Abstract
The resistance of Helicobacter pylori to the recently available antibiotic treatment regimens has been a growing problem. We investigated the prevalence of H. pylori resistance to clarithromycin, metronidazole, and amoxicillin among 51 H. pylori isolates from Japanese children. In addition, the mutations of the corresponding gene were studied by PCR and restriction fragment length polymorphism analysis. Primary resistance to clarithromycin, metronidazole, and amoxicillin was detected in 29, 24, and 0% of strains, respectively. The eradication rates in clarithromycin-susceptible and -resistant strains were 89 and 56%, respectively (P < 0.05). The prevalence of strains with acquired resistance to clarithromycin (78%) was higher than that of strains with primary resistance (P < 0.01). Among the clarithromycin-resistant strains studied, 92% showed cross-resistance to azithromycin. No acquired resistance to amoxicillin was demonstrated. The A2144G mutation in the 23S rRNA gene was detected in 11 of 12 (92%) clarithromycin-resistant strains tested, whereas the mutation was not detected in any of the 15 susceptible strains. The deletion of the rdxA gene was not demonstrated in any of the strains. The results indicate that a high prevalence of clarithromycin-resistant strains is associated with eradication failure. Testing of susceptibility to clarithromycin is recommended.
In adults, Helicobacter pylori plays an important role in the pathogenesis of chronic gastritis, peptic ulcer disease, and possibly, gastric carcinoma. H. pylori infection is also associated with chronic gastritis and duodenal ulcer in children (5, 13). Eradication of the organism not only accelarates ulcer healing (14) but also prevents long-term ulcer relapse (12). Recently, proton pump inhibitor (PPI)-based eradication regimens containing two antibiotics have been demonstrated to have high eradication rates (greater than 90%) (3, 22). Amoxicillin, clarithromycin, and metronidazole are the most frequently used antibiotics for the treatment of H. pylori infection. However, antibiotic resistance frequently causes failure of eradication of H. pylori (1, 16, 21). The resistance of H. pylori to the recently available antibiotic treatment regimens has been a growing problem. In developed countries, metronidazole resistance is found in 10 to 50% of adult patients infected with H. pylori (1, 10, 11), whereas virtually all strains are resistant to the agent in developing countries (26). On the other hand, although the rates of clarithromycin resistance are relatively low, ranging from 2 to 15% (1, 2, 10, 11, 27), the rate of clarithromycin resistance has been increasing during recent years.
With regard to antibiotic resistance, however, there have been few reports of antibiotic resistance in children 2, 17, 23; M. Lopez-Brea, M. Martinez, D. Domingo, and T. Alarcon, abstract from the XIII International Workshop on Gastroduodenal Pathology and Helicobacter pylori, Gut 47(Suppl. 1):A95, 2000], and its clinical significance remains to be established. The purpose of the present study was to determine the prevalence of antibiotic resistance among H. pylori strains isolated from Japanese children. The mutations of the corresponding genes in clinical isolates were also investigated.
Patients and bacterial strains
From December 1999 to March 2001, a total of 51 H. pylori isolates from 48 pediatric patients (ages, 5 to 16 years; mean age, 11.9 years) were studied. Biopsy specimens were taken from the gastric antrum or body for the testing of H. pylori. The diagnoses included gastritis (n = 27), gastric ulcer (n = 2), and duodenal ulcer (n = 19). Among these patients, 42 patients had no prior history of eradication therapy; 36 patients received a 7- to 14-day course of a PPI, omeprazole or lansoprazole, plus amoxicillin and clarithromycin as the first-line therapy (18, 19). Three patients in whom eradication was unsuccessful during the period of the study and six patients with previous histories of eradication failure with amoxicillin and clarithromycin treatment received second-line therapy with a PPI and amoxicillin plus metronidazole (n = 6) or minocycline (n = 3); H. pylori was isolated from all nine patients before the start of the second-line therapy. Eradication was confirmed by tests with the biopsy specimens (histology, rapid urease test, and/or culture) and the [13C]urea breath test at 1 to 3 months after the therapy was completed.
