Abstract
Mitochondrial DNA (mtDNA) is highly polymorphic, and its variations in humans may contribute to individual differences in function as well as susceptibility to various diseases such as Parkinson disease, Alzheimer disease, bipolar disorder, and cancer. However, it is unclear whether and how mtDNA polymorphisms affect intracellular function, such as calcium signaling or pH regulation. Here we searched for mtDNA polymorphisms that have intracellular functional significance using transmitochondrial hybrid cells (cybrids) carrying ratiometric Pericam (RP), a fluorescent calcium indicator, targeted to the mitochondria and nucleus. By analyzing the entire mtDNA sequence in 35 cybrid lines, we found that two closely linked nonsynonymous polymorphisms, 8701A and 10398A, increased the basal fluorescence ratio of mitochondria-targeted RP. Mitochondrial matrix pH was lower in the cybrids with 8701A/10398A than it was in those with 8701G/10398G, suggesting that the difference observed by RP was mainly caused by alterations in mitochondrial calcium levels. Cytosolic calcium response to histamine also tended to be higher in the cybrids with 8701A/10398A. It has previously been reported that 10398A is associated with an increased risk of Parkinson disease, Alzheimer disease, bipolar disorder, and cancer, whereas 10398G associates with longevity. Our findings suggest that these mtDNA polymorphisms may play a role in the pathophysiology of these complex diseases by affecting mitochondrial matrix pH and intracellular calcium dynamics.
Synopsis
Mitochondria play important roles in energy production and regulation of intracellular calcium levels. Mitochondria have their own genetic material, mitochondrial DNA (mtDNA). In spite of its short length (16 kbp), mtDNA is highly variable among individuals and is thought to contribute to interindividual functional variability in energy-requiring activities such as intelligence and athletic performance. However, it is unclear whether mtDNA polymorphisms affect intracellular function and condition. Using transmitochondrial hybrid cells, the authors found two closely linked mtDNA polymorphisms, 10398A/G and 8701A/G, which cause alterations in mitochondrial pH and calcium concentration. Cytosolic calcium response to histamine tended to be different between transmitochondrial hybrid cells carrying these two mtDNA polymorphisms. It has been reported that the 10398A mtDNA polymorphism is a risk factor for Parkinson disease, Alzheimer disease, cancer, and bipolar disorder, whereas 10398G is associated with longevity. The present findings suggest that these mtDNA polymorphisms may play a role in the pathophysiology of these complex diseases by affecting mitochondrial matrix pH and intracellular calcium dynamics.
Introduction
The central importance of mitochondria in ATP production is well established [1,2]. The pH gradient across the mitochondrial membrane and the inner mitochondrial membrane potential make up the electrochemical gradient, which regulates the efficiency of ATP synthesis and other mitochondrial activity. Recent studies are also focusing on the roles of mitochondria in regulation of intracellular calcium dynamics. Mitochondrial calcium uptake affects various important cellular processes such as apoptosis [3–5], exocytosis [6,7], synaptic plasticity [8], and possibly spine dynamics [9].
Mitochondria have their own DNA, mitochondrial DNA (mtDNA), which encodes the genes of 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), and 13 subunits of enzymes related to oxidative phosphorylation [10]. Other subunits of mitochondrial proteins are encoded in the nuclear genome.
Mutations in mtDNA are known to cause various mitochondrial diseases such as mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS), which is caused by the 3243A/G mutation in mtDNA [11,12]. The mechanisms by which these mtDNA mutations cause functional impairment are well studied [13,14]. On the other hand, mtDNA is highly polymorphic, and certain polymorphisms are thought to be risk factors in complex diseases such as diabetes mellitus [15], Alzheimer disease [16,17], Parkinson disease [18–21], bipolar disorder [22,23], and some kinds of cancer [24,25]. It has also been reported that mtDNA polymorphisms are related to interindividual functional variability in human cognition [26], personality [27], athletic performance [28], and longevity [29]. However, these associations are solely dependent on population genetics. It is difficult to draw a definite conclusion from genetic association analyses alone because the high variability of mtDNA among individuals makes such analysis susceptible to confounding effects of population stratification; in addition, the effects of other polymorphisms in mitochondrial or nuclear genes are difficult to control. Although functional analyses of these polymorphisms are needed, to date there are few reports that identify functional effects of mtDNA polymorphisms. This is mainly due to methodological difficulties, as conventional molecular biological techniques are not readily applicable to mtDNA.
We examined the phenotypic effect of mitochondrial DNA without interference from nuclear genes by analysing transmitochondrial hybrid cells (cybrids), which were made by fusing a cell line lacking mtDNA, called ρ0 (rho zero) cells [30], with platelets from humans. Although the consequences of pathogenic mtDNA mutations have been examined using this technique, the functional significance of mtDNA polymorphisms has not been well investigated yet.
In this study, we searched for functional mtDNA polymorphisms using the following strategies: (1) by targeting a calcium indicator, ratiometric Pericam (RP) [31], to mitochondria and the nucleus in the same cell, mitochondrial and cytosolic calcium levels were monitored simultaneously; (2) to reduce cellular variability, a ρ0 cell line was subcloned before generation of cybrids; (3) by sequencing the whole mtDNA genome in 35 cybrids, functional mtDNA polymorphisms were comprehensively analyzed, and two nonsynonymous mtDNA polymorphisms, 10398A/G and 8701A/G, were identified; and (4) using a pH indicator, we confirmed that these mtDNA polymorphisms altered both mitochondrial matrix pH and intracellular calcium levels.
Results
Generation and Confirmation of Cybrid Cell Lines
First, we established a 143B.TK− ρ0206 cell line that stably expresses two RPs. RP is a ratiometric fluorescent protein developed as a calcium indicator [31]. The fluorescence ratio of 510 nm emission at 480 nm excitation to that at 410 nm excitation is reported to reflect calcium concentration at a constant pH. Mitochondria-targeted RP is also sensitive to pH because there is a higher pH [32,33] in the mitochondrial matrix (approximately 7.7–8.0) [34,35] than in the cytosol. In this study, two RPs, one targeted to mitochondria (mtRP) and the other to the nucleus (nucRP), were expressed in the same living cell. Thus, we were able to simultaneously monitor mitochondrial and cytosolic calcium concentrations [31]. Because calcium levels in the nucleus were reported to be similar to those in cytosol, nuclear calcium levels were used to indicate cytosolic calcium level.
We subcloned several ρ0 cell lines carrying RPs and selected one that showed a reproducible calcium response. We confirmed by mtDNA-specific PCR and Southern blot analysis that this cell line lacked mtDNA. We fused this cell line with platelets taken from 35 human volunteers and confirmed the integration of mtDNA from the volunteers by Southern blot analysis.
Measurements of Fluorescence Ratio of mtRP in all Cybrid Cell Lines
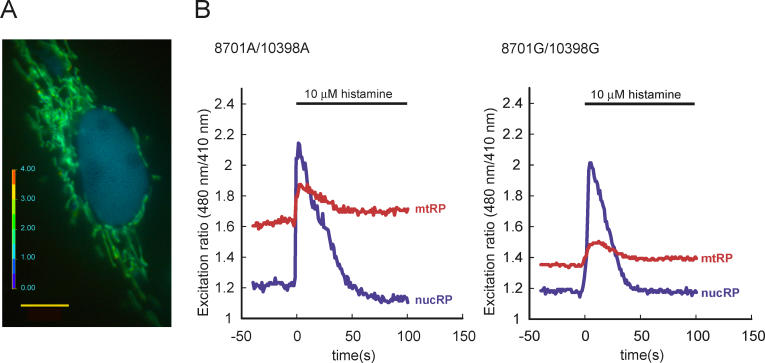
Using these cybrids, we measured fluorescence ratios of mtRP and nucRP. In fluorescent images of cybrids, mitochondria had a higher 480 nm/410 nm ratio than the nucleus (Figure 1A), possibly reflecting higher calcium levels and pH in mitochondrial matrix than in cytosol. This result is consistent with previous observations in HeLa cells [32]. At first, baseline fluorescence ratios were recorded for 2 minutes. Subsequently, cells were stimulated by 10 μM histamine, which elicits inositol trisphosphate–mediated calcium release from the endoplasmic reticulum [36]. In the first 60 s following stimulation, the fluorescence ratio of mtRP and nucRP initially increased and then returned to basal levels (Figure 1B). The mtRP fluorescence ratio was measured in 11–42 cells for each of the 35 cell lines by fluorescent microscopy.
