Abstract
We have cloned and sequenced a Listeria welshimeri DNA fragment homologous to the previously described fibronectin-binding protein-encoding gene (fbp) of Listeria monocytogenes (P. Gilot, Y. Jossin, and J. Content, J. Med. Microbiol., 49:887-896, 2000). This L. welshimeri DNA fragment expresses a 24.8-kDa protein that binds to human fibronectin. Based on the fbp sequences, we developed novel PCR assays for the identification of L. welshimeri and L. monocytogenes.
The genus Listeria comprises six species: L. monocytogenes, L. ivanovii, L. seeligeri, L. innocua, L. welshimeri, and L. grayi. All of these species are widespread in the environment, but only L. monocytogenes is considered to be a significant human and animal pathogen. Human disease due to L. monocytogenes usually occurs in pregnant women, the elderly, and immunocompromised patients. Clinical manifestations range from mild flu-like symptoms and gastroenteritis to septicemia, central nervous infections, and feto-maternal infections with abortion, premature labor, or birth of an infected child. Besides L. monocytogenes, however, occasional human infections due to L. ivanovii and L. seeligeri have also been reported. L. ivanovii is nevertheless mainly responsible for abortion in sheep (4, 8, 11, 24, 28).
All Listeria species have been isolated from soil, decaying vegetable matter, silage, sewage, water, animal feed, fresh and processed meats, raw milk, cheese, slaughterhouse waste, and asymptomatic human and animal carriers. Because of their widespread occurrence, Listeria species have many opportunities to enter food production and processing environments. Due to their psychrotrophic nature, they are then able to grow in food, even at temperatures such as those of refrigerators (1, 2, 4, 9, 11). Consequently, outbreaks and sporadic cases of listeriosis have been traced to different foodstuffs, such as pasteurized milk, cheese, coleslaw, and meat products (8, 11, 15). The coexistence of several Listeria species on the same food is not unusual, and often the incidence of Listeria species other than Listeria monocytogenes is higher than the incidence of L. monocytogenes itself. Since all Listeria species are potential food contaminants, the presence on foodstuffs of any of these species can be considered to be an indicator of their contamination and of the potential presence of L. monocytogenes (19). However, because the threats to public health posed by contamination of foods by each of these Listeria species are not similar, it is very important that all of them be rapidly and reliably detected and identified.
The identification of Listeria species has long been hampered by the small number of tests allowing the differentiation between these closely related species. Hemolysis, a major characteristic of Listeria species identification, may be, in some cases (especially for environmental and food isolates), difficult to read on blood agar, and tests monitoring the acid production from carbohydrates are time-consuming (4, 28). To avoid these drawbacks, faster PCR identification procedures targeting the 16S rRNA gene, the 16S/23S spacer, and genes encoding proteins such as aminopeptidase, invasion-associated protein, listeriolysin O, internalin A, LmA antigen, or the PrfA regulator have been developed (7, 16, 17, 20, 22, 25, 29, 30). L. welshimeri remains, nevertheless, one of the less known species of the genus Listeria, with only genes coding for rRNA and for the invasion-associated protein (Iap) already sequenced and deposited in nucleotide databanks. Apart from species-specific internal iap gene sequences, the only known specific markers for L. welshimeri among the members of the genus are the capacity to ferment d-tagatose (although shared by only 89% of the tested strains) and obtaining of characteristic proteic or genomic fingerprints by multilocus enzyme electrophoresis, repetitive element sequence-based PCR, random amplification of polymorphic DNA, and pulsed-field gel electrophoresis (3, 5, 6, 10, 18, 23). Consequently, whereas L. welshimeri was found to be present in the environment and has been isolated from animal and human carriers, studies monitoring its ecology are still rare (2, 26)
We previously reported that L. monocytogenes binds to human fibronectin, a 450-kDa dimeric glycoprotein found in body fluids, on the surface of eukaryotic cells, and in an insoluble component of the extracellular matrix. The binding of fibronectin to L. monocytogenes appeared to be saturable and dependent on proteinaceous receptors. Several of these fibronectin-binding proteins were identified in cell lysates and cell-wall extracts of the bacterium (12). An L. monocytogenes DNA library was screened with fibronectin, and a gene encoding a 24.6-kDa fibronectin-binding protein (Fbp) was isolated, sequenced, and proved to be transcribed in L. monocytogenes. The fbp gene was found to be present in all tested isolates of the species L. monocytogenes, and a homologous DNA fragment could be amplified from L. welshimeri chromosomal DNA. Restriction endonuclease-PCR (RE-PCR) showed that the fbp gene displays a degree of allelic variation among isolates of L. monocytogenes, whereas the corresponding amplified fragment of L. welshimeri is monomorphic among all tested isolates of this species. RE-PCR targeting the fbp gene produced a specific DNA banding profile for each of these two species, indicating that a specific PCR assay could be developed for L. welshimeri as well as for L. monocytogenes (13).
