR methods. Conversely, the lower limit of sensitivity for the four methods was different when a C. pneumoniae culture was serially diluted in medium containing HEp-2 cells. The limit of detection of the VD4 PCR assay was the same as that of the 16S rRNA nested PCR assay and 10-fold lower (more sensitive) than that of the VD2 or ompA PCR assay (0.014 versus 0.14 IFU).
PCR is a technique theoretically capable of detecting a single copy of purified target DNA, but the procedure often lacks sensitivity, reproducibility, and specificity when applied to direct testing of clinical material (10). This unsatisfactory performance of PCR is believed to be due to the presence of inhibitors of the polymerase in clinical material, low numbers of DNA molecules in clinical specimens, degradation of DNA molecules (false negative), or contamination by previously amplified DNA (false positive). The sensitivities of the four PCR assays were similar when acutely challenged animal tissues were used as DNA templates. This result was partly due to the high concentration of C. pneumoniae DNA in the lungs of the intranasally inoculated mouse. In contrast, performances were slightly different when clinical specimens from stroke patients or controls subjects were tested. The VD4 real-time PCR assay and the 16S rRNA nested PCR assay were both able to detect C. pneumoniae in a single but different PBMC specimen, but the results of the VD2 assay and the ompA nested PCR assay were negative. These results were consistent with the higher sensitivity of the VD4 and 16S rRNA PCR assays than of the VD2 and ompA PCR assays when mixtures of C. pneumoniae elementary bodies and HEp-2 cell DNA were tested. In contrast to our findings, the ompA nested PCR assay (45) was found to be the most sensitive assay in another study of five gel-based PCR assays for C. pneumoniae detection in PBMC specimens (29). In that study, the results of three single-step PCR assays (7, 11, 28) were all negative for 148 PBMC specimens tested, even though 11 specimens were found positive by an ompA PCR assay (P < 0.001).
The detection of C. pneumoniae DNA in PBMC specimens is the subject of some controversy. While some investigators have reported a high prevalence of C. pneumoniae DNA in patients with cardiovascular disease and in middle-aged blood donors (5, 27), others have found C. pneumoniae DNA in PBMC specimens from only a small proportion of patients with coronary artery disease (29, 51). It has been hypothesized that some of the differences in the performances of PCR assays for detecting C. pneumoniae in PBMC are attributable to differences between assays, the presence of PCR inhibitors in the blood, sampling variability, or low concentrations of C. pneumoniae DNA in PBMC (29, 42). Smieja et al. (42) found that performing 5 or 10 replicates considerably increased sensitivity and reproducibility by demonstrating that the proportion of replicates that were positive would increase with the concentration of C. pneumoniae in the PBMC sample. We confirmed the low amount of C. pneumoniae DNA in the positive PBMC sample by using a standard curve in the VD4 real-time assay. Only two of five replicates of the VD4 reaction were positive, consistent with the observations for low amounts of DNA reported by Smieja et al. (42).
Among the 179 OP specimens tested by PCR and culturing, two of four culture-positive specimens were found positive by the VD2 and VD4 real-time PCR assays. In contrast, all four culture-positive samples were found negative by both nested PCR assays. Although nucleic acid amplification techniques can detect the presence of C. pneumoniae in clinical specimens and have been considered more sensitive than culturing (4), discrepant results for culturing and PCR have been reported. In a study to assess the utility of PCR for the diagnosis of acute-phase infection with C. pneumoniae in symptomatic and asymptomatic patients (13), only 23 of 31 culture-positive specimens were PCR positive. The authors suggested that one possible explanation for this discrepancy was the presence of PCR inhibitors in some samples. We tested PCR inhibition by amplification of the β-globin gene and found complete inhibition in only two OP specimens, which did not correspond to any of the culture-positive specimens. Some level of inhibition may be present in all specimens. For some specimens, the inhibition may be limited so that higher concentrations of human DNA are amplified but low levels of C. pneumoniae DNA are not detected. This notion is consistent with the low number of inclusions visualized in the positive cultures of OP specimens, as well as by the low DNA copy numbers or CT values of ≥35 detected in all positive OP specimens by the real-time PCRs. We found a better correlation between the real-time PCR assays and culturing than between the nested PCR assays and culturing.
All positive specimens were repeat tested at least once by using undiluted extracts and 1:5 dilutions of the same DNA extracts. A few specimens were not repeatedly positive, suggesting DNA degradation due to thaw-freeze cycles or sampling variability (42). Another possibility for samples being found positive by a single assay is a false-positive reaction. However, the presence of contamination is not likely, as none of the negative controls was positive.