Figure 1. Typical Excitation Ratio Image of a Cybrid and Histograms Showing Responses to Histamine.
(A) Image of a typical cybrid showing fluorescence ratios of excitation at 480 nm to 410 nm in pseudocolors. Mitochondria had a higher 480 nm/410 nm ratio than the nucleus, possibly reflecting higher calcium levels and pH in mitochondrial matrix than in cytosol. Scale bar, 10 μm.
(B) Representative fluorescence ratio responses of 8701A/10398A cybrids (left) and 8701G/10398G cybrids (right) evoked by 10 μM histamine. The y axis shows the ratio of the 525 nm fluorescence at 480 nm to that at 410 nm. Blue traces show the ratio in the nucleus and red traces show the ratio in mitochondria. Histamine (10 μM) was added at time 0 s and was not washed out during the experiment.
Using a one-way analysis of variance, we tested whether or not interindividual variation of mtDNA causes differences in basal mtRP fluorescence ratios. A statistically significant variation of basal mtRP fluorescence ratios was found among these cybrid lines (p < 0.001, F = 6.328).
Entire Sequences of mtDNA in All Cybrid Cell Lines
To identify the mtDNA polymorphisms that cause heterogeneity among cybrids, we determined the entire 16.5-kbp sequence of mtDNA in each of the 35 cybrids. mtDNA polymorphisms were selected in comparison with the revised Cambridge Reference Sequence [10,37]. A total of 216 polymorphic sites, including 13 novel polymorphisms, were identified. All cybrids had different sequences.
Identification of mtDNA Polymorphisms Altering the Fluorescence Ratio of mtRP
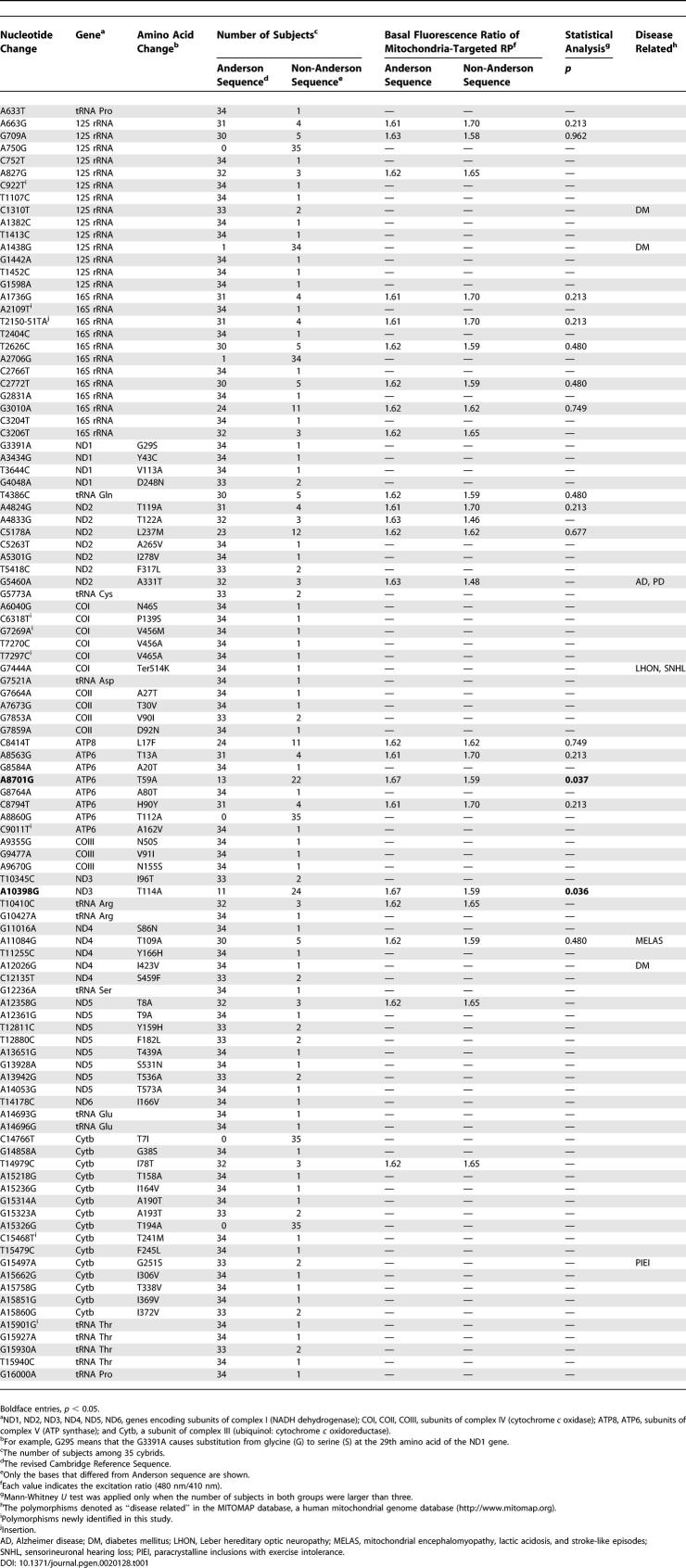
To identify the polymorphisms with functional significance, all nonsynonymous polymorphisms or polymorphisms in tRNA or rRNA found in more than three cybrids (n = 16) were selected (Table 1). For each polymorphism, the average mtRP fluorescence ratios for two genotypes at the particular polymorphic site were compared (Table 1). The polymorphisms at only two positions, 8701 and 10398, showed nominally significant differences of basal mtRP fluorescence ratios. Basal mtRP fluorescence ratios in cybrids with mtDNA 10398A were significantly higher than those with 10398G (10398A, 1.67 ± 0.09 [mean ± SD], n = 11; 10398G, 1.59 ± 0.12 [mean ± SD], n = 24; p < 0.05, Mann-Whitney U test). Basal mtRP fluorescence ratios in the cybrids with 8701A were also significantly higher than those with 8701G (8701A, 1.67 ± 0.1 [mean ± SD], n = 13; 8701G, 1.59 ± 0.11 [mean ± SD], n = 22; p < 0.05). Because 8701A was linked to 10398A in all but two cybrids, it was difficult to assess the independent effects of these polymorphisms. Since these two cybrids showed intermediate values (average fluorescence ratio, 1.64), it suggests that both of these polymorphisms may contribute to the observed difference. Because there were only two subjects with 8701G/10398A, further analyses were performed using only 8701A/10398A and 8701G/10398G cybrids.
Table 1.
The mtDNA Polymorphisms in 35 Cybrids
Basal mtRP fluorescence ratios in cybrids with 8701A/10398A mtDNA were significantly higher than those with 8701G/10398G mtDNA (8701A/10398A, 1.67 ± 0.09 [mean ± SD], n = 11; 8701G/10398G, 1.59 ± 0.11 [mean ± SD], n = 22; p < 0.05, Mann-Whitney U test; Figures 1 and 2). Peak mtRP fluorescence ratios after the histamine stimulation also showed a similar but nonsignificant trend (8701A/10398A, 1.86 ± 0.11 [mean ± SD]; 8701G/10398G, 1.78 ± 0.14 [mean ± SD]; p = 0.05).