In this work, the L. welshimeri DNA fragment homologous to the fbp gene of L. monocytogenes was cloned, sequenced, and characterized with the aim of developing specific PCR assays for the identification of L. welshimeri and L. monocytogenes.
Cloning and sequencing of the L. welshimeri fragment homologous to the fbp gene.
We previously shown that the complete 648-bp open reading frame (ORF) of the fbp gene could be amplified with primers G296 (5"-CGGGATCCTGAAAGAGTTTATCGAGCCATACC-3"; nucleotides [nt] 2 to 25) (Fig. 1) and G297 (5"-GGAATTCTTATTTACGTTTCTTAACAACCTC-3"; nt 648 to 625) (Fig. 1) from all tested strains of L. monocytogenes. With the same primers, a homologous DNA fragment of the same molecular weight was also amplified from all tested isolates of L. welshimeri, but not from strains of L. grayi, L. innocua, L. seeligeri, and L. ivanovii (13). At their 5" extremities, primers G296 and G297 possess an oligonucleotide tail containing restriction sites for BamHI and EcoRI, respectively. This property was used to easily clone the 648-bp amplified fragment of L. welshimeri in a plasmid vector. For that aim, this DNA fragment was amplified from chromosomal DNA of L. welshimeri P.P. by using the primers described above and Pfu DNA polymerase, according the manufacturer's instructions (Stratagene, La Jolla, Calif.). Amplification was performed with a Perkin-Elmer thermocycler (Gene Amp PCR system 2400) with the same cycling conditions previously described with Taq DNA polymerase, but with only 25 cycles of amplification (13). The PCR product was then successively incubated (1 h, 37°C) with self-digested pronase at a final concentration of 0.4 mg/ml, extracted with a phenol-chloroform-isoamyl alcohol solution, precipitated with ethanol, suspended in water, and incubated overnight at 37°C with EcoRI and BamHI, following the manufacturer's instructions (Promega). The hydrolyzed 648-bp DNA fragment was finaly electrophoresed on a 0.7% agarose gel, eluted from the gel (GenElute agarose spin column; Suppelco), and inserted by standard cloning techniques into the pGex-5X-3 vector (Pharmacia) previously hydrolyzed with BamHI and EcoRI (27). The ligation reaction was used to transform Escherichia coli DH5α, and a positive clone was selected for sequencing. The sequence of the cloned L. welshimeri DNA fragment was determined by fluorescent dye-primer cycle sequencing of both strands of the insert with pGex reverse and forward Texas red-labeled primers (5"-GGGCTGGCAAGCCACGTTTGG-3" and 5"-CCGGGAGCTGCATGTGTCAGA-3"), internal Texas red-labeled primers (5"-GCTCCATACGATTACTTCAAC-3" and 5"-AGATTTTGTTTTCGCCATAA-3"), the Thermo Sequenase premixed cycle sequencing kit from Amersham, and the Vistra DNA Sequencer 725 apparatus of Amersham (Fig. 1). The DNA insert has a G+C content of 34.6%, which is close to the 36% G+C content of all L. welshimeri chromosomal DNA (28). The cloned fragment is composed of an uninterrupted ORF and possesses 88.4% identical nucleotides to the L. monocytogenes Fbp ORF (Fig. 1). Nucleic database searches (BLAST N) found homology only with the L. monocytogenes ORF cited above and with an L. innocua nucleotidic sequence. Analysis of the L. welshimeri sequence confirmed the presence or absence of the MseI, RsaI, SacI, HhaI, DdeI, and TaqI restriction sites previously localized on the published physical map of the amplified fragment (13).