H. pylori strains were cultured under a microaerophilic atmosphere (5% O2, 15% CO2, 80% N2) at 37°C for 3 to 7 days, and the isolates were identified by Gram staining and biochemical tests for catalase, oxidase, and urease activities. For the PCR assay, 27 of 51 H. pylori strains were stored at −70°C in brucella broth with 5% fetal bovine serum.
Antimicrobial susceptibility testing
The susceptibilities of the H. pylori isolates to clarithromycin, metronidazole, amoxicillin, and azithromycin were examined by a microdilution method (20). The bacteria were subcultured on Mueller-Hinton agar supplemented with 10% defibrinated horse blood under the same microaerophilic atmosphere mentioned above (5% O2, 15% CO2, 80% N2) at 37°C for 48 h. The bacterial suspension was adjusted to a final concentration of a McFarland no. 1 standard. A 96-well microplate (Eiken, Tokyo, Japan) was prepared with twofold increments of antibiotic concentrations and by inoculation of 100 μl of the bacterial suspension into each well. After 72 h of incubation under a microaerophilic atmosphere (5% O2, 5% CO2, 90% N2) at 37°C, the MIC of each antibiotic was determined. Quality control was performed with H. pylori ATCC 43504. According to Glupczynski et al. [Y. Glupczynski, F. Mégraud, L. P. Andersen, and M. Lopez-Brea, abstract from the XII International Workshop on Gastroduodenal Pathology and Helicobacter pylori, Gut 45(Suppl. 3):A3, 1999], resistance breakpoints for metronidazole and amoxicillin were defined as >8 and >0.5 μg/ml, respectively. The breakpoint for clarithromycin was defined as ≥1.0 μg/ml (28). The breakpoint for azithromycin was provisionally defined as ≥1.0 μg/ml.
PCR and restriction fragment length polymorphism analysis
The extraction of H. pylori genomic DNA was performed as reported previously (33). The DNA extracts were stored at 4°C for no longer than 14 days until the PCR assay. For the detection of mutation of the gene for clarithromycin resistance, oligonucleotide primers 18 (5"-AGTCGGGACCTAAGGCGAG-3") and 21 (5"-TTCCCGCTTAGATGCTTTCAG-3") were used for PCR amplification of the 23S rRNA gene (34). For the detection of metronidazole resistance, primers RdxA1 (5"-AATTTGAGCATGGGGCAGA-3") and RdxA2 (5"-GAAACGCTTGAAAACACCCCT-3") were used for determination of deletion of the rdxA gene (4). Each PCR amplification was performed in a thermal cycler (PE Applied Biosystems, Chiba, Japan), as described previously (4, 34).
Approximately 1.4-kb amplicons of the 23S rRNA gene were digested with MboII (Takara, Shiga, Japan) for 14 h at 37°C for the detection of an adenine-to-guanine mutation at position 2143 and were digested with BsaI (Takara) for 14 h at 55°C for the detection of the A2144G mutation (33). Digested fragments were separated on a 1% agarose gel containing ethidium bromide (0.5 μg/ml) and were viewed on a UV transilluminator. The sizes of the PCR products of the rdxA gene were analyzed by 1.5% agarose gel electrophoresis containing ethidium bromide (0.5 μg/ml).
Statistical analysis
The difference between the rates of primary and acquired resistance and the difference in eradication rates between patients infected with clarithromycin-susceptible and -resistant strains were analyzed by Fisher's exact test. A P value of <0.05 was regarded as statistically significant.
