Abstract
Ninety beta-hemolytic Escherichia coli isolates associated with diarrhea in neonatal pigs from multiple farms in Oklahoma were investigated for known associated disease serotypes, virulence factors, ribotypes, and antimicrobial susceptibility phenotypes. Fifteen different serotypes were observed, with 58% of isolates belonging to groups that produce one of three major enterotoxins: O149, O147, and O139. Thirty percent of the swine E. coli isolates possessed a combination of F4 fimbriae and the heat-labile toxin and heat-stable toxin B enterotoxins. Seventy-three percent of the E. coli isolates were resistant to five or more antibiotics. Interestingly, 53% of swine E. coli isolates exhibited resistance to chloramphenicol (CHL), an antibiotic whose use in food animals has been prohibited in the United States since the mid-1980s. The cmlA gene, which encodes a putative CHL efflux pump, was detected by PCR in 47 of the 48 CHL-resistant isolates, and 4 of these also possessed the cat2 gene, which encodes a chloramphenicol acetyltransferase. The one CHL-resistant isolate that did not contain either cmlA or cat-2 possessed the flo gene, which confers resistance to both florfenicol and CHL. To determine whether CHL-resistant swine E. coli isolates represented dissemination of a clonal strain, all 90 isolates were analyzed by ribotyping. Seventeen distinct E. coli ribogroups were identified, with CHL resistance observed among the isolates in all except one of the major ribogroups. The identification of the cmlA gene among diverse hemolytic enterotoxigenic E. coli strains demonstrates its broad dissemination in the swine production environment and its persistence even in the absence of CHL selection pressure.
Antimicrobials are valuable tools that animal producers use to quickly address clinical disease and to maintain healthy and productive animals, but the treatment of whole herds and flocks with antimicrobials for disease prevention and growth promotion is a controversial practice (13, 16, 20, 23, 28). Broad use of antimicrobials in agriculture selects for resistant bacteria that may enter the food chain and potentially result in food-borne illness in humans that is less responsive to treatment with conventional antibiotics. In addition to the human health concerns, antimicrobial-resistant pathogens also pose a severe and costly animal health problem, as they prolong illness and decrease productivity through higher morbidity and mortality rates.
Escherichia coli is the most common etiologic agent of neonatal diarrhea in pigs aged 0 to 4 days (7, 13). Causative strains are usually enterotoxigenic E. coli (ETEC) isolates that colonize the small intestine and that produce one or more enterotoxins. Clinical signs of ETEC infection may first be observed within hours after birth, resulting in increased rates of mortality during the first few days of life. Treatment typically consists of a broad-spectrum antimicrobial, although resistance to such drugs has greatly increased over the last several years (1, 4, 11, 17, 22, 25).
Chloramphenicol (CHL) is a broad-spectrum antibiotic that was used extensively in veterinary medicine until concerns over its toxicity emerged (26). Human exposure to CHL has been linked to aplastic anemia, a type of bone marrow suppression that is usually irreversible and often fatal. Interestingly, development of the disease does not appear to be dependent on the dose or duration of exposure to CHL. The possibility that trace residues of CHL in food products may induce the disease led the U.S. Food and Drug Administration to ban its use in food animals in the 1980s (14). Currently, only a fluorinated derivative of CHL, florfenicol (FFN), is approved for veterinary use in food animals, but FFN is not approved for use in swine in the United States.
Resistance to CHL may be mediated either enzymatically through the chemical inactivation of the drug or nonenzymatically through drug efflux. Chloramphenicol acetyltransferase catalyzes the acetylation of the 3"-OH of CHL and is responsible for most enzymatic resistance to CHL (27, 29). The cmlA gene confers nonenzymatic resistance to CHL. Although its mechanism has yet to be characterized, the similarity of the primary structure of the cmlA protein to those of bacterial transport proteins suggests that it functions as a drug efflux pump (6, 32). The flo gene, whose product shares 57% amino acid sequence identity to the product of cmlA, also encodes a putative efflux pump that confers resistance to both CHL and FFN (8, 9, 12, 21). Additionally, Cloeckaert et al. (10) recently reported on a new flo gene variant that was identified on the R55 IncC plasmid isolated from Klebsiella pneumoniae and that also confers nonenzymatic CHL resistance.