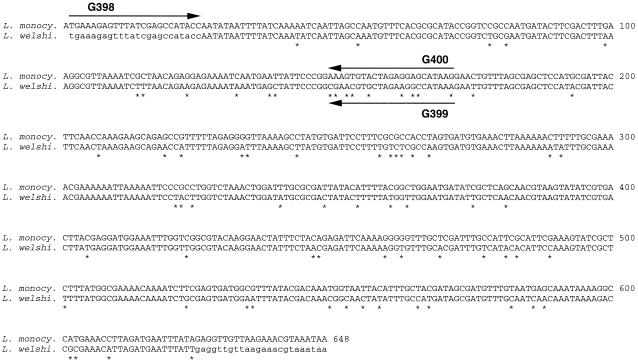
FIG. 1.
Alignment of the nucleotide sequence of the fbp gene of Listeria monocytogenes with the homologous DNA fragment of Listeria welshimeri. The sequence of the fbp gene of L. monocytogenes (EMBL accession no. AJ132543 ) was aligned with the sequence of the amplified DNA fragment of L. welshimeri (accession no. AJ293985). The 24 first and 24 last nucleotides of the L. welshimeri DNA fragment (lowercase letters) are those of primers G296 and G297, used for the amplification and cloning of the gene, but are not necessarily entirely those of the L. welshimeri sequence. The presence of nonidentical nucleotides between the two sequences is indicated by an asterisk. Arrows indicate the regions of hybridization of primers G398, G399, and G400.
The protein expressed by the L. welshimeri DNA fragment binds human fibronectin.
The predicted 215-amino-acid sequence encoded by the cloned fragment possess 90.7% identical amino acids to the Fbp protein of L. monocytogenes and conserves notable features of Fbp, such as a richness in lysine (11.6%), distributed throughout the molecule, an AKTK repeated amino acid block in the middle of the protein (positions 99 to 102 and 170 to 173), and a cytochrome c family heme-binding signature (Prosite PD0C00169) at positions 156 to 161. Protein database searches (BLAST P) found only similarities to the Fbp protein of L. monocytogenes, to an L. innocua protein (accession no. NP_464248), and to YwfA (40.3% identity), a protein of unknown function of Lactococcus lactis subsp. lactis (accession no. AAK06272).
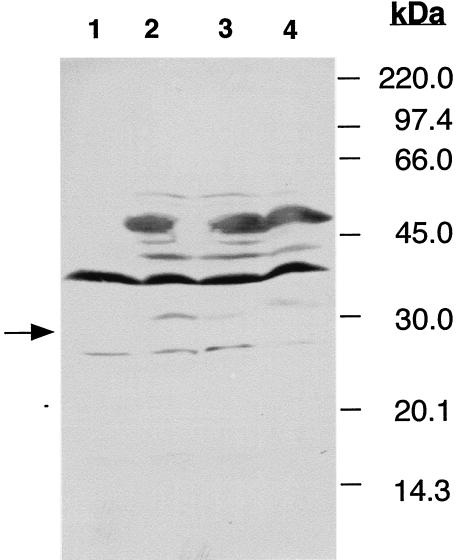
The high degree of similarity between the protein encoded by the amplified DNA fragment of L. welshimeri and the Fbp of L. monocytogenes suggests that both proteins may be able to bind human fibronectin. As in pGex-5X-3, the ORF of the L. welshimeri DNA fragment was cloned in phase with the gene encoding the 26-kDa glutathione-S-transferase (GST) from the parasite Schistosoma japonicum. We were able to test if the 51-kDa recombinant fusion protein binds fibronectin. As a control, the complete Fbp ORF of L. monocytogenes was similarly cloned in phase with the GST-encoding gene. Recombinant Escherichia coli DH5α cells were then grown in the presence of isopropyl-β-d-thiogalactopyranoside (IPTG), as described by the producer of the expression vector (Pharmacia). After sonication, E. coli proteins were separated by electrophoresis on a sodium dodecyl sulfate (SDS)-12% polyacrylamide gel and transblotted to polyvinylidene difluoride (PVDF) membranes, which were incubated with human fibronectin, as previously described (12). Figure 2 shows that both the L. welshimeri and the L. monocytogenes 51-kDa fusion proteins bind human fibronectin (lanes 2 to 4), whereas the 26-kDa GST produced by E. coli DH5α containing nonrecombinant pGex-5x-3 failed to bind fibronectin (arrow, lane 1). This indicates that the binding of fibronectin to the fusion protein is not due to the GST moiety and that the amino acids that differ between the two Listeria fibronectin-binding proteins are not important for the binding to human fibronectin.
