Abstract
Background
The development and testing of functions for the modeling of protein energetics is an important part of current research aimed at understanding protein structure and function. Knowledge-based mean force potentials are derived from statistical analyses of interacting groups in experimentally determined protein structures. Current knowledge-based mean force potentials are developed at the atom or amino acid level. The evolutionary information contained in the profiles is not investigated. Based on these observations, a class of novel knowledge-based mean force potentials at the profile level has been presented, which uses the evolutionary information of profiles for developing more powerful statistical potentials.
Results
The frequency profiles are directly calculated from the multiple sequence alignments outputted by PSI-BLAST and converted into binary profiles with a probability threshold. As a result, the protein sequences are represented as sequences of binary profiles rather than sequences of amino acids. Similar to the knowledge-based potentials at the residue level, a class of novel potentials at the profile level is introduced. We develop four types of profile-level statistical potentials including distance-dependent, contact, Φ/Ψ dihedral angle and accessible surface statistical potentials. These potentials are first evaluated by the fold assessment between the correct and incorrect models generated by comparative modeling from our own and other groups. They are then used to recognize the native structures from well-constructed decoy sets. Experimental results show that all the knowledge-base mean force potentials at the profile level outperform those at the residue level. Significant improvements are obtained for the distance-dependent and accessible surface potentials (5–6%). The contact and Φ/Ψ dihedral angle potential only get a slight improvement (1–2%). Decoy set evaluation results show that the distance-dependent profile-level potentials even outperform other atom-level potentials. We also demonstrate that profile-level statistical potentials can improve the performance of threading.
Conclusion
The knowledge-base mean force potentials at the profile level can provide better discriminatory ability than those at the residue level, so they will be useful for protein structure prediction and model refinement.
Background
The development and evaluation of new energy functions is critical to the accurate modeling of the properties of biological macromolecules [1]. A potential that can discriminate between the native and miss-folded structures is crucial for any protein structure prediction protocol to be fully successful. Toward this end, two different types of potential functions are currently in use [2-4]. The first class of potentials, the so-called physical-based potential, is based on the fundamental analysis of forces between atoms [5-7]. The second class, the so-called knowledge-based potentials, extracts parameters from experimentally solved protein structures [8-11]. The advantage of the first class of potentials is that, in principle, they can be derived from the laws of physics. The disadvantage is that the calculation of free energy is very difficult because the computation should include an atomic description of the protein and the surrounding solvent. Currently this type of computation is generally too expensive for protein folding [12]. While, with today's computer resources, knowledge-based potentials can be quite successful at fold recognition [13] and ab initio structure prediction [14,15].
Much can be learned through statistical analysis of interacting groups in experimentally determined protein structures. Such analysis provides the basis for knowledge-based potentials of mean force. Generally, knowledge-based potentials have used a simple one- or two-point-per-residue representation, which results in the potentials at the residue level. Each residue in a protein sequence is represented by one or two points in three-dimensional space. These points are usually located at the coordinates of each residue's Cα atoms, Cβ atoms or at the coordinates of the center of each side chain. Discrimination is based on each residue's preference to be buried or exposed [16], its preference for a particular secondary structure conformation [17], its preference for the contact number with other residues [18] and its preference to be in contact at a particular distance and sequence separation from other residues [19,20]. However, to capture the finer details of atom-atom interactions in proteins, a more detailed description is necessary. Each heavy atom either at the main-chain or side-chain is represented by an independent point, which results in the knowledge-based potentials at the atom level. A number of potentials at the atom level have been designed [21-24]. Because of its atom level definition, the knowledge-based potentials at the atom level can provide better discriminatory power than obtained at the residue level [25].
Although the knowledge-based mean force potentials at the residue level are based on the coarse description of protein structures, they are easier to be used in fold recognition or threading than those at the atom level [22]. Many fold-recognition methods use knowledge-based potentials to interpret probabilistic scoring functions. Sequence-template alignments are evaluated in terms of a scoring function and the score of the alignment is interpreted as a "free energy" of the sequence in the conformation imposed by the alignment [26]. This interpretation indicates that the most probable sequence-structure alignment is the one with the lowest "free energy". The 123D method [27] applies the pairwise sequence alignment and contact capacity potentials to fast protein fold recognition. The SPARK method [18] combines the sequence-profile alignment and single-body knowledge-based energy score for fold recognition. The GenTHREADER method [28] apply neural network to evaluate the compatibility of the sequence and the template with pairwise potentials and solvation potentials as input. In addition to the fold recognition or threading, knowledge-based potentials are widely used in selection of native structures of proteins [29,30], estimation of protein stability [31], ab initio protein structure prediction [32-34], etc.
The aim of this paper is to develop a class of novel knowledge-based mean force potentials at the profile level, which uses the evolutionary information of the profile [35]. Such potentials can provide better discriminatory power than those at the residue level and can be incorporated into the process of fold recognition or threading. Multiple sequences alignments of protein sequences may contain much information regarding evolutionary processes. This information can be detected by analyzing the output of PSI-BLAST [35,36]. The frequency profiles are directly calculated from the multiple sequence alignments and then converted into binary profiles with a cut-off probability for usage. Such binary profiles make up of a new alphabet for protein sequences. Similar to the knowledge-based potentials at the residue level, a class of novel potentials at the profile level is introduced. We developed four types of profile-level statistical potentials including distance-dependent, contact, Φ/Ψ dihedral angle and accessible surface statistical potentials. These potentials are first evaluated by the fold assessment between the correct and incorrect models generated by comparative modeling. They are then used to recognize the native structure from the well-constructed decoy set. Experimental results show that all the knowledge-base mean force potentials at the profile level outperform those at the residue level.
Results
Fold assessment on test models
To evaluate the performance of the statistical potentials at the profile level and those at the residue level, the first experiment is made to discriminate between the good models and the bad models on our structure models. The parameters of various potentials are selected as the optimal values as suggested by others [11,18]. For the distance-dependent potentials, the interaction center is set as Cβ atom. The distance range is 30 Å with distance interval of 1 Å. The sequence separation k varies from 3 to 9. The rare situation with sequence separation larger than 9 is included in the last bin. For the contact potential, the number of contact bin is set to 25. In the rare occasions of more than 25 contacts, the statistics are included in the bin for 25 contacts. All the contacts with sequence separation larger than 1 are computed. For the Φ/Ψ dihedral angle potential, each of the torsion is divided into 36 bins. There are total 1296 bins. For the accessible surface potential, the interaction center is set as Cβ atom. The distance range (the radius of the sphere) is set as 9 Å. The burial range varies from 0 to 40 atoms with burial interval of 2 atoms. The atoms within the same residues are not considered for statistics.
The statistics of propensity of various potentials are performed on the PDB25 dataset. These potentials are then calculated to discriminate between good models and bad models for each of the sequence. The fraction of correctly predicted case (CP), the success rates, the Z-scores and the ROC curve are employed to evaluate the performance. The results are shown in table 1 and Fig. 2. In the ROC curve, a lower plot corresponds to a better discriminative power.