The present study examined the antimicrobial susceptibility patterns and genetic relatedness of beta-hemolytic E. coli strains isolated from neonatal swine with diarrhea. We hypothesized that a high percentage of isolates would be clonal in nature and resistant to antimicrobials commonly used in swine production. Although we do report a high rate of multiple-drug-resistant phenotypes, a surprisingly high incidence of resistance to CHL and FFN was observed, despite the lack of an obvious selection pressure in swine production. We further investigated the mechanisms of CHL resistance since there is limited information regarding the molecular mechanisms of resistance to this drug among hemolytic swine ETEC isolates.
MATERIALS AND METHODS
Materials.
Materials purchased from commercial sources included antimicrobial susceptibility plates and JustOne strips (Trek Diagnostic Systems, Westlake, Ohio), AmpliTaq Gold DNA polymerase and deoxynucleoside triphosphates (Applied Biosystems, Foster City, Calif.), and RiboPrinter reagents (Qualicon, Inc., Wilmington, Del.). The oligonucleotides used as primers in the PCR were synthesized by Biosynthesis, Inc. (Lewisville, Tex.). PCR products were submitted for DNA sequencing at the DNA Core Facility in the Department of Veterinary Pathobiology, Texas A&M University, College Station.
Bacterial strains.
The present study focuses on 90 beta-hemolytic E. coli isolates recovered from neonatal pigs with diarrhea from multiple farms in Oklahoma from 1998 to 1999. E. coli was isolated from swine intestines upon necropsy by spread plating on blood and MacConkey agar plates. Indole and oxidase tests were performed for lactose-positive colonies. API 20E test strips (bioMérieux Vitek, Hazelwood, Mo.) were also used to confirm the identification of the isolate as E. coli. Swine E. coli isolates displaying decreased susceptibilities to CHL were subsequently collected for further analysis to determine the mechanism of resistance. Isolates were stored as 10% glycerol stocks at −80°C until analysis. Isolates were submitted to the E. coli Reference Center located at the Pennsylvania State University for O-antigen serotyping and virulence factor analysis.
Determination of antimicrobial susceptibility.
The MICs of the antimicrobials were determined by broth microdilution according to the methods described by the National Committee for Clinical Laboratory Standards (NCCLS) (24). Susceptibility testing was performed with the Sensititre automated antimicrobial susceptibility system according to the manufacturer's instructions (Trek Diagnostic Systems). The following antimicrobials were assayed: amikacin, amoxicillin-clavulanic acid, ampicillin, apramycin, ceftiofur, ceftriaxone, cephalothin, CHL, ciprofloxacin, FFN, gentamicin, kanamycin, nalidixic acid, streptomycin, sulfamethoxazole, tetracycline, and trimethoprim-sulfamethoxazole. E. coli ATCC 25922, E. coli ATCC 35218, and Pseudomonas aeruginosa ATCC 27853 were used as quality control strains for broth microdilution susceptibility testing.
PCR.
Genes encoding antimicrobial resistance determinants were detected by PCR. The primer sets used for amplification of cmlA, flo, cat-1, cat-2, and cat-3 were the same as those described previously (19, 29). Templates of total DNA from each isolate were prepared as follows. Bacteria were streak plated onto tryptic soy agar plates containing 5% sheep's blood, and the plates were incubated overnight at 35°C. Three to five bacterial colonies were lifted from the plate and resuspended in 0.5 ml of sterile water. The suspension was heated to 95°C for 10 min, and then the cell debris was removed by centrifugation. The supernatant (10 μl) was used as the template in the PCRs. Each reaction mixture (50 μl) also contained 1× AmpliTaq Gold DNA polymerase reaction buffer, 2.5 mM MgCl2, 1 mM deoxynucleoside triphosphates, 1 pmol of each respective oligonucleotide primer per μl, and 1 U of AmpliTaq Gold DNA polymerase. All reaction mixtures were first heated to 95°C for 10 min to activate the AmpliTaq Gold polymerase. The denaturation, annealing, and extension conditions for PCR were the same as those described previously for each primer set (19, 29). Sequence comparisons were made with the BLAST program of the National Center for Biotechnology Information (3).
Ribotyping.