Prevalence of antibiotic resistance and MICs
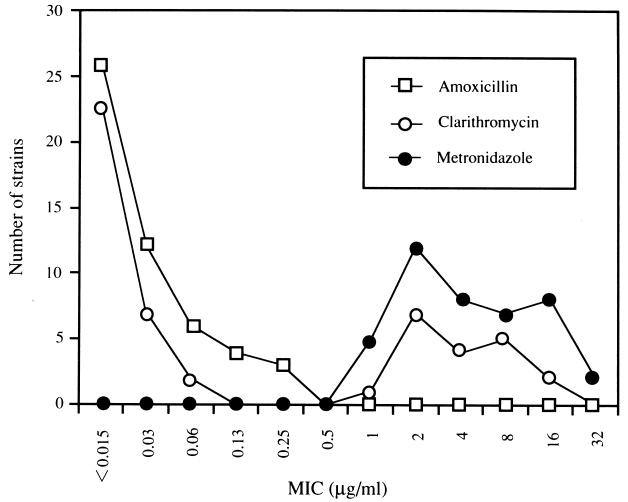
Primary resistance to clarithromycin, metronidazole, and amoxicillin was demonstrated in 29, 24, and 0% of the strains, respectively. Ten percent of the strains were resistant to both clarithromycin and metronidazole. Strains isolated from seven (78%) of nine patients with first-line treatment failure were resistant to clarithromycin. The prevalence of strains with acquired resistance to clarithromycin was higher than that of strains with primary resistance (P < 0.01). No strains that acquired resistance to amoxicillin were detected. Among 12 clarithromycin-resistant strains studied, 11 strains (92%) showed cross-resistance to azithromycin. For all 15 clarithromycin-susuceptible strains, azithromycin MICs were <1 μg/ml. The clarithromycin MICs showed a bimodal distribution (Fig. 1): the MICs for clarithromycin-resistant strains ranged between 1 and 16 μg/ml. In contrast, the distribution of the metronidazole MICs demonstrated a continuous spectrum and ranged between 1 and 32 μg/ml.
FIG. 1.
Distribution of antibiotic MICs for 51 H. pylori strains. In the present study, resistance breakpoints for clarithromycin, metronidazole, and amoxicillin were defined as ≥1.0, >8, and >0.5 μg/ml, respectively.
Clinical outcome
In the present study, first-line therapy with clarithromycin and amoxicillin was successful in eradicating H. pylori from 29 of 36 patients (81%): the rates of eradication were 89% for the clarithromycin-susceptible strains and 56% for the clarithromycin-resistant strains (P < 0.05). Second-line therapy with amoxicillin and metronidazole or minocycline was successful for six of seven patients (86%) infected with clarithromycin-resistant strains; H. pylori was eradicated from two patients infected with clarithromycin-susceptible strains.
Detection of gene mutations
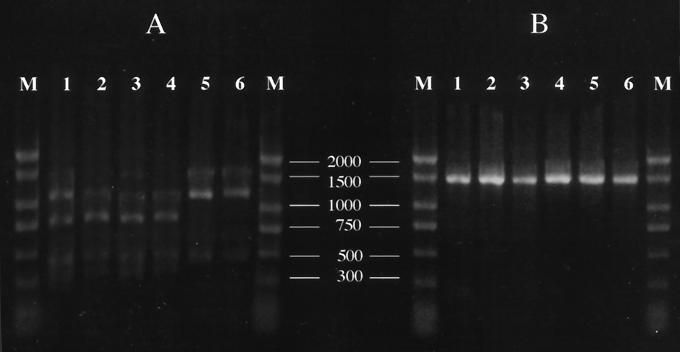
On the basis of an analysis with BsaI digestion, the A2144G mutation in the 23S rRNA gene were detected in 11 of 12 (92%) clarithromycin-resistant strains tested (Fig. 2). In contrast, this mutation was not detected in the remaining 15 strains for which clarithromycin MICs were ≤0.06 μg/ml. The PCR products from none of the strains were digested with MboII. For all 27 strains studied, the rdxA amplicon was approximately 800 bp. No deletion of the rdxA gene has been suggested because the PCR amplicon in metronidazole-resistant strains with the mutational deletion is 200 bp smaller than expected from the published sequences (4).
FIG. 2.
Restriction endonuclease analysis of 23S rRNA amplicons. (A) Digestion with BsaI; (B) digestion with MboII. Lanes 1 to 5, clarithromycin-resistant H. pylori strains for which MICs are 8, 4, 2, 1, and 4 μg/ml, respectively; lanes 6, clarithromycin-susceptible H. pylori strain for which the MIC is <0.015 μg/ml; lanes M, DNA size markers (the numbers between the panels are in base pairs). The A2144G mutation was found in lanes 1 to 4 but was not found in lane 5. Note that the A2143G mutation detected by digestion with MboII was not detected in any of the strains studied.