Table 1.
Comparative results of potentials at our structure models
| Potentials | CP | Success rates | Z-scores | Potentials | CP | Success rates | Z-scores |
| Distance | 0.86 | 400/431 | 2.86 | Dihedral | 0.81 | 256/431 | 1.92 |
| Distance_profile | 0.91 | 422/431 | 3.26 | Dihedral_profile | 0.82 | 270/431 | 2.08 |
| Contact | 0.81 | 221/431 | 1.84 | Surface | 0.85 | 309/431 | 2.33 |
| Contact_profile | 0.83 | 232/431 | 1.96 | Surface_profile | 0.90 | 335/431 | 2.78 |
The distance, contact, dihedral and surface refer to the four kinds of potentials at the residue level. The potentials with _profile suffix indicate the corresponding potentials at the profile level. In the success rates columns, the first number is the number of native structures ranked number one; the second number is the total number of proteins in the decoy set.
Figure 2.
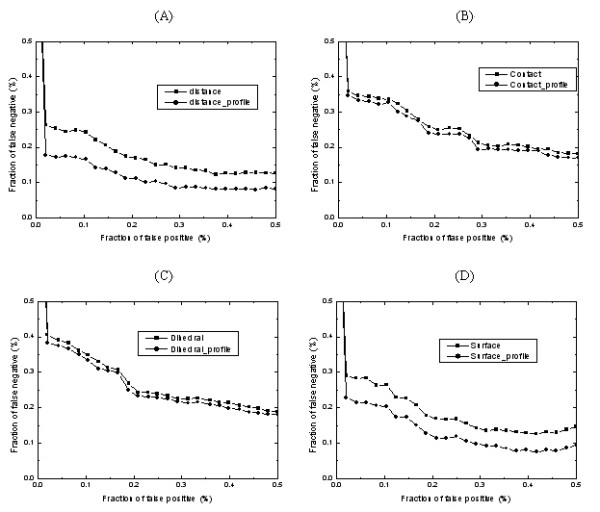
ROC curves of various potentials tested on our structure models. The lower the curve, the better the discrimination between the good and bad models. Subfigure (A), (B), (C) and (D) show the performance of residue-level and profile-level potentials of distance-dependent, contact, Φ/Ψ dihedral angle and accessible surface statistical potentials respectively. The potentials with _profile suffix indicate the corresponding potentials at the profile level.
As can be seen, all the knowledge-based mean force potentials at the profile level outperform those at the residue level. The improvements of various potentials at the profile level are different from those at the residue level. Significant improvements of CP are obtained for the distance-dependent and accessible surface potentials (5–6%). The contact and Φ/Ψ dihedral angle potential only get a slight improvement of CP (1–2%).
Tested on Baker's set
The Baker's set [37] is a well-constructed decoy set that is obtained by large-scale comparative modeling. The dataset consists of 41 single domain proteins and each protein is attached with about 1400 decoy structures. The decoy structures are classified into good models and bad models by the same criterion as used by our structure models. Models with >30% structural overlap with the experimentally determined structures are grouped into good models. Models with <15% structural overlap with the experimentally determined structures are grouped into bad models. The fold assessment results are shown in table 2.
Table 2.
Comparative fold assessment results of potentials at the Baker's set
| Potentials | CP | Success rates | Z-scores | Potentials | CP | Success rates | Z-scores |
| Distance | 0.77 | 25/41 | 2.58 | Dihedral | 0.75 | 17/41 | 1.41 |
| Distance_profile | 0.81 | 30/41 | 2.74 | Dihedral_profile | 0.77 | 20/41 | 1.58 |
| Contact | 0.74 | 15/41 | 1.36 | Surface | 0.74 | 18/41 | 2.47 |
| Contact_profile | 0.75 | 17/41 | 1.27 | Surface_profile | 0.78 | 22/41 | 2.54 |
See the footnote of table 1 for the name of the potentials.
Overall the knowledge-based mean force potentials at the profile level still outperform those at the residue level. Significant improvements are obtained for the distance-dependent and accessible surface potentials. The CP scores of all potentials on the Baker's set are lower than those on our structure models. There are two reasons for this phenomenon. The first one is that the Baker's set is inherently difficult to discriminate. Such dataset is carefully constructed and satisfies the so-called four criteria listed in their introduction [37]. The second one is that the number and distribution of good models and bad models in this dataset are different from those in our dataset. In Baker's dataset, the total models for a sequence are very large (more than one thousand) and the distribution between the number of good models and that of bad models is different. For example, the sequence 1ptq has only 8 good models and 1647 bad models, while the sequence 1res has 1722 good models and only one bad model. In our dataset, each sequence has about thirty models and the good models and bad models are equally distributed (about fifteen respectively).
PROSTAR decoy set evaluation
All the decoy sets from PROSTAR website [38] are well-constructed and widely used for evaluation of all kinds of newly developed potentials [21,24]. Three subsets including MISFOLD [39], IFU [40] and PDBERR [38] are selected for testing. The IFU dataset contains a set of models for small peptides rather than the whole protein chains. Since direct generation of profiles for such small peptides may not be reliable, we first generate the profiles of the whole protein chains and extract the corresponding profiles for such small peptides. There are two proteins (3SNS, 1ILB) that are not found in the PDB database [41], the corresponding decoy models are removed (3SNS_16-29, 3SNS_6-21, 1ILB_99-110). The results of decoy set evaluation are given in table 3. When the energy Z-scores of the native structure are lower than those of the decoy models, a correct discrimination is obtained.
Table 3.
The results of PROSTAR decoy set evaluation
| Decoy set | MISFOLD | IFU | PDBERR |
| Number of decoy pair | 25 | 41 | 3 |
| Distance | 25 | 28 | 3 |
| Distance_profile | 25 | 35 | 3 |
| Contact | 24 | 25 | 1 |
| Contact_profile | 25 | 28 | 3 |
| Dihedral | 25 | 26 | 3 |
| Dihedral_profile | 25 | 29 | 3 |
| Surface | 25 | 21 | 1 |
| Surface_profile | 25 | 24 | 3 |
Given in the table are the number of decoy pair and correctly recognized decoy pair for all potentials on the three decoy sets. See the footnote of table 1 for the name of the potentials.
All the knowledge-based mean force potentials get good results on the MISFOLD and PDBERR dataset and acceptable results on the IFU dataset. The IFU dataset is more challenging than the other two dataset, because this dataset contains the decoy models for small peptides and fold assessment by statistical potentials is most difficult for the very small models [11]. Small models are difficult to assess because of the relatively small number of pairwise interactions by which they are judged, not because of their incompleteness. Overall, the potentials at the profile level still outperform those at the residue level on the IFU dataset. The best discrimination is achieved by the distance-dependent potentials at the profile level, which correctly recognize 35 out of 41 decoy pairs, corresponding to accuracy of 85%. Such results outperform other atom-level potentials such as the Residue specific all-Atom Probability Discriminatory Function (RAPDF) [21] and the atomically detailed potentials of T32S3 [24]. These two potentials get 100% accuracy on the MISFOLD dataset as done by the profile-level distance-dependent potentials. They correctly identified 73% and 80% of the decoy pair on the IFU dataset respectively [24], while the profile-level distance-dependent potentials correctly identified 85% of the decoy pair on the same dataset.