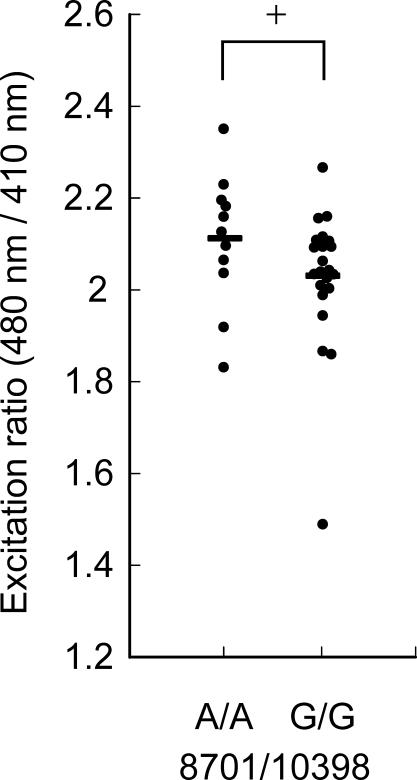
Figure 2. Effects of 10398A/G on Basal Fluorescence Ratios of Mitochondria-Targeted RP.
Differences between the basal fluorescence ratios in 10398A and 10398G cybrids are shown. All cybrids with 8701A have 10398A (left). While most cybrids with 8701G had 10398G (right), two cybrids had 8701A and 10398G (the data of these two cybrids were 1.52 and 1.77). For each cybrid, data taken from 11–42 cells (average, 26 cells) were averaged. The y axis shows the ratio of the 525 nm fluorescence at 480 nm to that at 410 nm. Horizontal bars indicate the mean values for each type of cybrid. *p < 0.05 (Mann-Whitney U test).
Replication Study Using Cybrid Cell Lines Stably Expressing RP
Because the nominally significant effects of polymorphisms at positions 10398 and 8701 on basal mtRP fluorescence ratio were found after multiple statistical analyses, we performed a second study to ensure it was not a false-positive finding. Three cybrid lines with 8701A/10398A mtDNA and three with 8701G/10398G mtDNA were chosen from among the 35 cybrids for the replication study (Table S1). The results confirmed our initial observation of a higher basal mtRP fluorescence ratio in cybrids with mtDNA 8701A/10398A (8701A/10398A, 1.78 ± 0.04 [mean ± SD], three cell lines; 8701G/10398G, 1.67 ± 0.01 [mean ± SD], three cell lines). Peak mtRP fluorescence ratios after the histamine stimulation also showed a similar trend (8701A/10398A, 1.99 ± 0.04 [mean ± SD], three cell lines; 8701G/10398G, 1.88 ± 0.02 [mean ± SD], three cell lines).
Replication Study Using Independently Established Cybrid Cell Lines
To further confirm that the observed difference is caused by the difference of mtDNA sequence, we performed another replication study using independently established cybrid cell lines. In this experiment, the native 143B.TK− ρ0206 cell line was used without any further subcloning. We established independent cybrid cell lines by using platelets derived from a subgroup of the original 35 donors (Table S1). By transiently transfecting the cybrids with ratiometric coxIV-Pericam cDNA, fluorescence imaging was performed. Moreover, cells were stimulated by 10 μM histamine, and in vivo calibration was performed using calibration buffers with ionomycin, which equilibrate intramitochondrial and extracellular calcium. To minimize the cell-to-cell variability, an index for calcium level, (R − R min ) / (R max − R) (see Materials and Methods), was analyzed in cells responded to histamine. As a result, our initial observation in cybrids with mtDNA 8701A/10398A was confirmed (8701A/10398A, 1.07 ± 0.40 [mean ± SD], three cell lines; 8701G/10398G, 0.51 ± 0.09 [mean ± SD], three cell lines; Figure S1).
Generation of pH Indicator for Measurements of Mitochondrial Matrix pH
It has been reported that the fluorescence ratio of RP is dependent not only on calcium levels but also on pH [32,33], especially at the higher pH range found in the mitochondrial matrix. To test the relative contributions of these factors to the observed finding, we attempted to measure mitochondrial pH using mitochondria-targeted, pH-sensitive green fluorescent protein (mt pH-GFP) [38,39] and mitochondria-targeted red fluorescent protein from Discosoma (mt DsRed) expressed under the control of a bidirectional promoter [40–42]. Because the pKa of pH-GFP is approximately 8.0, it is useful for measuring mitochondrial matrix pH. Since DsRed is a pH-insensitive protein, it can be used as a reference [43].
We confirmed that this pH indicator, mt pH-GFP/DsRed, could be successfully used to measure mitochondrial pH.
Measurements of Mitochondrial Matrix pH in Cybrids with mtDNA 8701A/10398A and 8701G/10398G
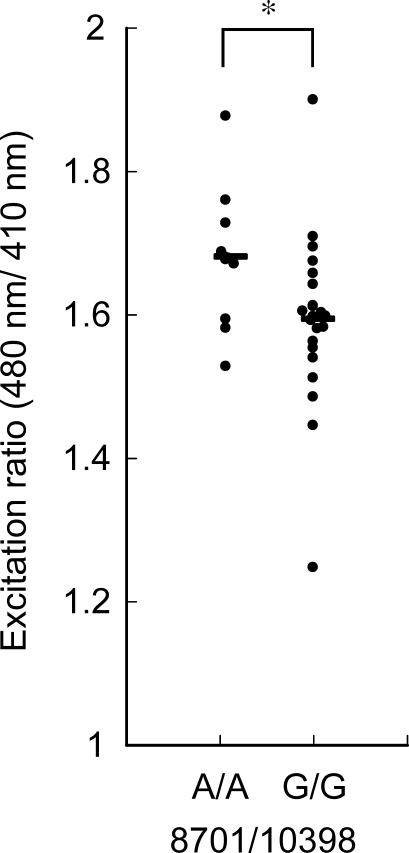
For the measurements of mitochondrial matrix pH, we used the above-mentioned independently established lines of cybrids generated from the native 143B.TK− ρ0 206 cell line that does not carry RPs (four with 8701A/10398A and four with 8701G/10398G). By transiently cotransfecting the mt pH-GFP/DsRed and Tet-Off vectors, basal mitochondrial matrix pH was recorded for several minutes in each cybrid cell line. We performed in vivo calibration of mitochondrial basal matrix pH in each cell by using four calibration buffers (pH 7.0, 7.5, 8.0, and 8.5) with the ionophores nigericin and monensin, which equilibrate intramitochondrial and extracellular pH. The calculated basal mitochondrial matrix pH was about 8.0, which is consistent with previous reports using HeLa cells and ECV304 cells [34,35]. Compared with the 8701G/10398G cybrids, the basal mitochondrial matrix pH was significantly lower in the 8701A/10398A cybrids (8701A/10398A, 8.03 ± 0.03 [mean ± SD], four cell lines; 8701G/10398G, 8.13 ± 0.07 [mean ± SD], four cell lines; p < 0.05, t test; Figure 3).
Figure 3. Basal Mitochondrial Matrix pH in the Cybrids with 8701A/10398A or 8701G/10398G .
Data from 11–18 cells in a triplicate experiment were averaged for each of four cell lines in each group. The y axis shows mitochondrial matrix pH calculated by calibration in vivo. Horizontal bars indicate the mean values for each type of cybrid. *p < 0.05 (t test).
Cytosolic Calcium Dynamics
To test whether or not these mtDNA polymorphisms affect cytosolic calcium dynamics, we examined the effects of 8701A/10398A and 8701G/10398G genotypes on the cytosolic calcium levels.