Ribotyping was performed with the RiboPrinter Microbial Characterization System (Qualicon, Inc.) and the standard EcoRI DNA preparation kit, as described in the manufacturer's operations and analytical guides. Bacterial DNA was digested with EcoRI, and gel electrophoresis was used to separate the restriction fragments into distinctive patterns. DNA from isolates with the different DNA patterns was hybridized with a chemiluminescent E. coli rRNA probe. The characterization system automatically placed the pattern for each isolate into common ribogroups (clusters) on the basis of the similarity of the band positions to the band positions and the intensities of patterns in the RiboPrinter's database. The ribogroups were analyzed and visually refined by the manufacturer's standard procedure. A dendrogram was constructed from the ribogroup patterns on the basis of the Pearson correlation coefficient by using an optimization coefficient of 1.56%. Similarity coefficients were calculated on the basis of both band positions and relative intensity.
RESULTS AND DISCUSSION
Antimicrobial susceptibility patterns of swine E. coli isolates.
Beta-hemolytic E. coli is the most common bacterial etiologic agent of diarrhea in neonatal and postweaning pigs. Treatment of enteric E. coli infection in swine commonly includes the use of broad-spectrum antibiotics (13, 16, 23). We characterized the patterns of susceptibility of 90 E. coli isolates from diarrheic neonatal pigs to 17 antimicrobial agents of human and veterinary therapeutic significance. The rates of resistance, as determined by measuring the MICs and comparing them to the resistance breakpoints established by NCCLS, are listed in Table 1. The highest rates of resistance were to tetracycline (96%), sulfamethoxazole (89%), kanamycin (84%), streptomycin (82%), FFN (64%), and CHL (53%). All isolates were susceptible to nalidixic acid, ciprofloxacin, amikacin, and ceftriaxone. Resistance to multiple drugs was frequently observed, with 66 of 90 (73%) of the E. coli isolates resistant to five or more antibiotics (data not shown). The swine E. coli isolates were similar to other clinical veterinary E. coli strains in terms of their decreased susceptibilities to tetracycline, gentamicin, streptomycin, and sulfamethoxazole (1, 11, 17, 19, 22, 25, 30, 31). These bacterial isolates also exhibited a rate of resistance to kanamycin similar to that reported previously for bovine E. coli isolates (30), a rate of resistance to ampicillin similar to that seen for avian E. coli isolates (5), levels of susceptibility to cephalosporins similar to those seen for avian E. coli isolates (5), and levels of susceptibility to fluoroquinolones similar to those seen for bovine E. coli isolates (30). These similarities and differences in antibiotic resistance exhibited by these three distinct veterinary groups of E. coli may reflect therapeutic use or the availability of certain antimicrobial agents for the treatment of infections in poultry, cattle, and swine as well as a shared ecology of drug resistance genes among the farm microbiota (5, 15, 20, 30).
TABLE 1.
Antimicrobial resistance phenotypes of swine E. coli isolates
| Class and antimicrobial | % Resistant strainsa (n = 90) |
|---|---|
| Phenicols | |
| FFNb | 64 |
| CHL | 53 |
| Penicillins | |
| Ampicillin | 34 |
| Amoxicillin-clavulanic acid | 4 |
| Cephalosporins | |
| Cephalothin | 13 |
| Ceftiofur | 1 |
| Ceftriaxone | 0 |
| Tetracycline | 96 |
| Aminoglycosides | |
| Amikacin | 0 |
| Apramycin | 14 |
| Gentamicin | 14 |
| Kanamycin | 84 |
| Streptomycin | 82 |
| Sulfonamides and potentiated sulfonomides | |
| Sulfamethoxazole | 89 |
| Trimethoprim-sulfamethoxazole | 22 |
| Quinolones or fluoroquinolones | |
| Nalidixic acid | 0 |
| Ciprofloxacin | 0 |
MICs were determined by microdilution methods according to NCCLS standards.
Resistance was based on the NCCLS breakpoint for bovine respiratory pathogens, ≥8 μg/ml.