FIG. 2.
The Listeria welshimeri DNA fragment expresses a fibronectin-binding protein. The entire L. monocytogenes Fbp ORF (lane 4) and the homologous ORF of L. welshimeri (lanes 2 and 3) were cloned in phase with the GST gene in pGex-5X-3. Recombinant plasmids were then inserted into E. coli DH5α cells, which were grown in the presence of IPTG. E. coli DH5α cells transformed by pGex-5X-3 were treated in a similar manner and used as a control (lane 1). Total sonicates of the bacteria (50 μg of protein) were fractionated by SDS-polyacrylamide gel electrophoresis (12% polyacrylamide) and transferred to a PVDF membrane, which was incubated with human fibronectin. Membrane-bound fibronectin was revealed by peroxidase-labeled rabbit anti-human fibronectin, with α-chloronaphthol as the substrate. The arrow indicates the presence of GST in lane 1. The heavily stained protein of around 40 kDa present in lanes 1 to 4 is an E. coli fibronectin-binding protein. Localization of molecular mass markers is indicated to the right of the panel.
Selection of primers and specificity of the PCR amplifications.
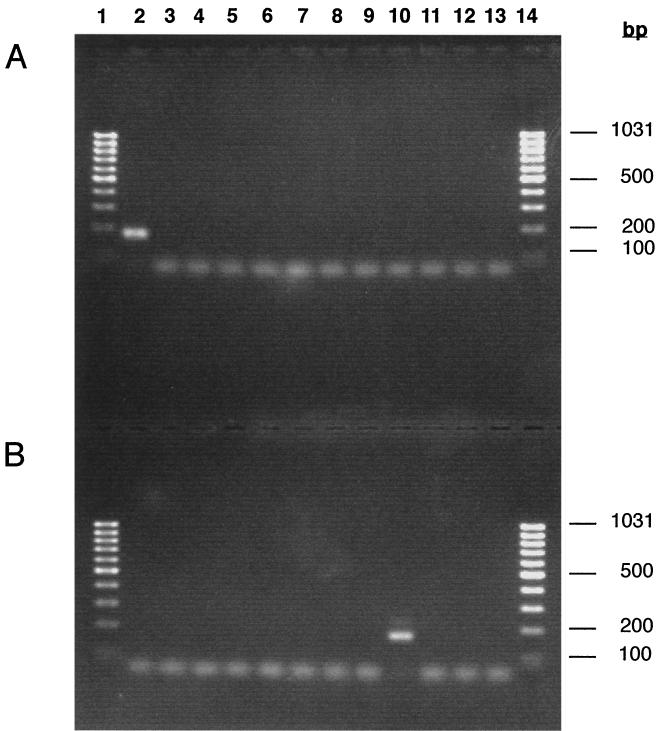
Based on L. welshimeri and L. monocytogenes Fbp ORF DNA sequences, primer pairs G398 (5"-TGAAAGAGTTTATCGAGCCATACC-3") and G399 (5"-TTTATGGCCTTCTAGCACGTTCG-3") and primer pairs G398 and G400 (5"-CTTATGCTCCTCTAGTACACTTT-3") were selected to specifically amplify a 170-bp DNA fragment from chromosomal DNA of L. welshimeri and L. monocytogenes, respectively (Fig. 1). To test the specificity of the primers described above, PCR amplification from chromosomal DNA (200 ng) purified from strains belonging to the six species of the genus Listeria was performed with the primers described above (final concentration, 1 μM) and Taq DNA polymerase (final concentration, 0.025 U/μl [Promega]) in 50 μl of a buffer containing 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates (dNTPs), 50 mM KCl, 0.1% Triton X-100, and 10 mM Tris-Cl (pH 9.0). Amplifications were performed with a Perkin-Elmer thermocycler (Gene Amp PCR system 2400) through the following temperature program: one cycle of 45 s at 95°C; then 35 cycles of 45 s at 95°C, 60 s at 55°C, and 60 s at 72°C; and finally 1 cycle of 10 min at 72°C. When using, on the one hand, primers G398 and G400, a unique DNA fragment of the expected molecular weight was amplified from the chromosomal DNA of L. monocytogenes (Fig. 3A, lane 2), but not from the chromosomal DNA of the other species of the genus Listeria, including L. welshimeri (Fig. 3A, lanes 3 to 13). On the other hand, primers G398 and G399 allow the amplification of a unique DNA fragment from the chromosomal DNA of L. welshimeri (Fig. 3B, lane 10), but not from the chromosomal DNA of the other Listeria species, including L. monocytogenes (Fig. 3B, lanes 2 to 9 and 11 to 13). Furthermore, whereas BLAST P searches found similarities between L. monocytogenes and L. welshimeri Fbps and YwfA of L. lactis subsp. lactis, PCR with primer couples G398-G399 and G398-G400 did not amplify any DNA fragment from the chromosomal DNA of L. lactis subsp. lactis IL-1403 (results not shown). The above results confirm the specificity of both PCR assays for L. monocytogenes and L. welshimeri, respectively.