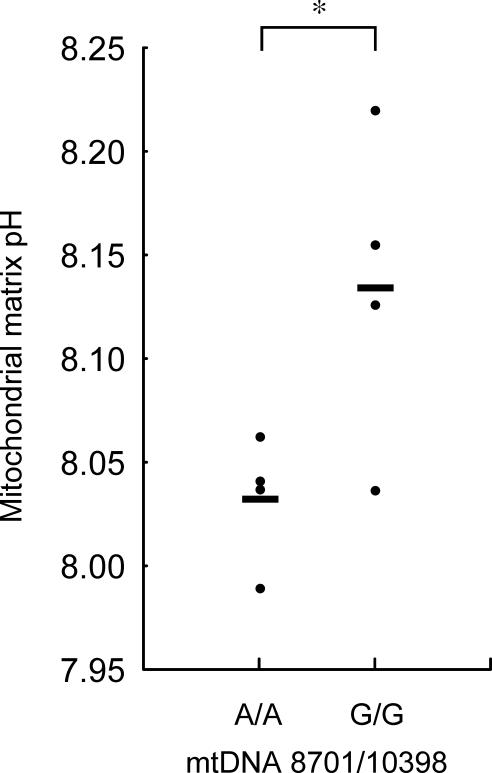
The nucRP data from the first experiments indicated that the 8701A/10398A cybrids tended to have higher peak cytosolic calcium levels after the histamine stimulation compared with the 8701G/10398G cybrids (8701A/10398A, 2.10 ± 0.14 [mean ± SD], 11 cell lines; 8701G/10398G, 2.02 ± 0.15, 22 cell lines, [mean ± SD]; p = 0.08, Mann-Whitney U test; Figure 4). There was no significant difference of basal cytosolic calcium level (8701A/10398A, 1.29 ± 0.07 [mean ± SD], 11 cell lines; 8701G/10398G, 1.25 ± 0.07 [mean ± SD], 22 cell lines; p > 0.10, Mann-Whitney U test). This result suggests that cytosolic calcium response may be enhanced in the 8701A/10398A cybrids.
Figure 4. Peak Cytosolic Calcium Level after Histamine Stimulation in Cybrids with 8701A/10398A and 8701G/10398G .
The values of two cybrids with 8701A/10398G were 1.92 and 2.20. For each cybrid, data taken from 11–42 cells (average, 26 cells) were averaged. The y axis shows the ratio of the 525 nm fluorescence at 480 nm to that at 410 nm. Horizontal bars indicate the mean values for each type of cybrid. +p = 0.08 (Mann-Whitney U test).
The nucRP data from the replication study using cybrid cell lines stably expressing RP confirmed that peak cytosolic calcium levels after histamine stimulation were higher in the 8701A/10398A cybrids than in 8701G/10398G cybrids (8701A/10398A, 2.56 ± 0.03 [mean ± SD], three cell lines; 8701G/10398G, 2.26 ± 0.16 [mean ± SD], three cell lines).
Discussion
In the present study, we comprehensively searched for mtDNA polymorphisms that alter the fluorescence ratios of mtRP, and we identified two mtDNA single nucleotide polymorphisms (mtSNPs), 10398A and 8701A, that increase the basal fluorescence ratios of mtRP.
In the 10398A/G mtSNP, threonine is substituted by alanine at the C terminus of ND3, a subunit of complex I (NADH: ubiquinone oxidoreductase) [10]. In the 8701A/G mtSNP, there is an amino acid substitution from threonine to alanine at ATPase6, the F0 subunit 6 of complex V (ATP synthase) [10]. Complex I generates a proton gradient across mitochondrial inner membrane by exporting protons, whereas ATP synthase produces ATP by using this proton gradient. The proton gradient is also the driving force of calcium uptake by mitochondria. Therefore, both polymorphisms can affect the proton gradient across mitochondrial inner membrane and calcium levels.
During the course of our study, it was reported that the fluorescence ratio of RP is dependent not only on calcium levels but also on pH at the higher pH ranges in mitochondrial matrix [32,33]. Thus, we measured basal mitochondrial matrix pH in cybrids with 8701A/10398A and 8701G/10398G. Basal mitochondrial matrix pH was significantly lower in cybrids with 8701A/10398A than in those with 8701G/10398G. Alteration in mitochondrial pH has also been hypothesized to modulate ATP production, apoptosis, and the opening of the mitochondrial membrane permeability transition pore [44]. Transport of protons is coupled with electron transport by a respiratory chain consisting of complexes I, III, and IV. As described above, the mtDNA 10398 polymorphism affects the amino acid composition in a subunit of complex I. When the activity of complex I in the electron-transport chain was measured using citrate synthase activity as a reference [45], the activity of 8701A/10398A cybrids tended to be lower than that of 8701G/10398G cybrids (8701A/10398A, 5.77 ± 1.41 [mean ± SD], four cell lines; 8701G/10398G, 7.58 ± 1.27 [mean ± SD], four cell lines; p = 0.10, t test). This finding supports the above-mentioned difference in mitochondrial matrix pH.
The difference in basal mitochondrial matrix pH could not contribute to the initial finding of higher mtRP fluorescence ratio in the cybrids with 8701A/10398A because lower pH could only decrease the mtRP fluorescence rate, not increase it. Therefore, the most probable interpretation for the difference in basal mtRP fluorescence ratio is higher mitochondrial calcium levels in cybrids carrying 8701A/10398A. In addition, the calcium responses to histamine stimulation tended to be higher in the cytosol of 8701A/10398A cybrids than 8701G/10398G cybrids, both in the initial experiment and the replication study using cybrid cell lines stably expressing RP. These findings suggest that alterations in mitochondrial function cause the enhanced cytosolic calcium response and higher basal mitochondrial calcium levels in cybrids with 8701A/10398A.
Mitochondria have several calcium transport mechanisms. Calcium uptake is attributable to a mitochondrial calcium uniporter in inner mitochondrial membrane. Calcium efflux from mitochondria is mediated by the Na+/Ca2+ exchanger and the H+/Ca2+ antiporter. The opening of the membrane permeability transition pore also allows efflux of calcium from mitochondria. Clarification of mitochondrial calcium transport systems at the molecular level would facilitate the understanding of biochemical mechanism of how the 10398 polymorphism alters mitochondrial calcium dynamics. Further study is needed to reveal how these two mtSNPs alter mitochondrial matrix pH and calcium levels.
Among the two polymorphisms we found to have functional significance, 10398A/G has previously been associated with human health and diseases. The 10398G polymorphism is reportedly associated with a reduced risk of Parkinson disease [19], whereas 10398A was reported to increase the risk of Alzheimer disease in men [17], invasive breast cancer in African-American women [24], and prostate cancer in African-American men [25]. In addition, 10398A is reportedly associated with an increased risk of bipolar disorder [22,23,46]. According to the mtSNP database (http://www.giib.or.jp/mtsnp/index_e.html), most Europeans have 8701A, but it is polymorphic in the Japanese population. In contrast, 10398A is polymorphic both in Europeans and Japanese. The 10398A/G characterizes the European haplogroup I, J, and K [17], and Asian-specific super haplogroup M [47]. Thus, the present findings can be translated that Asian-specific super haplogroup M is associated with different mitochondrial pH and calcium from other Asian-specific haplogroups.