Tetracyclines, aminoglycosides, and sulfonamides are widely used in swine production for the treatment and prevention of disease and for growth promotion, and therefore, a high rate of resistance to drugs in these antimicrobial classes was not unexpected. The phenicols, however, are not approved for use in swine in the United States. CHL has been banned from use in food animals since the mid-1980s, and FFN is approved for use only in cattle (14). Despite the apparent lack of selection pressure, high rates of resistance to these two antibiotics were identified in swine E. coli isolates, with 64% of isolates resistant to FFN and 53% of isolates resistant to CHL. For the CHL-resistant E. coli isolates, the CHL MIC ranged from 32 to 256 μg/ml and the FFN MIC ranged from 8 to 256 μg/ml (Table 2). Most isolates (47 of 48) were resistant to CHL at 32 μg/ml and FFN at 8 to 16 μg/ml. For one isolate the MICs of both CHL and FFN were 256 μg/ml.
TABLE 2.
Prevalence of cmlA, cat-2, and flo genes in CHL-resistant swine E. coli
| Resistance genotype | MIC (μg/ml)
|
No. of isolates positive for resistance gene/no. of isolates tested | |
|---|---|---|---|
| CHL | FFN | ||
| cmlA | ≥32 | 8-≥16 | 43/48 |
| cmlA, cat-2 | 32 | 8-16 | 4/48 |
| flo | 256 | 256 | 1/48 |
The cmlA gene is widely disseminated among swine E. coli isolates.
We next investigated the genetic mechanisms for resistance to CHL and FFN by assaying the swine E. coli isolates for the presence of five genes known to confer resistance to these antimicrobials: cmlA, cat-1, cat-2, cat-3, and flo. Using total genomic DNA from each of the 48 CHL-resistant isolates as the template in a PCR, we found that 47 were positive for the cmlA gene, with 4 of these isolates also possessing one of the chloramphenicol acetyltransferase genes (Table 2). CHL MICs were not higher for the isolates with a cmlA+ cat-2+ genotype than for those with the cmlA gene alone, suggesting that there is no additive effect from the two resistance mechanisms.
The one CHL-resistant isolate (isolate CVM873) that did not possess either cmlA or cat-2 was positive for the flo gene and was identified as belonging to the O147 serogroup. The flo gene has been described previously and confers resistance to both FFN and CHL. The FFN and CHL MICs for this swine isolate were 256 μg/ml. The high level of resistance to the phenicols in this E. coli isolate is similar to the levels of resistance exhibited by bovine E. coli isolates that possess the flo gene (30). Although FFN is approved for use only in cattle in the United States, the presence of the flo gene has previously been reported in CHL-resistant E. coli strains isolated from chickens (19). Only one isolate possessed the flo genotype, yet 47 of 48 isolates that were negative for flo were resistant to FFN (MICs, ≥8 μg/ml). The CHL resistance gene, cmlA, does not confer resistance to FFN (12), suggesting that a gene reservoir for FFN resistance already exists in swine E. coli isolates and involves a gene(s) other than flo and cmlA. This may present a clinical obstacle for expanded veterinary use of this drug in the treatment of E. coli-related swine enteric diseases.
CHL-resistant swine E. coli isolates do not represent expansion of a single clone.
The persistence of CHL resistance in swine E. coli isolates may have resulted from continual colonization with one or a few clonal strains from a common environmental reservoir that was selected earlier when CHL was used therapeutically in the 1980s. We therefore examined the relatedness of all 90 strains according to their serotypes, ribotypes, and the presence of five virulence factors (pathotypes). The E. coli pathoypes and O serotypes are listed in Table 3. Fifteen different serotypes were observed, with 58% of the isolates belonging to groups that produce one of three major enterotoxins, O149, O147, and O139 (7, 13). Seventy-nine percent (15 of 19) of O147 E. coli isolates and 72% (18 of 25) of O149 E. coli isolates were CHL resistant. Fifty-three percent (48 of 90) of swine E. coli isolates, of which 75% (36 of 48) were CHL resistant, belonged to one of two E. coli pathotypes that possessed either heat-labile toxin, heat-stable toxin B (STb), and F4 fimbriae or heat-stable toxin A, STb, Shiga-like toxin 2, and F107 pili (Table 3). Sixty-four percent (58 of 90) of the swine E. coli isolates possessed the gene for STb, and 36% (32 of 90) possessed the stx-2 gene. The most common fimbrial antigens detected were F107 (43 of 90 isolates) and F4 (33 of 90 isolates). All isolates were negative for the stx-1 or the cnf-2 gene (data not shown). These data demonstrate that CHL resistance does not exclusively belong to any one swine E. coli pathotype or O serogroup.