Multiple decoy sets evaluation
To give an un-bias result and fair comparison with other potentials, we use five out of seven multiple decoy sets as used by Zhang et al. [42]. They include the 4state_reduce set [43], lmds set [44], fisa set [14], fisa_casp3 set [45], lattice_ssfit set [46]. Totally, there are 32 multiple decoy sets available (listed at Table 1 of Zhang et al. [42]). No decoy structures in the original decoy sets are omitted in this study. The diverse and comprehensive decoy sets ensure the fair evaluation of the overall quality of the potentials. We also compare our potentials with DFIRE-SCM [42], which is one of the most recent residue-level potentials. The results are evaluated in terms of success rates in native discriminations and Z-score for different decoy sets. The performances of different potentials are shown in Table 4.
Table 4.
The success rates and the average Z-scores of different potentials on the multiple decoy sets
| Source | 4state | Lattice_ssfit | Lmds | Fisa | Fisa_casp3 | Summary |
| DFIRD-SCM | 6/7 (3.94)a | 8/8 (6.19) | 3/10 (2.56) | 3/4 (4.70) | 3/3 (6.05) | 23/32 (4.68) |
| Distance | 5/7 (2.48) | 6/8 (4.97) | 2/10 (1.78) | 2/4 (3.06) | 1/3 (1.93) | 16/32 (2.84) |
| Distance_profile | 7/7 (3.53) | 8/8 (5.72) | 3/10 (2.45) | 2/4 (3.32) | 2/3 (2.94) | 22/32 (3.59) |
| Contact | 3/7 (1.38) | 4/8 (2.32) | 1/10 (0.83) | 0/4 (0.65) | 0/3 (1.69) | 8/32 (1.37) |
| Contact_profile | 3/7 (1.52) | 5/8 (2.96) | 1/10 (1.15) | 0/4 (0.72) | 0/3 (1.73) | 9/32 (1.61) |
| Dihedral | 7/7 (2.69) | 6/8 (3.51) | 2/10 (1.62) | 1/4 (1.05) | 1/3 (1.72) | 17/32 (2.12) |
| Dihedral_profile | 7/7 (2.72) | 7/8 (3.88) | 3/10 (1.55) | 1/4 (1.22) | 2/3 (2.58) | 20/32 (2.39) |
| Surface | 4/7 (1.80) | 4/8 (3.15) | 3/10 (1.21) | 1/4 (1.28) | 2/3 (2.26) | 14/32 (1.94) |
| Surface_profile | 4/7 (2.07) | 4/8 (3.57) | 5/10 (2.68) | 2/4 (1.89) | 2/3 (2.96) | 17/32 (2.64) |
aThe first number is the number of native structures ranked as number one; the second number is total number of proteins in the decoy set. The numbers in parentheses are the average Z-scores. The results of DFIRD-SCM method are directly taken from Zhang et al., Protein Sci. 2004, 13: 400–411.
As can be seen, all the profile-level statistical potentials outperform those at the residue-level. Overall, the success rates of profile-level potentials are better than those of residue-level potentials. Even with the same success rates on some datasets, the Z-scores of profile-level potentials are higher than those of residue-level. The distance-dependent knowledge-based potential [19] in this paper is the ProsaII potential as mentioned by Zhang et al. [42], which is inferior to DFIRE-SCM according to Zhang et al. [42]. The distance-dependent potential at the profile level is comparable with the DFIRE-SCM potential. The former correctly recognizes 22 out of 32 decoy structures, while the latter correctly recognizes 23 out of 32 decoy structures. The contact, Φ/Ψ dihedral angle and accessible surface statistical potentials are single-body residue-level statistical potentials, which are based on the coarse descriptions of protein structures. Such potentials get lower performance in comparison with other two-body atom-level statistical potentials in many experiments [21,25]. These simple potentials at the profile-level still outperform those at the residue-level according to our experiments. These results suggest that the binary profiles are smarter representations of protein structures than residues.
Discussion
The probability threshold has not significant influence on the profile-level statistical potentials
The frequency profiles are calculated from the multiple sequence alignments outputted by PSI-BLAST [35] and converted into binary profiles by a probability threshold Ph. The total number of binary profiles is dependent on the size of the database and the value of probability threshold Ph. Since each combination of the twenty amino acids corresponds to a binary profile and vice versa, the total number of binary profiles is 2^20. In fact, only a small fraction of binary profiles appear. These binary profiles substitute for novel alphabets of protein sequences to develop a class of novel profile-level statistical potentials. Since the probability threshold Ph is a parameter, it needs to be optimized. The results are shown in table 5. We surprisingly found that the probability threshold Ph has not significant influence on all the profile-level statistical potentials. When the probability threshold is larger than 0.28, the number of binary profiles is very small and the discriminative power of all the profile-level statistical potentials drops quickly. Since the decrease in the number of residue types reduces the discriminative ability of the potentials [11], we can draw a similar conclusion that an increase in the number of alphabets of protein sequences can improve the discriminative power of the potentials. This study provides a method for increasing the number of alphabets of protein sequences, that is, the profile method.
Table 5.
The optimized results of probability threshold.
| Probability threshold | Number of profiles | Distance_profile | Contact_profile | Dihedral_profile | Surface_profile |
| 0.04 | 21355 | - | 0.828263 | 0.815943 | 0.899127 |
| 0.05 | 19868 | - | 0.826101 | 0.815291 | 0.900184 |
| 0.06 | 15935 | - | 0.825473 | 0.816991 | 0.900127 |
| 0.08 | 7444 | - | 0.825815 | 0.815693 | 0.898678 |
| 0.10 | 3145 | - | 0.827828 | 0.815039 | 0.900998 |
| 0.12 | 1442 | - | 0.826786 | 0.814627 | 0.899441 |
| 0.14 | 759 | 0.907069 | 0.82626 | 0.816084 | 0.899387 |
| 0.16 | 404 | 0.909359 | 0.826889 | 0.815488 | 0.899437 |
| 0.17 | 303 | 0.906705 | 0.82597 | 0.81585 | 0.899063 |
| 0.18 | 235 | 0.909907 | 0.824466 | 0.816644 | 0.899639 |
| 0.20 | 186 | 0.908468 | 0.828051 | 0.815918 | 0.899407 |
| 0.22 | 138 | 0.906744 | 0.823012 | 0.811962 | 0.896226 |
| 0.24 | 81 | 0.907125 | 0.825444 | 0.81052 | 0.895877 |
| 0.26 | 46 | 0.904767 | 0.823669 | 0.809967 | 0.896212 |
| 0.28 | 28 | 0.892552 | 0.816189 | 0.799421 | 0.887908 |
| 0.30 | 21 | 0.873879 | 0.782432 | 0.777818 | 0.867548 |
| 0.32 | 21 | 0.872684 | 0.781907 | 0.779257 | 0.866134 |
Given in the table are the average CP scores of profile-level statistical potentials at different probability threshold. The discrimination is performed on our structure models. The distance_profile, contact_profile, dihedral_profile and surface_profile refer to the profile-level statistical potentials of distance-dependent, contact, Φ/Ψ dihedral angle and accessible surface respectively. Note that for small Ph value (<0.12), the profile-level distance-dependent potentials cannot produce efficient output, because the parameters of this potential are proportional to the square of the number of profiles.