Several parameters were optimized to enable the VD2 and VD4 real-time assays to detect C. pneumoniae with relatively high sensitivity and specificity. The novel primer sets generate short PCR products (smaller than 200 bp); in contrast, products are larger than 400 bp in a majority of PCR methods previously described for C. pneumoniae detection (6). Therefore, enzyme and other reaction components do not become limiting factors, and poor template integrity in crude DNA preparations will not compromise the yield of amplified products. In addition, the primers are designed to target a species-specific region, avoiding cross-reactions with other Chlamydia species. The ompA PCR target is more species specific than the 16S rRNA target. The rRNA genes are some of the most highly conserved genes in nature, making them poor specific targets for PCR. In contrast to those of some bacterial species, the 16S rRNA gene of C. pneumoniae is present only as a single copy in the genome (21); therefore, its use as a target does not increase the possibility of detection of small numbers of C. pneumoniae.
The VD4 assay appears to be sensitive and specific for C. pneumoniae detection. Our results suggest that testing several replicates of recently extracted DNA before freeze-thaw cycles by using a sensitive test such as the VD4 real-time PCR is the best approach for detecting C. pneumoniae in clinical specimens. This approach is particularly relevant when low copy numbers of C. pneumoniae DNA are present. In addition, the practice of diluting DNA extracts will increase the chances of positive reactions if PCR inhibitors are present.
Acknowledgments
We thank the individuals listed in Table 1 for supplying the C. pneumoniae isolates. We also thank Valerie Stevens, Ericka Christian, Jan Pruckler, and Annette Schwartz for technical assistance.
REFERENCES
- 1.Apfalter, P., F. Blasi, J. Boman, C. A. Gaydos, M. Kundi, M. Maass, A. Makristathis, A. Meijer, R. Nadrchal, K. Persson, M. L. Rotter, C. Y. Tong, G. Stanek, and A. M. Hirschl. 2001. Multicenter comparison trial of DNA extraction methods and PCR assays for detection of Chlamydia pneumoniae in endarterectomy specimens. J. Clin. Microbiol. 39:519-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balin, B. J., H. C. Gerard, E. J. Arking, D. M. Appelt, P. J. Branigan, J. T. Abrams, J. A. Whittum-Hudson, and A. P. Hudson. 1998. Identification and localization of Chlamydia pneumoniae in the Alzheimer's brain. Med. Microbiol. Immunol. (Berlin) 187:23-42. [DOI] [PubMed] [Google Scholar]
- 3.Black, C. M., P. I. Fields, T. O. Messmer, and B. P. Berdal. 1994. Detection of Chlamydia pneumoniae in clinical specimens by polymerase chain reaction using nested primers. Eur. J. Clin. Microbiol. Infect. Dis. 13:752-756. [DOI] [PubMed] [Google Scholar]
- 4.Boman, J., A. Allard, K. Persson, M. Lundborg P. Juto, and G. Wadell. 1997. Rapid diagnosis of respiratory Chlamydia pneumoniae infection by nested touchdown polymerase chain reaction compared with culture and antigen detection by EIA. J. Infect. Dis. 175:1523-1526. [DOI] [PubMed] [Google Scholar]
- 5.Boman, J., S. Soderberg, J. Forsberg, L. S. Birgander, A. Allard, K. Persson, K., E. Jidel, U. Kumlin, P. Juto, A. Waldenstrom, and G. Wadell. 1998. High prevalence of Chlamydia pneumoniae DNA in peripheral blood mononuclear cells in patients with cardiovascular disease and in middle-aged blood donors. J. Infect. Dis. 78:274-277. [DOI] [PubMed] [Google Scholar]