The frequency of 10398A is much smaller in the Japanese population (approximately 29% in the mtSNP database) compared with Europeans (approximately 74% in the mtSNP database). The average life expectancy in Japan is the highest in the world, and its biological basis is not yet well understood. Tanaka et al. sequenced the whole mtDNA sequence in 11 Japanese centenarians and reported that a subset of haplogroup M characterized by several mtDNA polymorphisms is more frequently seen in centenarians [48]. Among these polymorphisms, they have mainly focused on the 5178A polymorphism that characterizes haplogroup D in super haplogroup M. The 5178C/A is associated with several common diseases such as Parkinson disease [28], atherosclerosis in patients with diabetes mellitus [49], and myocardial infarction [50]. A recent study, however, suggested that association of 5178A with longevity is ethnicity dependent [51]. Therefore, it might be possible that these apparent effects of 5178C/A in the Japanese population may be mediated by the other linked polymorphisms that cause functional change. Although 5178A did not affect the fluorescence ratio of mtRP in this study, most subjects with 5178A have 10398G. Thus, it could be possible that 10398G mediates these apparent associations of 5178C/A with complex diseases and longevity. In fact, based on the mtSNP database, 10398G was significantly more frequently seen in centenarians (76 of 96 [79%]) compared with young Japanese (129 of 192 [67%]; p = 0.03). A recent study by Niemi et al. also suggested that 10398G affects longevity [52].
To date, there has been no study on the association of 8701A/G with health and disease, and information on 8701A/G is limited. Most of the above-noted associations with the 5178 polymorphism observed in the Japanese population could also be mediated by 8701, since it is closely linked with 10398 and 5178. In fact, 8701G is also more frequency seen in centenarians (68 of 96 [71%]) compared with young Japanese (111 of 192 [58%]; p = 0.03).
It was difficult to assess the functional effects of these two mtSNPs separately, because there were only two subjects carrying 8701A/10398G in our sample set. Although it is still an open question whether or not 8701A/G causes some functional change, at least 10398A/G seems to have some functional significance. The tendency of altered complex I activity between the 8701A/10398A and 8701G/10398G cybrids suggest that the observed difference between 8701A/10398A and 8701G/10398G is mediated at least partly by the functional alteration of complex I by 10398A/G.
Because there was considerable overlap, including an outlier, with regard to the basal mtRP fluorescence between 8701A/10398A and 8701G/10398G, there might be a concern that the observed difference may be caused by those outliers. Thus, we applied the Smirnov-Grubbs test for the detection of outliers. One cell line, having the lowest value in the 8701G/10398G group, was found to be an outlier (T = 3.17; p < 0.05). Even after this cell line was omitted, basal mtRP fluorescence ratios in cybrids with 8701A/10398A were significantly higher than those with 8701G/10398G (8701A/10398A, 1.67 ± 0.09 [mean ± SD], n = 11; 8701G/10398G, 1.60 ± 0.09 [mean ± SD], n = 21; p < 0.05, Mann-Whitney U test). Thus, the observed difference was not due to the effect of the outlier. One might suspect that observed variation of basal mtRP fluorescence ratios might not be caused by mtDNA sequence variation but resulted from an experimental variation. However, the interassay variance within one cybrid cell line having the same mtDNA sequence was significantly smaller than the variation among cybrid cell lines having different mtDNA sequences. This indicates that observed variation of mtRP fluorescence ratios among cybrid cell lines were not caused by experimental variation but caused by the difference of mtDNA sequence.
There might be the other concern that the present finding is somehow affected by the process of subcloning, and such a finding cannot be generalized. However, in our replication study using independently established cybrid cell lines, it was confirmed that the observed finding was not specific to this particular subcloned cybrids, but was actually caused by differences of mtDNA sequences.
In summary, we identified two mtSNPs that were suggested to alter mitochondrial matrix pH and intracellular calcium dynamics. To our knowledge, this is the first report of mtDNA polymorphisms affecting these intracellular functions. Among the two mtSNPs examined, the 10398A/G polymorphism was previously reported to be associated with Parkinson disease, Alzheimer disease, some kinds of cancer, bipolar disorder, and longevity. A drug that affects mitochondrial matrix pH or calcium levels might be a promising strategy for health and disease.
Materials and Methods
Subjects.
Cybrids were generated from platelets derived from 35 volunteers, 17 patients with bipolar disorder (mean age, 48 years; 9 men and 8 women) and 18 healthy controls (mean age, 48 years; 10 men and 8 women). All were Japanese. Patients with bipolar disorder were included in this study because these blood samples were collected as a part of a project to study the genetics of bipolar disorder. None of the experimental parameters reported in this paper showed a significant difference between patients and controls. Diagnoses were made by the consensus of two senior psychiatrists using the DSM-IV criteria. Written informed consent was obtained from all volunteers. The Ethics Committees of the RIKEN Brain Science Institute and other participating institutes approved the study. For the replication study using cybrid cell lines stably expressing RP, we selected cybrids from three subjects with 8701A/10398A mtDNA (mean age, 48 years; 1 man and 2 women) and three subjects with 8701G/10398G mtDNA (mean age, 51 years; 1 man and 2 women). Cybrids for replication studies using independently established cybrid cell lines and measurements of mitochondrial matrix pH were generated using platelets from four 8701A/10398A subjects (mean age, 52 years; 2 men and 2 women) and four 8701G/10398G subjects (mean age, 56 years; 2 men and 2 women). Cybrids for replication studies and the measurement of mitochondrial matrix pH were derived from healthy controls. Although these cell lines have several mtDNA polymorphisms in addition to 8701 and 10398, they were carefully selected so that they have minimum difference in other positions of mtDNA.
Generation of the 143B.TK− ρ0206 cell line stably carrying calcium-sensitive probes.
The 143B.TK− ρ0206 cell line derived from the osteosarcoma cell line (143B.TK−) was established by Attardi and King [30]. The 143B.TK− ρ0206 cells were grown in DMEM (Invitrogen, Carlsbad, California, United States), supplemented with 10% FBS, 50 U penicillin, 50 μg/ml streptomycin, and 50 μg/ml uridine. The cells were grown on dishes 35 mm in diameter to 50% confluence. The cells were cotransfected with Lipofect AMINE PLUS (Invitrogen) with ratiometric Pericam-nuc (0.25 μg DNA/dish) and ratiometric coxIV-Pericam (0.75 μg DNA/dish) cDNAs. After 2 d, these cells were replated onto coverslips 14 mm in diameter and allowed to grow to 2.5% confluence with selection medium, supplemented with 10% FBS, 1,000 μg/ml geneticin (Invitrogen), and 50 μg/ml uridine. After 2 wk, a single colony with fluorescence in nuclei and mitochondria was picked up under a fluorescent microscope. mtDNA-specific PCR and Southern blot analysis were used to confirm that the cells lacked mtDNA. The cell lines were replated onto coverslips and stimulated by histamine (Sigma-Aldrich, Saint Louis, Missouri, United States) as described below. One of the ten cell lines had a reproducible calcium response without spontaneous calcium oscillation. This line was selected for further examination.
Generation of cybrid cell lines.
Peripheral blood was drawn into a 10 ml Vactainer tube containing 1.5 ml acid citrate dextrose (BD Biosciences, Palo Alto, California, United States). Platelets were separated by centrifugation at a speed of 1,200 rpm for 20 min. The 143B.TK− ρ0206 cells that had been stably expressing RPs were fused with platelets from individuals using a polyethylene glycol solution [53]. After 3 d of fusion, the medium was replaced by a selection medium without uridine. After 2 wk, surviving cells were collected. The integration of mtDNA was verified by Southern blot analysis as previously described with some modifications [54]. Cybrids for the replication study using independently established cybrid cell lines and measurements of mitochondrial matrix pH were generated by fusing the native 143B.TK− ρ0206 cells and platelets from individuals as described above.
Measurements of fluorescence ratios of RP.