The energy of profile-level statistical potentials correlates well with RMSD
Another measure of the potential quality and its global attraction is the dependence of the energy on the proximity to the native structure. The proper coordinate to measure proximity to the native structure is not obvious. However in numerous cases the RMSD is used [47]. In Fig. 3, the scatter plot of the energy as a function of the decoy Cα RMSD value is plotted. Since the potentials of different native structures are not comparable, only one of the sequence (1vcc) and its models are plotted. As can be seen, the energy of profile-level statistical potentials correlates well with the Cα RMSD up to quite large RMSDs. This suggests that the profile-level potentials can be useful in simulations that attempt to get closer to the native conformation starting from a distant conformation.
Figure 3.
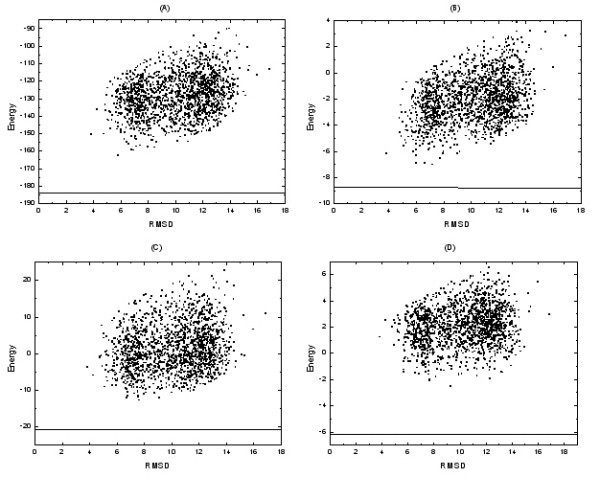
A scatter plot of energy versus RMSD. A horizontal line highlights the score of the native state. Subfigure (A), (B), (C) and (D) show the correlation of profile-level potentials of distance-dependent, contact, Φ/Ψ dihedral angle and accessible surface statistical potentials respectively. The total number of structure models included in each plot is 1858. Shown in the plot are the structure models of the sequence 1vcc from Baker's dataset.
Using evolutionary information can improve the discriminative power of knowledge-based mean force potentials
In the profile-level statistical potentials, the protein sequence is represented as a sequence of frequency profile rather than an amino acid sequence. The frequency profile contains the evolutionary information of protein sequences, which is the probabilities of the amino acids occurred in the specific position of the protein sequences. Such profiles are used to produce more discriminative potentials. As the best of our knowledge, this is the first usage of evolutionary information for developing more advanced potentials. The potentials at the profile level are prior to those at residue level according to the experiments. So evolutionary information can improve the discriminative power of knowledge-based mean force potentials. This conclusion is not surprising, since the evolutionary information is widely used in lots of biological problems such as the protein secondary structure prediction [48,49], remote homologue detection [50,51], sub-cellular localization [52,53], domain boundary prediction [54], fold recognition [55], protein-protein interaction prediction [56], function annotation [57], etc.
Profile-level statistical potentials can improve the performance of threading
Fold recognition or threading is another application of knowledge-based mean force potentials. Many methods combine the residue-level statistical potentials with sequence alignments for threading, such as the SPARKS method [18]. We have implemented a threading method that combines the profile-level statistical potentials with profile-profile alignments. Such profile-level threading method (referred as profile-threading) is compared with the threading method that uses the residue-level statistical potentials (referred as residue-threading).
Since the multi-body statistical potentials are hard to be used for threading, a combined potentials has been presented, which integrate the three single-body potentials of this study, that is, the Φ/Ψ dihedral angle, accessible surface and contact statistical potentials:
E(i) = Et(i, φi, ϕi) + wf Ef(i,Si) + wc Ec(i,Ni) (14)
where Et, Ef, Ec is the Φ/Ψ dihedral angle, accessible surface and contact statistical potentials respectively, i is amino acid for residue-level potentials and profile for profile-level potentials at the i-th position of the sequence, wf and wc are the weights of accessible surface and contact statistical potentials. The total potential for a protein is then obtained by summing the potentials of each of the amino acid or profile. Using the decoy set of PROSTAR, the optimal parameters of wf and wc for residue-level potential are selected as 0.5 and 3.375, which correctly identifies 59 out of 69 decoy pairs. The optimal parameters of wf and wc for profile-level potential are selected as 1 and 2.5, leading to correctly identify 62 out of 69 decoy pairs.
The profile-profile alignment method used here is the PICASSO3 method [58], which gives the best results of fold recognition [59]. The profile-profile score to align the position i of a sequence q and the position j of a template t is given by:
where fikq, fikt, Sikq and Sikt are the frequencies and the position-specific score matrix (PSSM) scores of amino acid k at position i of a sequence q and position j of a template t, respectively.
The profile-profile alignment is combined with the knowledge-based score for threading. The total score is given by:
utotal = mij + ws Ej (si)
where Ej(si) is the combined potentials score of the template at position j with the residue type (for residue-threading) or profile type (for profile-threading) si of the position i of the query sequence, ws is the weight factors for structure scores. The dynamic programming algorithm is employed to find the minimum of the total score of the sequence-template alignments.
The HOMSTRAD database [60] is selected to test the alignment accuracy of the two threading methods. Only families containing two single-chain sequences and with sequence identities less than 40% are considered. The resulting dataset contains 390 families and is randomly divided into training set and test set with ratio of 4:1. The genetic algorithm is used to find the optimal parameters on the training set including the structure factor ws, the gap-open penalty w0 and the gap-extension penalty w1. Such parameters are then applied to test the alignment accuracy on the test set. The results are shown in Table 6. The profile-level threading method outperforms the residue-level threading method, so profile-level statistical potentials can improve the performance of threading.
Table 6.
The results of two threading methods
| Method | W0 | W1 | Ws | Training accuracy | Test accuracy |
| Residue-threading | 4.5 | 0.5 | 0.175 | 78.2% | 75.6% |
| Profile-threading | 5 | 0.4 | 0.348 | 82.5% | 79.4% |
W0, W1 and Ws are the gap-open penalty, gap-extension penalty and the structure factor. The training accuracy and test accuracy are the alignment accuracy on the training set and test set.