- 6.Boman, J., C. A. Gaydos, and T. C. Quinn. 1999. Molecular diagnosis of Chlamydia pneumoniae infection. J. Clin. Microbiol. 37:3791-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, L. A., M. Perez Melgosa, D. J. Hamilton, C. C. Kuo, and J. T. Grayston. 1992. Detection of Chlamydia pneumoniae by polymerase chain reaction. J. Clin. Microbiol. 30:434-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dowell, S. F., R. W. Peeling, J. Boman, G. M. Carlone, B. S. Fields, J. Guarner, M. R. Hammerschlag, L. A. Jackson, C. C. Kuo, M. Maass, T. O. Messmer, D. F. Talkington, M. L. Tondella, S. R. Zaki, and The C. pneumoniae Workshop Participants. 2001. Standardizing Chlamydia pneumoniae assays: recommendations from the Centers for Disease Control and Prevention (USA) and the Laboratory Centre for Disease Control. Clin. Infect. Dis. 33:492-503. [DOI] [PubMed] [Google Scholar]
- 9.Espinola-Klein, C., H. J. Rupprecht, S. Blankenberg, C. Bickel, H. Kopp, G. Rippin, G. Hafner, U. Pfeifer, and J. Meyer. 2000. Are morphological or functional changes in the carotid artery wall associated with Chlamydia pneumoniae, Helicobacter pylori, cytomegalovirus, or herpes simplex virus infection? Stroke 31:2127-2133. [DOI] [PubMed] [Google Scholar]
- 10.Fredricks, D. N., and D. A. Relman. 1999. Application of polymerase chain reaction to the diagnosis of infectious diseases. Clin. Infect. Dis. 29:475-488. [DOI] [PubMed] [Google Scholar]
- 11.Gaydos, C. A., T. C. Quinn, and J. J. Eiden. 1992. Identification of Chlamydia pneumoniae by DNA amplification of the 16S rRNA gene. J. Clin. Microbiol. 30:796-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaydos, C. A., T. C. Quinn, L. D. Bobo, and J. J. Eiden. 1992. Similarity of Chlamydia pneumoniae strains in the variable domain IV region of the major outer membrane protein gene. Infect. Immun. 60:5319-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaydos, C. A., P. M. Roblin, M. R. Hammerschlag, C. L. Hyman, J. J.Eiden, J. Schachter, and T. C. Quinn. 1994. Diagnostic utility of PCR-enzyme immunoassay, culture, and serology for detection of Chlamydia pneumoniae in symptomatic and asymptomatic patients. J. Clin. Microbiol. 32:903-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gieffers, J., E. Reusche, W. Solbach, and M. Maass. 2000. Failure to detect Chlamydia pneumoniae in brain sections of Alzheimer's disease patients. J. Clin. Microbiol. 38:881-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grayston, J. T. 1992. Infections caused by Chlamydia pneumoniae strain TWAR. Clin. Infect. Dis. 155:757-761. [DOI] [PubMed] [Google Scholar]
- 16.Gupta, S., E. W. Leatham, D. Carrington, M. A. Mendall, J. C. Kaski, and A. J. Camm. 1997. Elevated Chlamydia pneumoniae antibodies, cardiovascular events, and azithromycin in male survivors of myocardial infarction. Circulation 96:404-407. [DOI] [PubMed] [Google Scholar]
- 17.Hammerschlag, M. R. 1999. Community-acquired pneumonia due to atypical organisms in adults: diagnosis and treatment. Infect. Dis. Clin. Practice 8:232-240. [Google Scholar]
- 18.Hammerschlag, M. R. 2000. The role of Chlamydia in upper respiratory tract infections. Curr. Infect. Dis. Rep. 2:115-120. [DOI] [PubMed] [Google Scholar]
- 19.Hammerschlag, M. R., Z. Ke, F. Lu, P. Roblin, J. Boman, and B. Kalman. 2000. Is Chlamydia pneumoniae present in brain lesions of patients with multiple sclerosis? J. Clin. Microbiol. 38:4274-4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jantos, C. A., A. Nesseler, W. Waas, W. Baumgartner, H. Tillmanns, and W. Haberbosch. 1999. Low prevalence of Chlamydia pneumoniae in atherectomy specimens from patients with coronary heart disease. Clin. Infect. Dis. 28:988-992. [DOI] [PubMed] [Google Scholar]
- 21.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 22.Kuo, C. C., A. S. M. Gown, E. P. Benditt, and J. T. Grayston. 1993. Detection of Chlamydia pneumoniae in aortic lesions of atherosclerosis by immunocytochemical stain. Arterioscler. Thromb. 10:1501-1504. [DOI] [PubMed] [Google Scholar]