TABLE 3.
Pathotypes and serotypes of swine E. coli
| Pathotypea | No. of isolates | No. of CHLr isolatesb | Serotype(s)c |
|---|---|---|---|
| None | 2 | 1 | O35 (1), O91 |
| Stx2 | 1 | 0 | O2 |
| EAE | 1 | 0 | NT |
| F4 | 2 | 2 | O149 (1), NT (1) |
| CNF1 | 9 | 0 | O4, O11, O75, O114, O127 |
| F107 | 12 | 6 | O138, O139 (4), O147 (1), NT (1) |
| Stx2, F107 | 4 | 1 | O2, O121, O147 (1), NT |
| STb, F4 | 1 | 0 | O98 |
| LT, F4 | 1 | 1 | O149 (1) |
| STa, STb, F4 | 1 | 0 | O8 |
| STa, STb, Stx2 | 1 | 0 | NT |
| STa, STb, F107 | 1 | 0 | O147 |
| STb, Stx2, F107 | 4 | 0 | O147, NT |
| LT, STb, F4 | 27 | 17 | O8, O149 (17), NT |
| STa, STb, Stx2, F107 | 21 | 19 | O147 (13), NT (6) |
| LT, STb, F4, F6 | 1 | 0 | O8 |
| STa, STb, Stx2, F5, F107 | 1 | 1 | NT (1) |
Isolates were positive for the indicated virulence factors: heat-labile toxin (LT), heat-stabile toxin A (STa), heat-stable toxin B (STb), Shiga-like toxin II (Stx2), cytotoxic necrotizing factor 1 (CNF1), K88(F4) fimbriae (F4), K99(F5) fimbriae (F5), 987P fimbriae (F6), F107 fimbriae (F107), and the E. coli attaching-and-effacing factor (EAE).
The number of CHL-resistant (CHL) isolates with the indicated pathotype.
Serotypes associated with the corresponding pathotype are listed with the number of CHL-resistant isolates of each serotype (in parentheses). NT, not typeable.
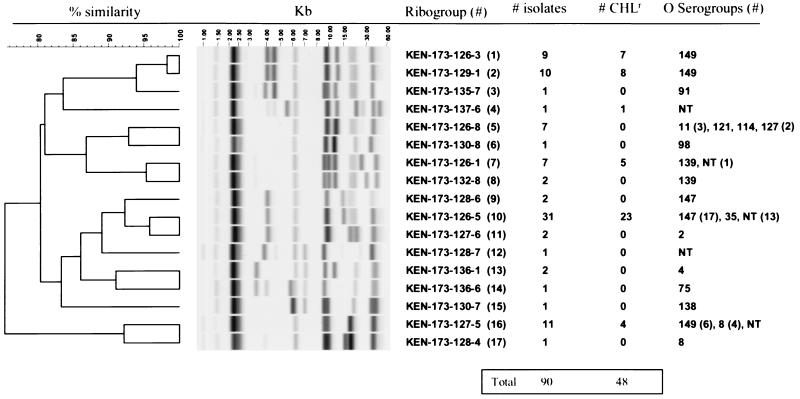
To determine whether CHL-resistant swine E. coli isolates represented dissemination of a clonal strain, all 90 isolates were analyzed by ribotyping. Seventeen distinct E. coli ribogroups were identified, with 83% of the isolates clustering into six major ribogroups (Fig. 1). CHL resistance was observed among the isolates in all except one of the major ribogroups, with the largest group containing 23 of 31 isolates resistant to CHL. Seventy-nine percent of the CHL-resistant E. coli isolates were found in ribogroups 1 (n = 7), 2 (n = 8), and 10 (n= 23). The majority of ribogroups contained E. coli isolates of one serotype; however, three ribogroups (ribogroups 5, 10, and 16) contained multiple serotypes. Ribogroup 10 was the largest cluster identified (n = 31) and was composed of isolates comprising primarily serotype O147 and 13 nontypeable isolates. Ribogroup 16 was the second largest cluster (n = 11) and was composed of isolates of either serogroup O149 or serogroup O8, suggesting a close evolutionary relationship between these two serotypes. Ribogroups 1 and 2 contained isolates that were only of the O149 serogroup and that were genetically similar to each other; however, they vastly differed from the other ribogroup with serogroup O149 strains (ribogroup 16) (Fig. 1). All ribogroups that possessed multiple strains (ribogroups 1, 2, 5, 7, 8, 9, 10, 11, 13, and 16) included isolates recovered from diseased swine in both 1998 and 1999 (data not shown), indicating the persistence of virulent bacterial clones. The potential for the use of ribotyping as a tool for subtyping within certain serotypes is supported by the clustering of E. coli isolates with common serotypes in different ribogroups. The different ribogroups of E. coli isolates are shown in Fig. 1 to share serotype O149 (ribogroups 1, 2, and 16), serotype O139 (ribogroups 7 and 8), and serotype O8 (ribogroups 16 and 17). Confirmation of the appropriateness of ribotyping as a subtyping tool for pathogenic swine E. coli will require testing of additional isolates from multiple sources, time periods, and geographical locations.