FIG. 3.
Specificity of the PCR assays for L. welshimeri and L. monocytogenes. Chromosomal DNAs of L. monocytogenes 90/636 (lane 2), L. ivanovii (lane 3), L. innocua sv 6a (lane 4), L. innocua sv 6b (lane 5), L. innocua 95/013 (lane 6), L. seeligeri 85/59/06 (lane 7), L. seeligeri 024/20 (lane 8), L. seeligeri 1140/09/03 (lane 9), L. welshimeri P.P. (lane 10), L. grayi CLIP 73019 (lane 11), L. grayi CLIP 14014 (lane 12), and L. grayi CLIP 640 (lane 13) were amplified with primer couple G398-G400 (A) or with primer couple G398-G399 (B). Amplification products (5 μl) were separated on a 2% agarose gel and visualized under UV light. Molecular weight markers (Gene Ruler 100-bp DNA ladder; MBI Fermentas) of the indicated size in base pairs (bp) are shown in lanes 1 and 14.
In a previous study, we have shown that the fbp gene of L. monocytogenes displays a degree of allelic variation among isolates. Among others, the RsaI site at position 154 to 157 (Fig. 1) is not always present in the fbp gene of all strains of L. monocytogenes isolated (13). Because primer G400 hybridizes to the Fbp ORF from nt 149 to 171, we tested whether our PCR assays are able to correctly identify other isolates of L. welshimeri and L. monocytogenes. For that aim, PCR amplifications with primer pairs G398-G399 and G398-G400 were performed with chromosomal DNA (100 ng) purified from 15 strains of L. welshimeri and from 20 strains of L. monocytogenes of nonclonal origin, including strains possessing or not the RsaI site at position 154. Fourteen of the L. welshimeri strains were isolated from different foodstuffs by the Belgian Reference Center for Listeriosis during the years 1996 to 1997; one isolate was a reference strain originating from the collection of the Reference Center for Listeriosis at the Pasteur Institute, Paris. Each of the L. monocytogenes strains tested belongs to 1 of the 20 different esterase types detected during a previously made analysis of L. monocytogenes populations isolated from foodstuffs and from human patients with listeriosis (14, 21). PCR with primers G398 and G400 correctly identified all allelic variants of the L. monocytogenes fbp gene and gave no false-positive reactions with any of the strains of L. welshimeri tested. Similarly, PCR with primers G398 and G399 correctly identified all strains of L. welshimeri and gave no false-positive reactions with any of the strains of L. monocytogenes tested (results not shown).