- 23.Kuo, C. C., L. A. Jackson, L. A. Campbell, and J. T. Grayston. 1995. Chlamydia pneumoniae (TWAR). Clin. Microbiol. Rev. 8:451-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kutlin, A., N. Tsumura, U. Emre, P. M. Roblin, and M. Hammerschlag. 1997. Evaluation of Chlamydia immunoglobulin M (IgM), IgG, and IgA rELISAs Medac for diagnosis of Chlamydia pneumoniae infection. Clin. Diagn. Lab. Immunol. 4:213-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak, K. J., S. J. Flood, J. Marmaro, W. Giusti, and K. Deetz. 1995. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 4:357-362. [DOI] [PubMed] [Google Scholar]
- 26.Maass, M., C. Bartels, P. M. Engel, U. Mamat, and H. H. Sievers. 1998. Endovascular presence of viable Chlamydia pneumoniae is a common phenomenon in coronary artery disease. J. Am. Coll. Cardiol. 31:827-832. [DOI] [PubMed] [Google Scholar]
- 27.Maass, M., J. Jahn, J. Gieffers, K. Dalhoff, H. A. Katus, and W. Solbach. 2000. Detection of Chlamydia pneumoniae within peripheral blood monocytes of patients with unstable angina or myocardial infarction. J. Infect. Dis. 181:S449-S451. [DOI] [PubMed] [Google Scholar]
- 28.Madico, G., T. C. Quinn, J. Boman, and C. A. Gaydos. 2000. Touchdown enzyme time release-PCR for detection and identification of Chlamydia trachomatis, C. pneumoniae, and C. psittaci using the 16S and 16S-23S spacer rRNA genes. J. Clin. Microbiol. 38:1085-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahony, J. B., S. Chong, B. K. Coombes, M. Smieja, and A. Petrich. 2000. Analytical sensitivity, reproducibility of results, and clinical performance of five PCR assays for detecting Chlamydia pneumoniae DNA in peripheral blood mononuclear cells. J. Clin. Microbiol. 38:2622-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maurin, M., J. Etienne, and D. Raoult. 1997. Serological cross-reactions between Bartonella and Chlamydia species: implications for diagnosis. J. Clin. Microbiol. 35:2283-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayr, M., S. Kiechl, J. Willeit, G. Wick, and Q. Xu. 2000. Infections, immunity, and atherosclerosis: associations of antibodies to Chlamydia pneumoniae, Helicobacter pylori, and cytomegalovirus with immune reactions to heat-shock protein 60 and carotid or femoral atherosclerosis. Circulation 102:833-839. [DOI] [PubMed] [Google Scholar]
- 32.Messmer, T. O., S. K. Skelton, J. F. Moroney, H. Daugharty, and B. S. Fields. 1997. Application of a nested, multiplex PCR to psittacosis outbreaks. J. Clin. Microbiol. 35:2043-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muhlestein, J. B., J. L. Anderson, E. H. Hammond, L. Zhao, S. Trehan, E. P. Schwobe, and J. F. Carlquist. 1998. Infection with Chlamydia pneumoniae accelerates the development of atherosclerosis and treatment with azithromycin prevents it in a rabbit model. Circulation 97:633-636. [DOI] [PubMed] [Google Scholar]
- 34.Normann, E., J. Naas, J. Gnarpe, H. Backman, and H. Gnarpe. 1999. Demonstration of Chlamydia pneumoniae in cardiovascular tissues from children with Kawasaki disease. Pediatr. Infect. Dis. J. 18:72-73. [DOI] [PubMed] [Google Scholar]
- 35.Peeling, R. W., S. P. Wang, J. T. Grayston, F. Blasi, J. Boman, A. Clad, H. Freidank, C. A. Gaydos, J. Gnarpe, T. Hagiwara, R. B. Jones, J. Orfila, K. Persson, M. Puolakkainen, P. Saikku, and J. Schachter. 2000. Chlamydia pneumoniae serology: interlaboratory variation in microimmunofluorescence assay results. J. Infect. Dis. 181(Suppl. 3):S426-S429. [DOI] [PubMed] [Google Scholar]
- 36.Pruckler, J. M., J. M. Pruckler, and E. W. Ades. 1995. Detection by polymerase chain reaction of all common Mycoplasma in a cell culture facility. Pathobiology 63:9-11. [DOI] [PubMed] [Google Scholar]
- 37.Pruckler, J. M., N. Masse, V. A. Stevens, L. Gang, Y. Yang, E. R. Zell, S. F. Dowell, and B. S. Fields. 1999. Optimizing culture of Chlamydia pneumoniae by using multiple centrifugations. J. Clin. Microbiol. 37:3399-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramirez, J. A. 1996. Isolation of Chlamydia pneumoniae from the coronary artery of a patient with coronary atherosclerosis. Ann. Intern. Med. 125:979-982. [DOI] [PubMed] [Google Scholar]