Cybrids in modified Krebs R buffer (125 mM NaCl, 5 mM KCl, 5.5 mM glucose, 20 mM HEPES, and 1 mM MgCl2 [pH 7.4]) were imaged at room temperature on an Olympus IX-70 (Olympus, Tokyo, Japan) with a CoolSNAPHQ CCD camera (Roper Scientific, Tucson, Arizona, United States) controlled by Universal Imaging Meta series 4.5/4.6 (Universal Imaging, Media, Pennsylvania, United States). Dual-excitation imaging with RP used two excitation filters (480DF10 and 410DF10), alternated by a Lambda 10–2 filter exchanger (Sutter Instruments, Novato, California, United States), a 505DRLP-XR dichroic mirror, and a 525AF45 emission filter. The cybrids were stimulated by bath-application of 10 μM histamine applied to the coverslip. The ratio of the 525 nm fluorescence at 480 nm to that at 410 nm was used for further analysis. The regions of interest for measuring calcium response in each cell were the mitochondria and nucleus.
For each cell line, 11–42 cells were measured. At first, the basal fluorescence ratios of mtRP obtained from each cell were used for statistical analysis by one-way analysis of variance. Using one-way analysis of variance (df = 34), we tested whether or not the variation among 35 cybrid cell lines having different mtDNA was larger than the variation within one cell line. Then, the 11–42 data points in each cell line were averaged to calculate the representative value for each cell line.
The coefficient of variation in fluorescence ratio of mtRP in each cell line was 12.2% ± 3.2% (mean ± SD in 35 cybrids).
For the replication study using eight independently established cybrid cell lines (four with 8701A/10398A mtDNA and four with 8701G/10398G mtDNA), cybrids were transiently transfected with ratiometric coxIV-Pericam (1 μg DNA/dish) cDNA using Lipofect AMINE PLUS. After 2 d, measurements were performed in these cell lines. Different from the case in the cells stably expressing RP, there is no assurance that the expression level of RP is constant across cells in this experiment, which may potentially affect the stability of fluorescence ratio. To avoid the effect of variability of expression levels of RP among cells, we performed in vivo calibration for each cell. To exactly estimate calcium levels, we applied histamine stimulation to the cells, and only those responded to histamine were further analyzed. Cells responded to calcium could be found in three of four cell lines for each group. Cells were stimulated by 10 μM histamine, and calibration was performed using calibration buffers. Calcium-free buffer consisted of modified Krebs R buffer with 10 μM ionomycin (Calbiochem, San Diego, California, United States), 50 μM BAPTA-AM (Dojindo Laboratories, Kumamoto, Japan), and 10 mM EGTA (Dojindo Laboratories). Calcium saturation buffer was made of modified Krebs R buffer with 10 μM ionomycin and 10 mM CaCl2. [Ca2+] measured by in situ calibration was calculated by the equation [Ca2+] = K′ d [(R − R min ) / (R max − R)](1/n), where K′ d is the apparent dissociation constant corresponding to the calcium concentration and n is the Hill coefficient [55]. Because K′ d and n are constants but K′ d of RP in mitochondria is not known, we simply used the value of (R − R min ) / (R max − R) for the assessment of mitochondrial calcium level. The condition for imaging is basically similar to those described above.
Analysis of entire sequences of mtDNA.
Total DNA was extracted from each cybrid using standard protocols. The entire mtDNA was sequenced as described [56]. A computer program, Sequencher version 4.1.1 (Gene Codes, Ann Arbor, Michigan, United States), was used to indicate possible SNP loci. For verification, visual inspection of each candidate SNP was carried out. At least two overlapping DNA templates amplified with different primer pairs were used for identification of each SNP. mtSNPs were identified by comparison with the revised Cambridge sequence (GI1944628) reported by Andrews et al. [37]. For all polymorphisms that resulted in changed amino acid sequence or those within rRNA or tRNA regions, averaged fluorescence ratios of mtRP were compared between genotypes by Mann-Whitney U test. This analysis was performed only when three or more cybrids had the same polymorphism. For statistical analysis, Mann-Whitney U tests were applied using SPSS software (SPSS, Tokyo, Japan).
Construction of pH indicator for measurements of mitochondrial matrix pH.
GFP variants were prepared as described [57]. As a template, the original GFP mutation was employed. This template incorporates the mutation [38,58]. Mutations were verified by sequencing the entire gene. The pH-GFP [39] with polyhistidine tag at the N terminus was expressed in Escherichia coli JM109 (DE3) (Promega, Madison, Wisconsin, United States), purified, and spectroscopically characterized as described [55]. The gene for mt pH-GFP was amplified by PCR using the GFP variants as a template, with a forward primer containing complex IV mitochondrial target sequences and an MluI site, and a reverse primer containing an EcoRV site. The gene for mt DsRed was amplified by PCR using pDsRed1–1 Mito as a template with a forward primer containing a NotI site and a reverse primer containing a SalI site. The restricted mt pH-GFP product was subcloned in-frame into multiple cloning site I, and mt DsRed was subcloned into multiple cloning site II of the pBI bidirectional Tet vector (Clontech Laboratories, Mountain View, California, United States).
Transient expression of pH indicator in cybrid cell lines using the Tet system.
To ensure the same expression levels of two genes, mt pH-GFP and mt DsRed, the bidirectional Tet expression vector was used in Tet-Off Gene expression systems [40,41]. The pBI bidirectional Tet expression vector contains a “bidirectional” promoter composed of a tetracycline-response element flanked by two minimal cytomegalovirus promoters in opposite orientations [42]. To induce these two genes, the other vector expressing the tetracycline-controlled transactivator under the cytomegalovirus promoter control was cotransfected.
Cybrids were grown in DMEM (Sigma-Aldrich) supplemented with 10% Tet system-approved FBS (Clontech), 100 U penicillin, and 100 μg/ml streptomycin. The cells were grown on dishes 35 mm in diameter to about 80% confluence. The cells were cotransfected with Tet-Off vector (Clontech) and pBI–(mt pH-GFP)–(mt DsRed) (total 1 μg DNA/dish) using Lipofect AMINE PLUS (Invitrogen). After 2 d, mitochondrial matrix pH was measured in these cells.
Mitochondrial matrix pH measurements and in vivo calibration.
Cybrids in modified Krebs R buffer were imaged at room temperature. The imaging system is described above. Imaging with mt pH-GFP/DsRed used two excitation filters (480DF10 and S565/25), a dichroic mirror for GFP/DsRed, and emission filters (525AF45 and S620/60).
It was confirmed that the fluorescence ratio (525 nm/620 nm) decreased after the treatment with FCCP (Sigma-Aldrich), suggesting that this set of fluorescent indicators is sensitive to the mitochondrial matrix pH.
For in situ calibration, cybrids were perfused with the following pH titration buffer (125 mM KCl, 20 mM NaCl, 0.5mM CaCl2, 0.5 mM MgCl2, and 25 mM pH buffer [MOPS was used for pH 7.0, HEPES for pH 7.5 and 8.0, and glycylglycine for pH 8.5]) [59]. All calibration experiments were carried out in the presence of 10 μM nigericin and 10 μM monensin (Sigma-Aldrich). After baseline measurements were recorded for several minutes, calibration was performed in vivo. The fluorescence ratio of 525 nm emission at 480 nm excitation to 620 nm emission at 565 nm excitation reflected the dynamics of mitochondrial matrix pH. Mitochondrial matrix pH was calculated by plotting the calibration curve of each cell.
Supporting Information
The y axis shows the basal mitochondrial calcium levels indicated by the value of (R − R min ) / (R max − R) measured by mtRP. In situ calibration for [Ca2+] uses the equation [Ca2+] = K′ d [(R − R min ) / (R max − R)](1/n). R is the basal ratio of the 525 nm fluorescence at 480 nm to that at 410 nm. R min is the ratio at free calcium. R max is the ratio at saturating calcium. K′ d is the apparent dissociation constant. n is the Hill coefficient. Bars indicate the standard error of mean for each cybrid cell line.