Conclusion
In this study, a class of novel knowledge-based mean force potentials at the profile level has been presented. The frequency profiles are directly calculated from the multiple sequence alignments outputted by PSI-BLAST and converted into binary profiles with a probability threshold. Such binary profiles make up of a new alphabet for protein sequence. Because the binary profiles contain evolutionary information, they provide better descriptions of protein structures than the residues. We develop a class of novel statistical potentials at the profile level. Fold assessment and decoy sets evaluation results show that the statistical potentials at the profile-level outperform those at the residue level. Future work will aim at application of the profile-level statistical potentials to protein structure prediction and exploring other applications of such binary profiles such as remote homology detection, prediction of protein class etc.
Methods
Dataset
To evaluate the usefulness of the statistical potentials, large sets of protein structure models are needed [61]. Three datasets are used in this study. The first one is our structure models generated by large-scale comparative modeling [62]. The second one is the Baker's models [37] that also produced by comparative modeling. The third one is the PROSTAR decoy set [38]. The three datasets are briefly described as follows.
The freely available software MODELLER [62] is used for comparative modeling. The protein chains of the Protein Data Bank (PDB) [41] are downloaded from the SCOP database [63]. The sequence set and the template set are taken from the ASTRAL compendium [64] with sequence identity less than 40% and 80% respectively. Two sets of models including good models and bad models are calculated by large-scale comparative modeling. The models are classified depending on their structural similarity to the actual structure of the target protein. The good models are built on the basis of the correct templates and the structure-structure alignments between the target sequences and the template structures. The correct templates mean that the target sequences and the template structures share the same fold. The structure-structure alignment method is the iterative least-squares superposition method implemented by the MODELLER package [62]. Models with <30% structural overlap with the actual experimentally determined structure are eliminated. Structural overlap [11] is defined as the fraction of the equivalent Cα atoms upon least-squares superposition of the two structures with the 3.5 Å cutoff. The final set contains 4207 good models. The bad models are built on the basis of templates with incorrect folds but correct alignments (the structure-structure alignment) or the templates with correct folds but incorrect alignments (the sequence-sequence alignments). Models with >15% structure overlap with the actual target structure are eliminated. The final set contains 7045 bad models.
The Baker set [37] currently consists of 41 single domain proteins with varying degrees of secondary structures and lengths from 25 to 87 residues. Each protein is attached with about 1400 decoy structures generated by ab initio protein structure prediction method of Rosetta [45]. This set provides a good challenge for scoring functions and selection schemes to test themselves against the local minima around the native state. The Baker set can be downloaded from http://depts.washington.edu/bakerpg/ using the link "Download the all atom decoys used by Tasi et al. (pdbs)".
The PROSTAR set contains a set of well-constructed decoy sets. Three subsets including MISFOLD, IFU and PDBERR are selected for testing the performance of potentials. Each decoy set contains one correct and one or more incorrect or approximate conformations. The MISFOLD decoy set [39] consists of 25 examples of pairs of proteins with the same number of residues in the chain, but different sequences and conformations. The IFU decoy set is based on a set of 44 peptides that are proposed to be independent folding units as determined by local hydrophobic burial and experimental evidence [40]. The PDBERR decoy set is comprised of three structures determined using X-ray crystallography which are later found to contains errors and the corresponding correct experimental conformations [38].
Known structures for calculating potentials
The database of proteins used for the statistical analysis of various potentials is a subset of PDB database [41] obtained from the PISCES [65] web-server. The representative structures are selected such that they share <25% sequence identity with each other and better than 2.5 Å resolutions. The structures that contain missing atoms and chain breaks are excluded. We also remove the overlapped protein chains that are used in the three decoy sets. The resulting database contains 2352 chains and refers to PDB25 dataset.
Generating and converting of profiles
The PSI-BLAST [35] is used to generate the profiles of amino acid sequences with the default parameter values except that the number of iterations is set to 10. The search is performed against the NR90 database that is obtained by culling the NR database of NCBI using the Perl script from EBI [66]. The redundant sequences with sequence identity larger than 90% are removed. The frequency profiles are directly obtained from the multiple sequence alignments outputted by PSI-BLAST. The target frequency reflects the probability of an amino acid occurrence in a given position of the sequences. The method of target frequency calculation is similar to that implemented in PSI-BLAST. The multiple sequence alignments are used to calculate the frequency profiles. The sequence weight is assigned by the position-based sequence weight method [67]. Since calculation of target frequencies from the multiple sequence alignments may be influenced by a lot of factors including small sample size [68] and prior knowledge of relation among the residues [69,70]. We have implemented the data-dependent pseudo-count method to estimate the target frequencies [69]. Given the observed frequency of amino acid i (fi) and the background frequency of amino acid i (pi), the pseudo-count for amino acid i is computed as follows:
where qij is the score of amino acid i being aligned to amino acid j in BLOSUM62 substitution matrix that is the default score matrix of PSI-BLAST.
The target frequency is then calculated as:
Qi = (αfi + βgi)/α + β (2)
where α is the number of different amino acids in a given column minus one and β is a free parameter set to a constant value of 10, the value initially used by PSI-BLAST.
Because the frequency profile is a matrix of frequencies for all amino acids, it cannot be used directly and need to be converted into a binary profile by a probability threshold Ph. When the frequency of an amino acid is larger than Ph, it is converted into an integral value of 1, which means that the specific amino acid can occur in a given position of the protein sequences during evolution. Otherwise it is converted into 0. A substring of amino acid combination is then obtained by collecting the binary profile with non-zero value for each position of the protein sequences. These substrings have approximately represented the amino acids that possibly occur at a given sequence position during evolution. Each combination of the twenty amino acids corresponds to a binary profile and vice versa. Fig. 1 has shown the process of generating and converting the profiles.
Figure 1.
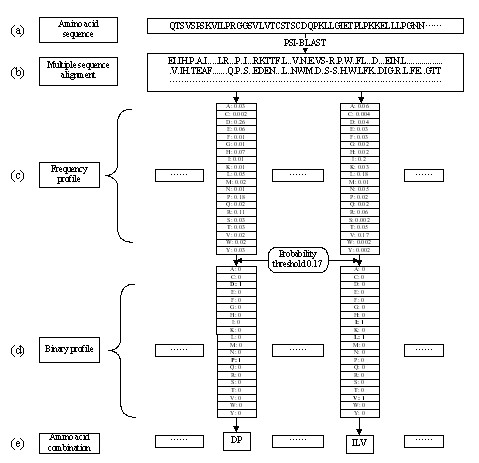
The process of calculating frequency profiles and converting it into binary profiles. (a) For a given amino acid sequence, (b) the multiple sequence alignment is obtained by PSI-BLAST. (c) The frequency profile is calculated on the multiple sequence alignment and (d) transforms into a binary profile with a probability threshold. (e) A substring of amino acid combination is then obtained by collecting the binary profile with non-zero value for each position of the protein sequences.