In an intention-to-treat analysis for adults, new triple-drug regimens containing clarithromycin achieve an overall H. pylori eradication rate of about 90% (16). However, it has been suggested that H. pylori resistance to clarithromycin is a major predictor of eradication failure with regimens containing the antibiotic [16; R. Cayla, F. Zerbib, P. Talbi, F. Mégraud, and H. Lamouliatte, abstract from the 4th United European Gastroenterology Week, Gut 37(Suppl. 2):A152, 1995]. The present study has shown that primary resistance to clarithromycin is frequently observed in Japanese children. In early studies with isolates from children, most H. pylori strains were susceptible to macrolides (23). On the other hand, recent studies have reported that clinical isolates from children are clarithromycin resistant, with rates of resistance ranging from 18 to 44.8% [2, 17; Lopez-Brea et al., Gut 47(Suppl. 1):A95, 2000]. The prevalence of clarithromycin resistance in H. pylori has increased during the periods studied (26). This new macrolide has commonly been used for the treatment of various diseases in the pediatric patient population, including respiratory tract infections. In Japan, according to its recent market share, clarithromycin accounts for more than 10% of oral antibiotics administered to children. In a Portuguese study, the rate of clarithromycin resistance was higher among H. pylori isolates from children than among those from adults (2). Furthermore, the present study has demonstrated that the cross-resistance between clarithromycin and azithromycin is nearly complete in H. pylori isolates. Glupczynski et al. (8) reported cross-resistance among three macrolides. It appears that a rise in the prevalence of clarithromycin resistance is associated with the increasing use of macrolides in clinical practice.
As in previous reports [1, 2, 8, 24; Cayla et al., Gut 37(Suppl. 2):A152, 1995] the prevalence of clarithromycin resistance increased significantly with treatment failure. Some patients can simultaneously harbor multiple H. pylori strains (15). Moreover, both clarithromycin-susceptible and -resistant strains have been isolated from some patients with no history of exposure to macrolides (25). Finally, in patients with eradication failure, strains recovered pretreatment and posttreatment had the same molecular typing patterns (7, 30). These findings suggest that administration of clarithromycin may select for the resistant strains. However, it remains unclear how clarithromycin resistance is induced in H. pylori. It has been known that acquired resistance to clarithromycin is stable (26). Regarding clarithromycin, in vitro resistance in H. pylori equates with in vivo resistance (11). This can be explained by the clear-cut bimodal distribution of clarithromycin MICs shown in the present study. Although susceptibility testing has not previously been recommended for H. pylori, we believe that in the case of clarithromycin, there is now mounting evidence in favor of such testing.
Clarithromycin-resistant strains frequently have mutations in the 23S rRNA gene (24). Versalovic et al. (33, 34) showed that A-to-G point mutations at positions 2143 and 2144 within domain V of the 23S rRNA gene are a cause of clarithromycin resistance. The A2143C mutation in the same gene was also reported, although it is rare (31). These mutations cause a decreased affinity of clarithromycin for the 23S ribosomal component, resulting in the impaired activity of clarithromycin against H. pylori (25). In a recent study, the A2143G mutation was consistently associated with high levels of clarithromycin resistance (MICs, >64 μg/ml [33] or >256 μg/ml [6]), whereas the mutants carrying the A2144G mutation showed low-level resistance. In the present study, all strains which had low-level resistance to clarithromycin demonstrated the A2144G mutation. Thus, our results support the fact that the A2144G mutation in the 23S rRNA gene of H. pylori is linked to clarithromycin resistance. In 2000, the National Committee for Clinical Laboratory Standards (28) defined the breakpoint for resistance to clarithromycin as an MIC of ≥1 μg/ml. The present study confirms the validity of this definition.
On the other hand, no strains were resistant to amoxicillin. Recently, amoxicillin resistance with a stable genetic feature has been reported (32). In Brazil, resistance to amoxicillin has been detected in 29% of strains recovered posttreatment (27). However, many studies have shown that all strains examined are susceptible to amoxicillin (1, 17). The clinical significance of amoxicillin susceptibility testing has not been documented (26).