(120 KB PDF)
(97 KB XLS)
Acknowledgments
We sincerely thank the members of the Laboratory for Cell Function and Dynamics at the RIKEN Brain Science Institute. We thank the members of the Research Resource Center at the RIKEN Brain Science Institute, especially Mr. Miyazaki, for technical assistance. We thank Ms. Bonnie Lee La Madeleine and Dr. S. Takeda, and Dr. T. Yoshida for valuable suggestions on the manuscript.
Author contributions. TK conceived and designed the experiments. AK and KM performed the experiments. AK and TK analyzed the data. AK, KM, TN, SS, MT, MY, NK, AM, and TK contributed reagents/materials/analysis tools. AK and TK wrote the paper.
Funding. This work was supported by an Independent Investigator Award from the National Alliance for Research on Schizophrenia and Depression (NARSAD) to T.K., a grant from the Japanese Ministry of Health, Labour and Welfare, and a grant from Japanese Ministry of Education, Culture, Sports, Science and Technology.
Competing interests. The authors have declared that no competing interests exist.
Abbreviations
- cybrid
transmitochondrial hybrid cell
- mt DsRed
mitochondria-targeted red fluorescent protein from Discosoma
- mt pH-GFP
mitochondria-targeted pH-sensitive green fluorescent protein
- mtDNA
mitochondrial DNA
- mtRP
ratiometric Pericam targeted to mitochondria
- mtSNPs
mitochondrial DNA single nucleotide polymorphisms
- nucRP
ratiometric Pericam targeted to nucleus
- RP
ratiometric Pericam
- rRNA
ribosomal RNA
- tRNA
transfer RNA
References
- Hansford RG, Zorov D. Role of mitochondrial calcium transport in the control of substrate oxidation. Mol Cell Biochem. 1998;184:359–369. [PubMed] [Google Scholar]
- Cortassa S, Aon MA, Marban E, Winslow RL, O'Rourke B. An integrated model of cardiac mitochondrial energy metabolism and calcium dynamics. Biophys J. 2003;84:2734–2755. doi: 10.1016/S0006-3495(03)75079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaurex N, Distelhorst C. Cell biology. Apoptosis—The calcium connection. Science. 2003;300:65–67. doi: 10.1126/science.1083628. [DOI] [PubMed] [Google Scholar]
- Hajnoczky G, Davies E, Madesh M. Calcium signaling and apoptosis. Biochem Biophys Res Commun. 2003;304:445–454. doi: 10.1016/s0006-291x(03)00616-8. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Pinton P, Ferrari D, Chami M, Szabadkai G, et al. Calcium and apoptosis: Facts and hypotheses. Oncogene. 2003;22:8619–8627. doi: 10.1038/sj.onc.1207105. [DOI] [PubMed] [Google Scholar]
- Kaftan EJ, Xu T, Abercrombie RF, Hille B. Mitochondria shape hormonally induced cytoplasmic calcium oscillations and modulate exocytosis. J Biol Chem. 2000;275:25465–25470. doi: 10.1074/jbc.M000903200. [DOI] [PubMed] [Google Scholar]
- Medler K, Gleason EL. Mitochondrial Ca(2+) buffering regulates synaptic transmission between retinal amacrine cells. J Neurophysiol. 2002;87:1426–1439. doi: 10.1152/jn.00627.2001. [DOI] [PubMed] [Google Scholar]
- Weeber EJ, Levy M, Sampson MJ, Anflous K, Armstrong DL, et al. The role of mitochondrial porins and the permeability transition pore in learning and synaptic plasticity. J Biol Chem. 2002;277:18891–18897. doi: 10.1074/jbc.M201649200. [DOI] [PubMed] [Google Scholar]
- Li Z, Okamoto K, Hayashi Y, Sheng M. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell. 2004;119:873–887. doi: 10.1016/j.cell.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Goto Y, Nonaka I, Horai S. A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Momoi MY, Tominaga K, Momoi T, Nihei K, et al. A point mutation in the mitochondrial tRNA(Leu)(UUR) gene in MELAS (mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes) Biochem Biophys Res Commun. 1990;173:816–822. doi: 10.1016/s0006-291x(05)80860-5. [DOI] [PubMed] [Google Scholar]
- Hess JF, Parisi MA, Bennett JL, Clayton DA. Impairment of mitochondrial transcription termination by a point mutation associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1991;351:236–239. doi: 10.1038/351236a0. [DOI] [PubMed] [Google Scholar]
- Smeitink J, van den Heuvel L, DiMauro S. The genetics and pathology of oxidative phosphorylation. Nat Rev Genet. 2001;2:342–352. doi: 10.1038/35072063. [DOI] [PubMed] [Google Scholar]
- Poulton J, Luan J, Macaulay V, Hennings S, Mitchell J, et al. Type 2 diabetes is associated with a common mitochondrial variant: Evidence from a population-based case-control study. Hum Mol Genet. 2002;11:1581–1583. doi: 10.1093/hmg/11.13.1581. [DOI] [PubMed] [Google Scholar]
- Hutchin T, Cortopassi G. A mitochondrial DNA clone is associated with increased risk for Alzheimer disease. Proc Natl Acad Sci U S A. 1995;92:6892–6895. doi: 10.1073/pnas.92.15.6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Walt JM, Dementieva YA, Martin ER, Scott WK, Nicodemus KK, et al. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci Lett. 2004;365:28–32. doi: 10.1016/j.neulet.2004.04.051. [DOI] [PubMed] [Google Scholar]
- Shoffner JM, Brown MD, Torroni A, Lott MT, Cabell MF, et al. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics. 1993;17:171–184. doi: 10.1006/geno.1993.1299. [DOI] [PubMed] [Google Scholar]
- van der Walt JM, Nicodemus KK, Martin ER, Scott WK, Nance MA, et al. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet. 2003;72:804–811. doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta C, Castro MG, Coto E, Blazquez M, Ribacoba R, et al. Mitochondrial DNA polymorphisms and risk of Parkinson's disease in Spanish population. J Neurol Sci. 2005;236:49–54. doi: 10.1016/j.jns.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Ghezzi D, Marelli C, Achilli A, Goldwurm S, Pezzoli G, et al. Mitochondrial DNA haplogroup K is associated with a lower risk of Parkinson's disease in Italians. Eur J Hum Genet. 2005;13:748–752. doi: 10.1038/sj.ejhg.5201425. [DOI] [PubMed] [Google Scholar]
- Kato T, Kunugi H, Nanko S, Kato N. Mitochondrial DNA polymorphisms in bipolar disorder. J Affect Disord. 2001;62:151–164. doi: 10.1016/s0165-0327(99)00173-1. [DOI] [PubMed] [Google Scholar]
- McMahon FJ, Chen YS, Patel S, Kokoszka J, Brown MD, et al. Mitochondrial DNA sequence diversity in bipolar affective disorder. Am J Psychiatry. 2000;157:1058–1064. doi: 10.1176/appi.ajp.157.7.1058. [DOI] [PubMed] [Google Scholar]
- Canter JA, Kallianpur AR, Parl FF, Millikan RC. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2005;65:8028–8033. doi: 10.1158/0008-5472.CAN-05-1428. [DOI] [PubMed] [Google Scholar]
- Mims MP, Hayes TG, Zheng S, Leal SM, Frolov A, et al. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2006;66:1880. doi: 10.1158/0008-5472.CAN-05-3774. author reply 1880–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuder P, Plomin R, McClearn GE, Smith DL, Vignetti S, et al. A polymorphism in mitochondrial DNA associated with IQ? Intelligence. 1995;21:1–11. [Google Scholar]