FIG. 1.
Ribogroup patterns of swine E. coli isolates. The image data for each lane were processed to normalize the band positions relative to the positions of the standards, to reduce background, and to scale the band intensity. The ribogroup name, the total number of isolates per E. coli ribogroup, the number of CHL-resistant isolates per E. coli ribogroup, and serotypes are shown on the right. The molecular mass scale (in kilobase pairs) and the similarity coefficient are shown at the top of the figure. NT, nontypeable.
Taken together with the serotyping and virulence gene data, we conclude that, unlike Salmonella enterica serovar Typhimurium DT104 (8), the high rate of CHL resistance in our isolates does not represent the clonal expansion of a single resistant strain but the dissemination of cmlA among genetically diverse E. coli isolates. We have made similar observations with regard to the FFN resistance gene, flo, in bovine and avian E. coli isolates (19, 30). Another possibility for the prevalence of the CHL resistance phenotype in swine E. coli isolates is that a plasmid carrying the cmlA gene is widely disseminated among these isolates. Bovine and avian E. coli isolates that are CHL resistant have been found to carry the flo gene on large plasmids that also contain genes for multiple drug resistance (19, 30).
All 48 of our CHL-resistant isolates from swine were also resistant to at least four other drugs (data not shown). CHL resistance may be coselected with other antimicrobial resistance phenotypes if a linkage exists between their respective genes. We examined the resistance phenotypes to determine whether CHL resistance can be coselected by other antimicrobials. There was no preferential selection of CHL-resistant strains by most antimicrobials. A statistically high level of significance for coselection, however, was observed with kanamycin (P = 0.0001), sulfamethoxazole (P = 0.0002), and tetracycline (P = 0.018), agents commonly used in swine in the United States. The use of these agents may serve to maintain plasmids on which CHL resistance determinants reside with other resistance genes. Also, cmlA may be linked to ETEC virulence plasmids (17, 18), creating a physical linkage that would ensure the persistence of the CHL resistance phenotype among swine ETEC isolates. Further analysis of the genetic location of cmlA is needed to address this issue.
In the early 1980s, studies reported rates of CHL resistance among E. coli isolates from commercial swine herds in South Dakota and Utah of 20 and 11%, respectively (11, 22). A survey of veterinarians at that time found that CHL was the preferred drug for the treatment of neonatal colibacillosis, a practice that presumably selected for CHL-resistant strains. The U.S. Food and Drug Administration has banned the use of CHL as a therapeutic agent in food animals since the mid-1980s. Since this ban has been actively enforced, the apparent selection pressure for resistance to this drug should have been removed. Thus, our report of a 53% rate of resistance to CHL is an unexpected, but not an unprecedented, finding. Several studies in Europe have also reported persistent rates of resistance years after withdrawal of CHL as a therapeutic drug for farm animals (1, 25). A recent report by Aarestrup et al. (2) indicated that it was possible to reduce the occurrence of antimicrobial resistance in enterococci isolated from food animals when the antimicrobial selection pressures were removed. They also demonstrated that antimicrobial resistance can persist, most likely as a consequence of coselection with other antimicrobials. The identification of the cmlA gene among diverse beta-hemolytic ETEC strains suggests the broad dissemination of this genotype in the swine production environment and suggests that CHL resistance can persist even in the absence of CHL selection pressure. This is most likely due to coselection of CHL resistance with either common swine ETEC virulence genes or other antimicrobial resistance phenotypes. Regardless, our data suggest that the withdrawal of antimicrobials from use in response to increased rates of resistance may not be an effective strategy for restoration of the therapeutic effectiveness of a specific drug. Simultaneous reductions in the selection pressures of coselecting agents may be required to reverse the emergence, spread, and persistence of antimicrobial resistance in the animal production environment.