Therefore, the PCR assays targeting the fbp gene could be considered as new valuable tools for the rapid identification of L. welshimeri and L. monocytogenes. The novel PCR assay specific for L. welshimeri could be particularly important for the study of the occurrence of this not very well known species in food, the environment, and animal and human carriers.
Acknowledgments
We are grateful to A. Genicot, the Institute of Hygiene and Epidemiology, Brussels, Belgium, and C. Jacquet and J. Rocourt, the Pasteur Institute, Paris, France, for providing strains of Listeria; to A. Bolotin, I. Poquet, and S. Kulakauskas, INRA, Jouy-en-Josas, France, for providing DNA of L. lactis subsp. lactis IL-1403; to I. Feck for help provided in DNA sequencing; and to M. Braibant for reading the manuscript.
REFERENCES
- 1.Abrahim, A., A. Papa, N. Soultos, I. Ambrosiadis, and A. Antoniadis. 1998. Antibiotic resistance of Salmonella spp. and Listeria spp. isolates from traditionally made fresh sausages in Greece. J. Food Prot. 61:1378-1380. [DOI] [PubMed] [Google Scholar]
- 2.André, P., and A. Genicot. 1987. Premier isolement de Listeria welshimeri chez l'homme. Zentbl. Bakteriol. Hyg. A 263:605-606. [PubMed] [Google Scholar]
- 3.Bille, J., B. Catimel, E. Bannerman, C. Jacquet, M.-N. Yersin, I. Caniaux, D. Monget, and J. Rocourt. 1992. API Listeria, a new and promising one-day system to identify Listeria isolates. Appl. Environ. Microbiol. 58:1857-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bille, J., J. Rocourt, and B. Swaminathan. 1999. Listeria, Erysipelothrix, and Kurthia, p. 346-356. In P. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken, (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 5.Boerlin, P., J. Rocourt, and J.-C. Piffaretti. 1991. Taxonomy of the genus Listeria by using multilocus enzyme electrophoresis. Int. J. Syst. Bacteriol. 41:59-64. [DOI] [PubMed] [Google Scholar]
- 6.Brosch, R., J. Chen, and J. B. Luchansky. 1994. Pulsed-field fingerprinting of Listeriae: identification of genomic divisions for Listeria monocytogenes and their correlation with serovar. Appl. Environ. Microbiol. 60:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bubert, A., I. Hein, M. Rauch, A. Lehner, B. Yoon, W. Goebel, and M. Wagner. 1999. Detection and differentiation of Listeria spp. by a single reaction based on multiplex PCR. Appl. Environ. Microbiol. 65:4688-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalton, C. B., C. C. Austin, J. Sobel, P. S. Hayes, W. F. Bibb, L. M. Graves, B. Swaminathan, M. E. Proctor, and P. M. Griffin. 1997. An outbreak of gastroenteritis and fever due to Listeria monocytogenes in milk. N. Engl. J. Med. 336:100-105. [DOI] [PubMed] [Google Scholar]
- 9.Encinas, J., J. Sanz, M. Garcia-Lopez, and A. Otero. 1999. Behaviour of Listeria spp. in naturally contaminated chorizo (Spanish fermented sausage). Int. J. Food Microbiol. 46:167-171. [DOI] [PubMed] [Google Scholar]
- 10.Farber, J., and C. Addison. 1994. RAPD typing for distinguishing species and strains in the genus Listeria. J. Appl. Bacteriol. 77:242-250. [DOI] [PubMed] [Google Scholar]
- 11.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilot, P., P. André, and J. Content. 1999. Listeria monocytogenes possesses adhesins for fibronectin. Infect. Immun. 67:6698-6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilot, P., Y. Jossin, and J. Content. 2000. Cloning, sequencing and characterisation of a Listeria monocytogenes gene encoding a fibronectin-binding protein. J. Med. Microbiol. 49:887-896. [DOI] [PubMed] [Google Scholar]