- 39.Ridker, P. M., R. B. Kundsin, M. J. Stampfer, S. Poulin, and C. H. Hennekens. 1999. Prospective study of Chlamydia pneumoniae IgG seropositivity and risks of future myocardial infarction. Circulation 99:1161-1164. [DOI] [PubMed] [Google Scholar]
- 40.Saikku, P., M. Leinonen, K. Mattila, M. R. Ekman, M. S. Nieminen, P. H. Makela, J. K. Huttunen, and V. Valtonen. 1988. Serological evidence of an association of a novel Chlamydia, TWAR, with chronic coronary heart disease and acute myocardial infarction. Lancet 2:983-986. [DOI] [PubMed] [Google Scholar]
- 41.Schrag, S. J., R. E. Besser, C. Olson, J. C. Burns, P. M. Arguin, F. Gimenez-Sanchez, V. A. Stevens, J. M. Pruckler, B. S. Fields, E. D. Belay, M. Ginsberg, and S. F. Dowell. 2000. Lack of association between Kawasaki syndrome and Chlamydia pneumoniae infection: an investigation of a Kawasaki syndrome cluster in San Diego County. Pediatr. Infect. Dis. J. 19:17-22. [DOI] [PubMed] [Google Scholar]
- 42.Smieja, M., J. B. Mahony, C. H. Goldsmith, S. Chong, A. Petrich, and M. Chernesky. 2001. Replicate PCR testing and probit analysis for detection and quantitation of Chlamydia pneumoniae in clinical specimens. J. Clin. Microbiol. 39:1796-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sriram, S., C. W. Stratton, S. Yao, A. Tharp, L. Ding, J. D. Bannan, and W. M. Mitchell. 1999. Chlamydia pneumoniae infection of the central nervous system in multiple sclerosis. Ann. Neurol. 46:6-14. [PubMed] [Google Scholar]
- 44.Thomas, M., Y. Wong, D. Thomas, M. Ajaz, V. Tsang, P. J. Gallagher, and M. E. Ward. 1999. Relation between direct detection of Chlamydia pneumoniae DNA in human coronary arteries at postmortem examination and histological severity (Stary grading) of associated atherosclerotic plaque. Circulation 99:2733-2736. [DOI] [PubMed] [Google Scholar]
- 45.Tong, C. Y., and M. Sillis. 1993. Detection of Chlamydia pneumoniae and Chlamydia psittaci in sputum samples by PCR. J. Clin. Pathol. 46:313-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tucker, R. A., E. R. Unger, B. P. Holloway, and D. C. Swan. 2001. Real-time PCR-based fluorescent assay for quantitation of human papillomavirus types 6, 11, 16, and 18. Mol. Diagn. 6:39-47. [DOI] [PubMed] [Google Scholar]
- 47.Tuuminen, T., S. Varjo, H. Ingman, T. Weber, J. Oksi, and M. Viljanen. 2000. Prevalence of Chlamydia pneumoniae and Mycoplasma pneumoniae immunoglobulin G and A antibodies in a healthy Finnish population as analysed by quantitative enzyme immunoassays. Clin. Diagn. Lab. Immunol. 7:734-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wald, N. J., M. R. Law, J. K. Morris, X. Zhou, Y. Wong, and M. E. Ward. 2000. Chlamydia pneumoniae infection and mortality from ischaemic heart disease: large prospective study. Br. Med. J. 321:204-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiss, S. M., P. M. Roblin, C. A. Gaydos, P. Cummings, D. L. Patton, N. Schulhoff, J. Shani, R. Frankel, K. Penney, T. C. Quinn, M. R. Hammerschlag, and J. Schachter. 1996. Failure to detect Chlamydia pneumoniae in coronary atheromas of patients undergoing atherectomy. J. Infect. Dis. 173:957-962. [DOI] [PubMed] [Google Scholar]
- 50.Wong, K. H., S. K. Skelton, and Y. K. Chan. 1992. Efficient culture of Chlamydia pneumoniae with cell lines derived from the human respiratory tract. J. Clin. Microbiol. 30:1625-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong, Y. K., K. D. Dawkins, and M. E. Ward. 1999. Circulating Chlamydia pneumoniae DNA as a predictor of coronary artery disease. J. Am. Coll. Cardiol. 34:1435-1439. [DOI] [PubMed] [Google Scholar]