Regarding the mechanism of metronidazole resistance in H. pylori, it has been hypothesized that mutations in the rdxA gene, which encode an oxygen-insensitive NADPH nitroreductase, are responsible for the resistance (9). However, some investigators have questioned the mutational inactivation of the rdxA gene (26). A continuous spectrum of metronidazole MICs suggests that there are many different pathways for the resistance (11). In the present study, the deletion of the rdxA gene was not found in any of the strains tested. The prevalence of metronidazole-resistant strains in children has been reported to be 19% in Portugal (2), 23% in Spain [Lopez-Brea et al., Gut 47(Suppl. 1):A95, 2000], and 43% in France (17). In contrast, a Canadian study has reported that all strains of H. pylori were susceptible to metronidazole (23). In Japan, the use of metronidazole in children is very rare because this antimicrobial agent is indicated only for trichomoniasis or Clostridium difficile-associated enteritis. Unlike clarithromycin, in vitro resistance to metronidazole does not always reflect in vivo resistance (11). Further studies are needed for standardization of the breakpoint for metronidazole in H. pylori.
Agar or broth dilution methods are well established for standard susceptibility testing for H. pylori but are difficult to perform routinely in the clinical laboratory. For this reason, the E test has been widely performed and has yielded results equivalent to those of the agar or broth dilution methods. Recently, it has been suggested that the results of the microdilution method show excellent concordance with those of these standard methods (20, 29). In the present study, the microdilution method was performed in an incubation atmosphere with 5% CO2. A higher concentration of CO2 may cause instability of the clarithromycin MICs. As a consequence, the results of clarithromycin MIC testing clearly correlated with the results of mutational analysis of the 23S rRNA gene. We believe that the microdilution method is an easy-to-use alternative for susceptibility testing for H. pylori. Further surveillance of antibiotic resistance in various communities is needed to establish the appropriate treatment of H. pylori infection.
Acknowledgments
We thank S. Toyoda, D. Abukawa, B. Yoshimura, A. Sawada, and M. Takahashi for help with the present study.
REFERENCES
- 1.Adamek, R. J., S. Suerbaum, B. Pfaffenbach, and W. Opferkuch. 1998. Primary and acquired Helicobacter pylori resistance to clarithromycin, metronidazole, and amoxicillin: influence on treatment outcome. Am. J. Gastroenterol. 93:386-389. [DOI] [PubMed] [Google Scholar]
- 2.Cabrita, J., M. Oleastro, R. Matos, A. Manhente, J. Cabral, R. Barros, A. I. Lopes, P. Ramalho, B. C. Neves, and A. S. Guerreiro. 2000. Feature and trends in Helicobacter pylori antibiotic resistance in Lisbon area, Portugal (1990-1999). J. Antimicrob. Chemother. 46:1029-1031. [DOI] [PubMed] [Google Scholar]
- 3.Chiba, N. 1996. Omeprazole and clarithromycin with and without metronidazole for the eradication of Helicobacter pylori. Am. J. Gastroenterol. 91:2139-2143. [PubMed] [Google Scholar]
- 4.Debets-Ossenkopp, Y. J., R. G. J. Pot, D. J. van Westerloo, A. Goodwin, C. M. J. E. Vandenbroucke-Grauls, D. E. Berg, P. S. Hoffman, and J. G. Kusters. 1999. Insertion of mini-IS 605 and deletion of adjacent sequences in the nitroreductase (rdxA) gene cause metronidazole resistance in Helicobacter pylori NCTC11637. Antimicrob. Agents Chemother. 43:2657-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drumm, B., P. Sherman, E. Cutz, and M. Karmali. 1987. Association of Campylobacter pylori on the gastric mucosa with antral gastritis in children. N. Engl. J. Med. 316:1557-1561. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Arata, M. I., F. Baquero, L. de Rafael, C. M. de Argila, J. P. Gisbert, F. Bermejo, D. Boixeda, and R. Canton. 