Knowledge-based mean force potentials
Similar to the knowledge-based potentials at the residue level, a class of novel potentials at the profile level is introduced. We developed four types of profile-level statistical potentials including distance-dependent, contact, Φ/Ψ dihedral angle and accessible surface statistical potentials. The difference between the potentials at the residue level and those at the profile level is that each residue is represented as a binary profile rather than a single residue. For the residue-level statistical potentials, the interaction types are the 20 standard amino acids. While for the profile-level statistical potentials, the interaction types are the binary profiles. Other parameters are same for the two kinds of statistical potentials. Such representation contains evolutionary information and provides more discriminative power than the single residue according to the experimental results.
Distance-dependent potential
The distance-dependent statistical potentials are calculated as described in [20,22]. The energy of two interaction types (ij) with sequence separation k and distance interval l is given by:
where Mijk is the number of occurrences for the interaction type pair ij separated by k residues in sequence:
where n is the number of classes of distances. σ is the weight given to each observation. σ = 1/50 is used for smoothing [19]. is the relative frequency of occurrence for the interaction center type pair ij at sequence separation k in the class of distance l:
is the relative frequency of occurrence for all the interaction center type pairs at sequence separation k in the class of distance l:
in which r is the number of different interaction center types and m is the number of classes for the sequence separation. The temperature T is set to 300 K, resulting in RT of 0.6 kcal/mole, where R is the gas constant.
Contact potential
The contact potential is obtained by the propensity of each of the interaction types for each of the contact number. The contact potential [18] is given by:
where i is the interaction types (amino acids or binary profiles), Ni is the contact number of the interaction center i. Nobs(i, k) is the number of observed contacts of interaction center i with other interaction centers at k'th bin and Ncbin is the number of contact bins. A contact is defined by the Ca-Ca distance of two interaction centers within 8 Å. The number of contact bins is set to 25. In the rare occasions of more than 25 contacts, the statistics is included in the bin for 25 contacts.
Φ/Ψ dihedral angle
The Φ/Ψ dihedral angle potential [18] is obtained by the propensity of each of the interaction types for each dihedral class. The Φ/Ψ dihedral angle potential is given by:
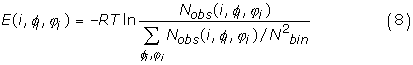
where i is the interaction type (amino acids or binary profiles), Φi, Ψi are the torsion angles at interaction center i. The torsion potential is the logarithm of the number of observed occurrence of the interaction center type i at torsion angles of Φi, Ψi [Nobs(i, Φi, Ψi)] normalized by the averaged occurrence. Each torsional angle is divided into 36 bins. That is, Nbin is equal to 36.
Accessible surface statistical potential
The accessible surface potential is calculated as described in [16,20]. The accessible surface of an interaction center is defined as the number of interaction centers within a sphere around the center interaction center. The radius of the sphere is the distance range of the potential. From these distributions, the statistical potential is calculated as follows:
where i is the interaction types (amino acids or binary profiles), Si is the number of the interaction center i. Nobs(i,s) is the observed occurrence of other interaction center with interaction center i at burial class s. Nsbin is the total number of burial classes.
Note that the last three potentials don't use the smoothing technique that is adopted by the distance-dependent potential, since the known structures for calculating potentials are very large. We find that the potentials without smoothing have the same discriminative power as those with smoothing (data not shown).
Energy and energy Z-score
For distance-dependent potentials, the energy of a protein structure model is the sum of the individual terms over all interaction type pair i and j, sequence separations k and distance classes l:
For the contact, accessible surface and Φ/Ψ dihedral angle potentials, the energy of the model is the sum of the terms for all of the residues (residue-level) or binary profiles (profile-level).
Before an energy is used to discriminate between the good and bad models, it is transformed into a Z-score of energy [20]:
where Em is the energy of the model, μr and σr are the average and standard deviation of the reference energy distribution respectively. Two different reference energy distributions are widely used. The first approach involves randomization of the order of residues in the tested model (sequence space reference). The second derivation of the reference energy distribution keeps the original sequence, but changes its conformation (structure space reference). Due to the similar performance [11] and the relative simplicity, the sequence space reference is applied here. The randomization procedure is repeated 200 times, generating 200 reference models.
Performance metrics
The fractions of the false positives (FP) and false negatives (FN) are defined as:
in which A is the number of true positives (good models predicted as good), B is the number of false positives (good models predicted as bad), C is the number of false negatives (bad models predicted as good) and D is the number of true negatives (bad models predicted as bad). The fraction of the Correctly Predicted (CP) cases or the correct classification rate at the optimal value of the energy Z-score cutoff is used to assess the performance of a given statistical potential in fold assessment as follows [11]:
in which the prime is used to indicated the corresponding values at the energy Z-score cutoff that results in the maximal correct classification rate.
Receiver operating characteristic (ROC) curves [71] are also used to assess the statistical potentials. An ROC plot is obtained by plotting the false negatives fraction against the corresponding false positives fraction for all cutoffs on the energy Z-score. The area under the ROC curve represents the probability of incorrect classification over the whole range of cutoffs, which ranges form 0 to 0.5. If it is 0.5, the scores for the good and bad models do not differ (no discrimination power), whereas a value of 0 indicates no overlap between the two sets of models (perfect discrimination).
For decoy set evaluation, two other performance metrics are adopted [42]. One is the success rate in native discriminations, which is defined as the overall ratio of native-rank as top 1 rank. The other is the Z-score of the decoy set, defined as:
where <> denotes the average over all decoy structures, and Enative is the energy of the native structure. Z-score is a measure of the bias toward the native structure.
Availability
The source code is included as an additional file [see Additional file 1], can be freely downloaded at http://www.insun.hit.edu.cn/news/view.asp?id=457 and is available upon request from the authors.
Authors' contributions
QD carried out the knowledge-based potential studies, participated in coding and drafted the manuscript. LL participated in the design of the study and performed the statistical analysis. XW conceived of the study, and participated in its design and coordination. All authors read and approved the final manuscript.
Supplementary Material
Source code. Source code for the profile-level statistical potentials
Acknowledgments
Acknowledgements
The authors would like to thank Xuan Liu for her comments on this work that significantly improve the presentation of the paper. Financial support is provided by the National Natural Science Foundation of China (60435020).
Contributor Information
Qiwen Dong, Email: qwdong@insun.hit.edu.cn.
Xiaolong Wang, Email: wangxl@insun.hit.edu.cn.
Lei Lin, Email: Linl@insun.hit.edu.cn.