- Kato C, Umekage T, Tochigi M, Otowa T, Hibino H, et al. Mitochondrial DNA polymorphisms and extraversion. Am J Med Genet. 2004;128B:76–79. doi: 10.1002/ajmg.b.20141. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Takeyasu T, Fuku N, Li-Jun G, Kurata M. Mitochondrial genome single nucleotide polymorphisms and their phenotypes in the Japanese. Ann N Y Acad Sci. 2004;1011:7–20. doi: 10.1007/978-3-662-41088-2_2. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Gong J, Zhang J, Yamada Y, Borgeld HJ, et al. Mitochondrial genotype associated with longevity and its inhibitory effect on mutagenesis. Mech Ageing Dev. 2000;116:65–76. doi: 10.1016/s0047-6374(00)00149-4. [DOI] [PubMed] [Google Scholar]
- King MP, Attardi G. Human cells lacking mtDNA: Repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ . Proc Natl Acad Sci U S A. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippin L, Magalhaes PJ, Di Benedetto G, Colella M, Pozzan T. Stable interactions between mitochondria and endoplasmic reticulum allow rapid accumulation of calcium in a subpopulation of mitochondria. J Biol Chem. 2003;278:39224–39234. doi: 10.1074/jbc.M302301200. [DOI] [PubMed] [Google Scholar]
- Frieden M, James D, Castelbou C, Danckaert A, Martinou JC, et al. Ca(2+) homeostasis during mitochondrial fragmentation and perinuclear clustering induced by hFis1. J Biol Chem. 2004;279:22704–22714. doi: 10.1074/jbc.M312366200. [DOI] [PubMed] [Google Scholar]
- Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc Natl Acad Sci U S A. 1998;95:6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcelli AM, Ghelli A, Zanna C, Pinton P, Rizzuto R, et al. pH difference across the outer mitochondrial membrane measured with a green fluorescent protein mutant. Biochem Biophys Res Commun. 2005;326:799–804. doi: 10.1016/j.bbrc.2004.11.105. [DOI] [PubMed] [Google Scholar]
- Brini M, Pinton P, King MP, Davidson M, Schon EA, et al. A calcium signaling defect in the pathogenesis of a mitochondrial DNA inherited oxidative phosphorylation deficiency. Nat Med. 1999;5:951–954. doi: 10.1038/11396. [DOI] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Elsliger MA, Wachter RM, Hanson GT, Kallio K, Remington SJ. Structural and spectral response of green fluorescent protein variants to changes in pH. Biochemistry. 1999;38:5296–5301. doi: 10.1021/bi9902182. [DOI] [PubMed] [Google Scholar]
- Matsuyama S, Llopis J, Deveraux QL, Tsien RY, Reed JC. Changes in intramitochondrial and cytosolic pH: Early events that modulate caspase activation during apoptosis. Nat Cell Biol. 2000;2:318–325. doi: 10.1038/35014006. [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, et al. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- Baron U, Freundlieb S, Gossen M, Bujard H. Co-regulation of two gene activities by tetracycline via a bidirectional promoter. Nucleic Acids Res. 1995;23:3605–3606. doi: 10.1093/nar/23.17.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Sawano A, Eli P, Hama H, Miyawaki A. Red fluorescent protein from Discosoma as a fusion tag and a partner for fluorescence resonance energy transfer. Biochemistry. 2001;40:2502–2510. doi: 10.1021/bi002263b. [DOI] [PubMed] [Google Scholar]
- Bernardi P. Modulation of the mitochondrial cyclosporin A–sensitive permeability transition pore by the proton electrochemical gradient. Evidence that the pore can be opened by membrane depolarization. J Biol Chem. 1992;267:8834–8839. [PubMed] [Google Scholar]
- Trounce IA, Kim YL, Jun AS, Wallace DC. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996;264:484–509. doi: 10.1016/s0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
- Kato T. DNA polymorphisms and bipolar disorder. Am J Psychiatry. 2001;158:1169–1170. doi: 10.1176/appi.ajp.158.7.1169-b. [DOI] [PubMed] [Google Scholar]
- Sudoyo H, Suryadi H, Lertrit P, Pramoonjago P, Lyrawati D, et al. Asian-specific mtDNA backgrounds associated with the primary G11778A mutation of Leber's hereditary optic neuropathy. J Hum Genet. 2002;47:594–604. doi: 10.1007/s100380200091. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Gong JS, Zhang J, Yoneda M, Yagi K. Mitochondrial genotype associated with longevity. Lancet. 1998;351:185–186. doi: 10.1016/S0140-6736(05)78211-8. [DOI] [PubMed] [Google Scholar]
- Matsunaga H, Tanaka Y, Tanaka M, Gong JS, Zhang J, et al. Antiatherogenic mitochondrial genotype in patients with type 2 diabetes. Diabetes Care. 2001;24:500–503. doi: 10.2337/diacare.24.3.500. [DOI] [PubMed] [Google Scholar]
- Takagi K, Yamada Y, Gong JS, Sone T, Yokota M, et al. Association of a 5178C→A (Leu237Met) polymorphism in the mitochondrial DNA with a low prevalence of myocardial infarction in Japanese individuals. Atherosclerosis. 2004;175:281–286. doi: 10.1016/j.atherosclerosis.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Dato S, Passarino G, Rose G, Altomare K, Bellizzi D, et al. Association of the mitochondrial DNA haplogroup J with longevity is population specific. Eur J Hum Genet. 2004;12:1080–1082. doi: 10.1038/sj.ejhg.5201278. [DOI] [PubMed] [Google Scholar]
- Niemi AK, Moilanen JS, Tanaka M, Hervonen A, Hurme M, et al. A combination of three common inherited mitochondrial DNA polymorphisms promotes longevity in Finnish and Japanese subjects. Eur J Hum Genet. 2005;13:166–170. doi: 10.1038/sj.ejhg.5201308. [DOI] [PubMed] [Google Scholar]
- Bentlage HA, Chomyn A. Immunoprecipitation of human mitochondrial translation products with peptide-specific antibodies. Methods Enzymol. 1996;264:218–228. doi: 10.1016/s0076-6879(96)64022-1. [DOI] [PubMed] [Google Scholar]
- Nishino I, Kobayashi O, Goto Y, Kurihara M, Kumagai K, et al. A new congenital muscular dystrophy with mitochondrial structural abnormalities. Muscle Nerve. 1998;21:40–47. doi: 10.1002/(sici)1097-4598(199801)21:1<40::aid-mus6>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Hayakawa M, Ozawa T. Automated sequencing of mitochondrial DNA. Methods Enzymol. 1996;264:407–421. doi: 10.1016/s0076-6879(96)64037-3. [DOI] [PubMed] [Google Scholar]
- Sawano A, Miyawaki A. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 2000;28:E78. doi: 10.1093/nar/28.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter RM, Elsliger MA, Kallio K, Hanson GT, Remington SJ. Structural basis of spectral shifts in the yellow-emission variants of green fluorescent protein. Structure. 1998;6:1267–1277. doi: 10.1016/s0969-2126(98)00127-0. [DOI] [PubMed] [Google Scholar]
- Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, et al. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The y axis shows the basal mitochondrial calcium levels indicated by the value of (R − R min ) / (R max − R) measured by mtRP. In situ calibration for [Ca2+] uses the equation [Ca2+] = K′ d [(R − R min ) / (R max − R)](1/n). R is the basal ratio of the 525 nm fluorescence at 480 nm to that at 410 nm. R min is the ratio at free calcium. R max is the ratio at saturating calcium. K′ d is the apparent dissociation constant. n is the Hill coefficient. Bars indicate the standard error of mean for each cybrid cell line.
(120 KB PDF)
(97 KB XLS)