1999. Mutation in 23S rRNA in Helicobacter pylori conferring resistance to erythromycin do not always confer resistance to clarithromycin. Antimicrob. Agents Chemother. 43:374-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson, J. R., E. Slater, J. Xerry, D. S. Tompkins, and R. J. Owen. 1998. Use of an amplified-fragment length polymorphism technique to fingerprint and differentiate isolates of Helicobacter pylori. J. Clin. Microbiol. 36:2580-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glupczynski, Y., and A. Burette. 1990. Drug therapy for Helicobacter pylori infection: problems and pitfalls. Am. J. Gastroenterol. 85:1545-1551. [PubMed] [Google Scholar]
- 9.Goodwin, A., D. Kersulyte, G. Sisson, S. J. O. Veldhuyzen van Zanten, D. E. Berg, and P. S. Hoffman. 1998. Metronidazole resistance in Helicobacter pylori is due to null mutation in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 28:383-393. [DOI] [PubMed] [Google Scholar]
- 10.Gotoh, A., Y. Kawakami, T. Akahane, T. Akamatsu, T. Shimizu, K. Kiyosawa, and T. Katsuyama. 1997. Susceptibility of Helicobacter pylori isolates against agents commonly administered for eradication therapy and the efficacy of chemotherapy. Microbiol. Immunol. 41:7-12. [DOI] [PubMed] [Google Scholar]
- 11.Graham, D. Y. 1998. Antibiotic resistance in Helicobacter pylori: implications for therapy. Gastroenterology 115:1272-1277. [DOI] [PubMed] [Google Scholar]
- 12.Graham, D. Y., G. M. Lew, P. D. Klein, D. G. Evans, D. J. Evans, Jr., Z. A. Saeed, and H. M. Malaty. 1992. Effect of treatment of Helicobacter pylori infection on the long-term recurrence of gastric or duodenal ulcer: a randomized, controlled study. Ann. Intern. Med. 116:705-708. [DOI] [PubMed] [Google Scholar]
- 13.Hassall, E., and J. E. Dimmick. 1991. Unique features of Helicobacter pylori disease in children. Dig. Dis. Sci. 36:417-423. [DOI] [PubMed] [Google Scholar]
- 14.Hentschel, E., G. Brandstätter, B. Dragosics, A. M. Hirschl, H. Nemec, K. Schütze, M. Taufer, and H. Wurzer. 1993. Effect of ranitidine and amoxicillin plus metronidazole on the eradication of Helicobacter pylori and the recurrence of duodenal ulcer. N. Engl. J. Med. 328:308-312. [DOI] [PubMed] [Google Scholar]
- 15.Hirschl, A. M., M. Richter, A. Makristathis, P. M. Prückl, B. Willinger, K. Schütze, and M. L. Rotter. 1994. Single and multiple strain colonization in patients with Helicobacter pylori-associated gastritis: detection by macrorestriction DNA analysis. J. Infect. Dis. 170:473-475. [DOI] [PubMed] [Google Scholar]
- 16.Huang, J. Q., and R. H. Hunt. 1999. Treatment after failure: the problem of “non-responders.” Gut 45(Suppl. 1):I40-I44. [DOI] [PMC free article] [PubMed]
- 17.Kalach, N., M. Bergeret, P. H. Benhamou, C. Dupont, and J. Raymond. 2001. High levels of resistance to metronidazole and clarithromycin in Helicobacter pylori strains in children. J. Clin. Microbiol. 39:394-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato, S., H. Ritsuno, K. Ohnuma, K. Iinuma, T. Sugiyama, and M. Asaka. 1998. Safety and efficacy of one-week triple therapy for eradicating Helicobacter pylori in children. Helicobacter 3:278-282. [DOI] [PubMed] [Google Scholar]
- 19.Kato, S., J. Takeyama, K. Ebina, and H. Naganuma. 1997. Omeprazole-based dual and triple regimens for Helicobacter pylori eradication in children. Pediatrics 100. [Online.] http//www.pediatrics.org/cgi/content/full/100./1/e3. [DOI] [PubMed]
- 20.Kawakami, Y., T. Akahane, A. Gotoh, Y. Okimura, K. Oana, and T. Katsuyama. 1997. Successful development of air-dried microplates (HP-Plates) for susceptibility testing against Helicobacter pylori isolates. Microbiol. Immunol. 41:703-708. [DOI] [PubMed] [Google Scholar]