References
- Sippl MJ. Knowledge-based potentials for proteins. Curr Opin Struct Biol. 1995;5:229–235. doi: 10.1016/0959-440X(95)80081-6. [DOI] [PubMed] [Google Scholar]
- Mirny L, Shakhnovich E. How to derive a protein folding potential?A new approach to an old problem. J Mol Biol. 1996;264:1164–1179. doi: 10.1006/jmbi.1996.0704. [DOI] [PubMed] [Google Scholar]
- Miyazawa S, Jernigan R. An empirical energy potential with a reference state for protein fold and sequence recognition. Proteins. 1999;36:357–369. doi: 10.1002/(SICI)1097-0134(19990815)36:3<357::AID-PROT10>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lazaridis T, Karplus M. Effective energy functions for protein structure prediction. Curr Opin Struct Biol. 2000;10:139–145. doi: 10.1016/S0959-440X(00)00063-4. [DOI] [PubMed] [Google Scholar]
- Fujitsuka Y, Takada S, Luthey-Schulten ZA, Wolynes PG. Optimizing physical energy functions for protein folding. Proteins. 2004;54:88–103. doi: 10.1002/prot.10429. [DOI] [PubMed] [Google Scholar]
- Stote R, Straub J, W tanabe M, WiorkiewiczKuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem. 1998;102:3586–3617. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Lii JH, Allinger NL. Directional Hydrogen Bonding in the MM3 Force Field. II. J Comp Chem. 1998;19:1001–1016. doi: 10.1002/(SICI)1096-987X(19980715)19:9<1001::AID-JCC2>3.0.CO;2-U. [DOI] [Google Scholar]
- Fang Q, Shortle D. Enhanced sampling near the native conformation using statistical potentials for local side-chain and backbone interactions. Proteins. 2005;60:97–102. doi: 10.1002/prot.20483. [DOI] [PubMed] [Google Scholar]
- Fang Q, Shortle D. A consistent set of statistical potentials for quantifying local side-chain and backbone interactions. Proteins. 2005;60:90–96. doi: 10.1002/prot.20482. [DOI] [PubMed] [Google Scholar]
- Loose C, Klepeis JL, Floudas CA. A new pairwise folding potential based on improved decoy generation and side-chain packing. Proteins. 2004;54:303–314. doi: 10.1002/prot.10521. [DOI] [PubMed] [Google Scholar]
- Melo F, Sanchez R, Sali A. Statistical potentials for fold assessment. Protein Sci. 2002;11:430–448. doi: 10.1110/ps.25502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Kollman P. Pathways to a protein folding intermediate observed in a 1-microsecond simulation in aqueous solution. Science. 1998;282:740–744. doi: 10.1126/science.282.5389.740. [DOI] [PubMed] [Google Scholar]
- Bowie JU, Luthy R, Eisenberg DA. a method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J Mol Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- Moult J, Fidelis K, Zemla A, Hubbard T. Critical Assessment of methods of protein structure prediction (CASP) - Round V. Proteins. 2003;53:334–339. doi: 10.1002/prot.10556. [DOI] [PubMed] [Google Scholar]
- Melo F, Feytmans E. Assessing protein structures with a non-local atomic interaction energy. J Mol Biol. 1998;277:1141–1152. doi: 10.1006/jmbi.1998.1665. [DOI] [PubMed] [Google Scholar]
- Gilis D, Rooman M. Identification and ab initio simulations of early folding units in proteins. Proteins. 2001;42:164–176. doi: 10.1002/1097-0134(20010201)42:2<164::AID-PROT30>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Zhou H, Zhou Y. Single-body residue-level knowledge-based energy score combined with sequence-profile and secondary structure information for fold recognition. Proteins. 2004;55:1005–1013. doi: 10.1002/prot.20007. [DOI] [PubMed] [Google Scholar]
- Sippl MJ. Calculation of conformational ensembles from potentials of mean force. An approach to the knowledge-based prediction of local structures in globular proteins. J Mol Biol. 1990;213:859–883. doi: 10.1016/s0022-2836(05)80269-4. [DOI] [PubMed] [Google Scholar]
- Sippl MJ. Boltzmann's principle, knowledge-based mean fields and protein folding. An approach to the computational determination of protein structures. J Comput Aided Mol Des. 1993;7:473–501. doi: 10.1007/BF02337562. [DOI] [PubMed] [Google Scholar]
- Samudrala R, Moult J. An all-atom distance-dependent conditional probability discriminatory function for protein structure prediction. J Mol Biol. 1998;275:895–916. doi: 10.1006/jmbi.1997.1479. [DOI] [PubMed] [Google Scholar]
- Melo F, Feytmans E. Novel knowledge-based mean force potential at atomic level. J Mol Biol. 1997;267:207–222. doi: 10.1006/jmbi.1996.0868. [DOI] [PubMed] [Google Scholar]
- Summa CM, Levitt M, Degrado WF. An atomic environment potential for use in protein structure prediction. J Mol Biol. 2005;352:986–1001. doi: 10.1016/j.jmb.2005.07.054. [DOI] [PubMed] [Google Scholar]
- Qiu J, Elber R. Atomically detailed potentials to recognize native and approximate protein structures. Proteins. 2005;61:44–55. doi: 10.1002/prot.20585. [DOI] [PubMed] [Google Scholar]
- Lu H, Skolnick J. A distance-dependent atomic knowledge-based potential for improved protein structure selection. Proteins. 2001;44:223–232. doi: 10.1002/prot.1087. [DOI] [PubMed] [Google Scholar]
- Berrera M, Molinari H, Fogolari F. Amino acid empirical contact energy definitions for fold recognition in the space of contact maps. BMC Bioinformatics. 2003;4:8. doi: 10.1186/1471-2105-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov NN, Nussinov R, Zimmer RM. Fast protein fold recognition via sequence to structure alignment and capacity: London, UK. 1996. pp. 53–72. [PubMed] [Google Scholar]
- Jones DT. GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J Mol Biol. 1999;287:797–815. doi: 10.1006/jmbi.1999.2583. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Luthy R, Bowie JU. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- Kunin V, A. OC. Clustering the annotation space of proteins. BMC Bioinformatics. 2005;6:24. doi: 10.1186/1471-2105-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein M, Sippl MJ. Protein sequence randomization: efficient estimation of protein stability using knowledge-based potentials. J Mol Biol. 2005;345:1199–1212. doi: 10.1016/j.jmb.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Chiu TL, Goldstein RA. How to generate improved potentials for protein tertiary structure prediction: a lattice model study. Proteins. 2000;41:157–163. doi: 10.1002/1097-0134(20001101)41:2<157::AID-PROT10>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Yang WY, Pitera JW, Swope WC, Gruebele M. Heterogeneous folding of the trpzip hairpin: full atom simulation and experiment. J Mol Biol. 2004;336:241–251. doi: 10.1016/j.jmb.2003.11.033. [DOI] [PubMed] [Google Scholar]
- Sander O, Sommer I, Lengauer T. Local protein structure prediction using discriminative models. BMC Bioinformatics. 2006;7:14. doi: 10.1186/1471-2105-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped Blast and Psi-blast: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SE, Zaragoza J, Rodriguez JR, Oliver MJ, Payton PR. Windows .NET Network Distributed Basic Local Alignment Search Toolkit (W.ND-BLAST) BMC Bioinformatics. 2005;6:93. doi: 10.1186/1471-2105-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J, Bonneau R, Morozov AV, Kuhlman B, Rohl CA, Baker D. An improved protein decoy set for testing energy functions for protein structure prediction. Proteins. 2003;53:76–87. doi: 10.1002/prot.10454. [DOI] [PubMed] [Google Scholar]