- 14.Gilot, P., A. Genicot, and P. André. 1996. Serotyping and esterase typing for analysis of Listeria monocytogenes populations recovered from foodstuffs and from human patients with listeriosis in Belgium. J. Clin. Microbiol. 34:1007-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilot, P., C. Hermans, M. Yde, J. Gigi, M. Janssens, A. Genicot, P. André, and G. Wauters. 1997. Sporadic case of listeriosis associated with the consumption of a Listeria monocytogenes-contaminated "Camembert' cheese. J. Infect. 35:195-197. [DOI] [PubMed] [Google Scholar]
- 16.Golsteyn Thomas, E. J., R. K. King, J. Burchak, and V. P. J. Gannon. 1991. Sensitive and specific detection of Listeria monocytogenes in milk and ground beef with the polymerase chain reaction. Appl. Environ. Microbiol. 57:2576-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham, T., E. Golsteyn-Thomas, V. Gannon, and J. Thomas. 1996. Genus- and species-specific detection of Listeria monocytogenes using polymerase chain reaction assays targeting the 16S/23S intergenic spacer region of the rRNA operon. Can. J. Microbiol. 42:1155-1162. [DOI] [PubMed] [Google Scholar]
- 18.Jersek, B., E. Tcherneva, N. Rijpens, and L. Herman. 1996. Repetitive element sequence-based PCR for species and strain discrimination in the genus Listeria. Lett. Appl. Microbiol. 23:55-60. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, J., M. Doyle, and R. Cassens. 1990. Listeria monocytogenes and other Listeria spp. in meat and meat products. A review. J. Food Prot. 53:81-91. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, W., S. Tyler, E. Ewan, F. Ashton, G. Wang, and K. Rozee. 1992. Detection of genes coding for listeriolysin and Listeria monocytogenes antigen A (lmA) in Listeria spp. by the polymerase chain reaction. Microbial. Pathog. 12:79-86. [DOI] [PubMed] [Google Scholar]
- 21.Malak, M., A. Vivier, P. André, J. Decallonne, and P. Gilot. 2001. RAPD analysis, serotyping, and esterase typing indicate that the population of Listeria monocytogenes strains recovered from cheese and from patients with listeriosis in Belgium are different. Can. J. Microbiol. 47:883-887. [DOI] [PubMed] [Google Scholar]
- 22.Manzano, M., L. Cocolin, C. Cantoni, and G. Comi. 2000. Temperature gradient gel electrophoresis of the amplified product of small 16S rRNA gene fragment for the identification of Listeria species isolated from food. J. Food Prot. 63:659-661. [DOI] [PubMed] [Google Scholar]
- 23.Mazurier, S., and K. Wernars. 1992. Typing of Listeria strains by random amplification of polymorphic DNA. Res. Microbiol. 143:499-505. [DOI] [PubMed] [Google Scholar]
- 24.McLauchlin, J. 1997. The pathogenicity of Listeria monocytogenes: a public health perspective. Rev. Med. Microbiol. 8:1-14. [Google Scholar]
- 25.Poyart, C., P. Trieu-Cuot, and P. Berche. 1996. The inlA gene required for cell invasion is conserved and specific to Listeria monocytogenes. Microbiology 142:173-180. [DOI] [PubMed] [Google Scholar]
- 26.Rocourt, J., and H. Seeliger. 1985. Distribution des espèces du genre Listeria. Zentbl. Bakteriol. Hyg. A 259:317-330. [PubMed] [Google Scholar]
- 27.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Seeliger, H., and D. Jones. 1986. Listeria, p. 1235-1245. In H. Sneath (ed.), Bergey's manual of systematic bacteriology. Williams and Wilkins, Baltimore, Md.
- 29.Wernars, K., K. Heuvelman, S. Notermans, E. Domann, M. Leimeister-Wächter, and T. Chakraborty. 1992. Suitability of the prfA gene, which encodes a regulator of virulence genes in Listeria monocytogenes, in the identification of pathogenic Listeria spp. Appl. Environ. Microbiol. 58:765-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winters, D., T. Maloney, and M. Johnson. 1999. Rapid detection of Listeria monocytogenes by a PCR assay specific for an aminopeptidase. Mol. Cell. Probes 13:127-131. [DOI] [PubMed] [Google Scholar]