- 21.Lind, T., F. Mégraud, P. Unge, E. Bayerdörffer, C. O'Morain, R. Spiller, S. Veldhuyzen van Zanten, K. D. Bardhan, M. Hellblem, M. Wrangstadh, L. Zeijlon, and C. Cederberg. 1999. The MACH2 study: role of omeprazole in eradication of Helicobacter pylori with 1-week triple therapies. Gastroenterology 116:248-253. [DOI] [PubMed] [Google Scholar]
- 22.Lind, T., S. Veldhuyzen van Zanten, P. Unge, R. Spiller, E. Bayerdörffer, C. O'Morain, K. D. Bardhan, M. Bradette, N. Chiba, M. Wrangstadh, C. Cederberg, and J. P. Idström. 1996. Eradication of Helicobacter pylori using one-week triple therapies combining omeprazole with two antimicrobials: the MACH I study. Helicobacter 1:138-144. [DOI] [PubMed] [Google Scholar]
- 23.Loo, V. G., P. Sherman, and A. G. Matlow. 1992. Helicobacter pylori infection in a pediatric population: in vitro susceptibilities to omeprazole and eight antimicrobial agents. Antimicrob. Agents Chemother. 36:1133-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maeda, S., H. Yoshida, K. Ogura, F. Kanai, Y. Shiratori, and M. Omata. 1998. Helicobacter pylori specific nested PCR assay for the detection of 23S rRNA mutation associated with clarithromycin resistance. Gut 43:317-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuoka, M., Y. Yoshida, K. Hayakawa, S. Fukuchi, and K. Sugano. 1999. Simultaneous colonisation of Helicobacter pylori with and without mutations in the 23S rRNA gene in patients with no history of clarithromycin exposure. Gut 45:503-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mégraud, F. 1998. Epidemiology and mechanism of antibiotic resistance in Helicobacter pylori. Gastroenterology 115:1278-1282. [DOI] [PubMed] [Google Scholar]
- 27.Mendonca, S., C. Ecclissato, M. S. Sartori, A. P. O. Godoy, R. A. Guerzoni, M. Degger, and J. Pedrazzoli, Jr. 2000. Prevalence of Helicobacter pylori resistance to metronidazole, clarithromycin, amoxicillin, tetracycline, and furazolidone in Brazil. Helicobacter 5:79-83. [DOI] [PubMed] [Google Scholar]
- 28.National Committee for Clinical Laboratory Standards. 2000. Approved standard M100-S10. MIC testing supplemental tables. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 29.Piccolomini, R., G. Di Bonaventura, G. Catamo, F. Carbone, and M. Neri. 1997. Comparative evaluation of E test, agar dilution, and broth microdilution for testing susceptibilities of Helicobacter pylori strains to 20 antimicrobial agents. J. Clin. Microbiol. 35:1842-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shortridge, V. D., G. G. Stone, R. K. Flamm, J. Beyer, J. Versalovic, D. Y. Graham, and S. K. Tanaka. 1997. Molecular typing of Helicobacter pylori isolates from a multicenter U.S. clinical trial by ureC restriction fragment length polymorphism. J. Clin. Microbiol. 35:471-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stones, G. G., V. D. Shortridge, J. Versalovic, J. Beyer, R. K. Flamm, D. Y. Graham, A. T. Ghoneim, and S. K. Tanaka. 1997. A PCR-oligonucleotide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin-resistant Helicobacter pylori. Antimicrob. Agents Chemother. 41:712-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Zwet, A. A., C. M. J. E. Vandenbrouke-Grauls, J. C. Thijs, E. J. van der Wouden, M. M. Gerrits, and J. G. Kusters. 1998. Stable amoxicillin resistance in Helicobacter pylori. Lancet 352:1595.. [DOI] [PubMed] [Google Scholar]
- 33.Versalovic, J., M. S. Osato, K. Spakovsky, M. P. Dore, R. Reddy, G. G. Stone, D. Shortridge, R. K. Flamm, S. K. Tanaka, and D. Y. Graham. 1997. Point mutations in the 23S rRNA gene of Helicobacter pylori associated with different levels of clarithromycin resistance. J. Antimicrob. Chemother. 40:283-286. [DOI] [PubMed] [Google Scholar]
- 34.Versalovic, J., D. Shortridge, K. Kibler, M. V. Griffy, J. Beyer, R. K. Flamm, S. K. Tanaka, D. Y. Graham, and M. F. Go. 1996. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 40:477-480. [DOI] [PMC free article] [PubMed] [Google Scholar]