- Braxenthaler M, Samudrala R, Pedersen J, Luo R, Milash B, Moult J. PROSTAR: The protein potential test site http://prostar.carb.nist.gov
- Holm L, Sander C. Evaluation of protein models by atomic solvation preference. J Mol Biol. 1992;225:93–105. doi: 10.1016/0022-2836(92)91028-N. [DOI] [PubMed] [Google Scholar]
- Pedersen JT, Moult J. Folding simulation with genetic algorithms and a detailed molecular description. J Mol Biol. 1997;269:240–259. doi: 10.1006/jmbi.1997.1010. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Liu S, Zhou H, Zhou Y. An accurate, residue-level, pair potential of mean force for folding and binding based on the distance-scaled, ideal-gas reference state. Protein Sci. 2004;13:400–411. doi: 10.1110/ps.03348304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B, Levitt M. Energy functions that discriminate X-ray and near native folds from well-constructed decoys. J Mol Biol. 1996;258:367–392. doi: 10.1006/jmbi.1996.0256. [DOI] [PubMed] [Google Scholar]
- Keasar C, Levitt M. A novel approach to decoy set generation: designing a physical energy function having local minima with native structure characteristics. J Mol Biol. 2003;329:159–174. doi: 10.1016/S0022-2836(03)00323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons KT, Bonneau R, Ruczinski I, Baker D. Ab initio protein structure prediction of CASP III targets using ROSETTA. Proteins. 1999;37:171–176. doi: 10.1002/(SICI)1097-0134(1999)37:3+<171::AID-PROT21>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Samudrala R, Xia Y, Levitt M, Huang ES. A combined approach for ab initio construction of low resolution protein tertiary structures from sequence. Pac Symp Biocomput. 1999:505–516. doi: 10.1142/9789814447300_0050. [DOI] [PubMed] [Google Scholar]
- Wang K, Fain B, Levitt M, Samudrala R. Improved protein structure selection using decoy-dependent discriminatory functions. BMC Struct Biol. 2004;4:8. doi: 10.1186/1472-6807-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Simossis VA, Taylor WR, Heringa J. A simple and fast secondary structure prediction method using hidden neural networks. BioInformatics. 2005;21:152–159. doi: 10.1093/bioinformatics/bth487. [DOI] [PubMed] [Google Scholar]
- Cao Y, Liu S, Zhang L, Qin J, Wang J, Tang K. Prediction of protein structural class with Rough Sets. BMC Bioinformatics. 2006;7:20. doi: 10.1186/1471-2105-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand B, Gowri VS, Srinivasan N. Use of multiple profiles corresponding to a sequence alignment enables effective detection of remote homologues. BioInformatics. 2005;21:2821–2826. doi: 10.1093/bioinformatics/bti432. [DOI] [PubMed] [Google Scholar]
- Casbon JA, Saqi MA. On single and multiple models of protein families for the detection of remote sequence relationships. BMC Bioinformatics. 2006;7:48. doi: 10.1186/1471-2105-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasson PM, Huppa JB, Davis MM, Brunger AT. A hybrid machine-learning approach for segmentation of protein localization data. Bioinformatics. 2005;21:3778–3786. doi: 10.1093/bioinformatics/bti615. [DOI] [PubMed] [Google Scholar]
- Lei Z, Dai Y. An SVM-based system for predicting protein subnuclear localizations. BMC Bioinformatics. 2005;6:291. doi: 10.1186/1471-2105-6-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim J, Kim SY, Lee J. PPRODO: prediction of protein domain boundaries using neural networks. Proteins. 2005;59:627–632. doi: 10.1002/prot.20442. [DOI] [PubMed] [Google Scholar]
- Zhou H, Zhou Y. Fold recognition by combining sequence profiles derived from evolution and from depth-dependent structural alignment of fragments. Proteins. 2005;58:321–328. doi: 10.1002/prot.20308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Recio J, Totrov M, Skorodumov C, Abagyan R. Optimal docking area: a new method for predicting protein-protein interaction sites. Proteins. 2005;58:134–143. doi: 10.1002/prot.20285. [DOI] [PubMed] [Google Scholar]
- Thibert B, Bredesen DE, Del Rio G. Improved prediction of critical residues for protein function based on network and phylogenetic analyses. BMC Bioinformatics. 2005;6:213. doi: 10.1186/1471-2105-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelman D, Sadreyev R, Grishin N. Probabilistic scoring measures for profile-profile comparison yield more accurate short seed alignments. Bioinformatics. 2003;19:1531–1539. doi: 10.1093/bioinformatics/btg185. [DOI] [PubMed] [Google Scholar]
- Ohlson T, Wallner B, Elofsson A. Profile-profile methods provide improved fold-recognition: a study of different profile-profile alignment methods. Proteins. 2004;57:188–197. doi: 10.1002/prot.20184. [DOI] [PubMed] [Google Scholar]
- Mizuguchi K, Deane CM, Blundell TL, Overington JP. HOMSTRAD: a database of protein structure alignments for homologous families. Protein Sci. 1998;7:2469–2471. doi: 10.1002/pro.5560071126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogolari F, Tosatto SC, Colombo G. A decoy set for the thermostable subdomain from chicken villin headpiece, comparison of different free energy estimators. BMC Bioinformatics. 2005;6:301. doi: 10.1186/1471-2105-6-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Andreeva A, Howorth D, Brenner SE, Hubbard TJP, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Research. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandonia JM, Hon G, Walker NS, Conte LL, Koehl P, Levitt M, Brenner SE. The ASTRAL Compendium in 2004. Nucleic acids research. 2004;32:189–192. doi: 10.1093/nar/gkh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Dunbrack RLJ. PISCES: a protein sequence culling server. Bioinformatics. 2003;19:1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Removing near-neighbour redundancy from large protein sequence collections. Bioinformatics. 1998;14:423–429. doi: 10.1093/bioinformatics/14.5.423. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Position-based sequence weights. J Mol Biol. 1994;243:574–578. doi: 10.1016/0022-2836(94)90032-9. [DOI] [PubMed] [Google Scholar]
- Schneider TS, Stormo GD, Gold L, Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J Mol Biol. 1986;188:415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Altschul SF, Koonin EV. Detection of conserved segments in proteins: iterative scanning of sequence databases with alignment blocks. Proc Natl Acad Sci USA. 1994;91:12091–12095. doi: 10.1073/pnas.91.25.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M, Hughey R, Krogh A, Mian IS, Sjölander K, Haussler D. Dirichlet Mixture priors to derive hidden Markov models for protein families: Menlo Park, CA. AAAI Press; 1993. pp. 47–55. [PubMed] [Google Scholar]
- Theodoridis S, Koutroumbas K. Pattern recognition. Academic Press.; 1999. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Source code. Source code for the profile-level statistical potentials