Acknowledgments
We thank Elisabeth Chaslus-Dancla for providing cat primer sequences and control strains, Laura Dye for providing E. coli isolates, and Lance Bolton for assistance with ribotyping.
REFERENCES
- 1.Aalbaek, B., J. Rasmussen, B. Nielsen, and J. E. Olsen. 1991. Prevalence of antibiotic-resistant Escherichia coli in Danish pigs and cattle. APMIS 99:1103-1110. [PubMed] [Google Scholar]
- 2.Aarestrup, F. M., A. M. Seyfarth, H. Emborg, K. Pedersen, R. S. Hendriksen, and F. Bager. 2001. Effect of abolishment on the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 45:2054-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baggesen, D. L., and F. M. Aarestrup. 1998. Characterization of recently emerged multiple antibiotic-resistant Salmonella enterica serovar Typhimurium DT104 and other multiresistant phage types from Danish pig herds. Vet. Rec. 143:95-97. [DOI] [PubMed] [Google Scholar]
- 5.Bass, L., C. A. Liebert, M. D. Lee, D. G. White, A. O. Summers, S. G. Thayer, and J. J. Maurer. 1999. The incidence and characterization of integrons, genetic elements associated with multiple drug resistance, in avian Escherichia coli. Antimicrob. Agents Chemother. 43:2925-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bissonnette, L., S. Champetier, J. Buisson, and P. H. Roy. 1991. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J. Bacteriol. 173:4493-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanco, J., M. Blanco, J. I. Garabal, and E. A. Gonzalez. 1991. Enterotoxins, colonization factors and serotypes of enterotoxigenic Escherichia coli from humans and animals. Microbiologia 7:57-73. [PubMed] [Google Scholar]
- 8.Bolton, L. F., L. C. Kelley, M. D. Lee, P. J. Fedorka-Cray, and J. J. Mauer. 1999. Detection of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 based on a gene which confers cross-resistance to florfenicol and chloramphenicol. J. Clin. Microbiol. 37:1348-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cannon, M., S. Harford, and J. Davies. 1990. A comparative study on the inhibitory actions of chloramphenicol, thiamphenicol, and some fluorinated derivatives. J. Antimicrob. Chemother. 26:307-317. [DOI] [PubMed] [Google Scholar]
- 10.Cloeckaert, A., S. Baucheron, and E. Chaslus-Dancla. 2001. Nonenzymatic chloramphenicol resistance mediated by IncC plasmid R55 is encoded by a floR gene variant. Antimicrob. Agents Chemother. 45:2381-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coates, S. R., and K. H. Hoopes. 1980. Sensitivities of Escherichia coli isolated from bovine and porcine enteric infections to antimicrobial antibiotics. Am. J. Vet. Res. 41:1882-1883. [PubMed] [Google Scholar]
- 12.Dorman, C. J., and T. J. Foster. 1982. Nonenzymatic chloramphenicol resistance determinants specified by plasmids R26 and R55-1 in Escherichia coli K-12 do not confer high-level resistance to fluorinated analogs. Antimicrob. Agents Chemother. 22:912-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fairbrother, J. M. 1999. Neonatal Escherichia coli diarrhea, p. 433-441. In Diseases of swine, 8th ed. Iowa State University Press, Ames.
- 14.Gilmore, A. 1986. Chloramphenicol and the politics of health. Can. Med. Assoc. J. 134:423-435. [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstein, C., M. D. Lee, S. Sanchez, C. R. Hudson, B. Phillips, B. Register, M. Grady, C. Liebert, A. O. Summers, D. G. White, and J. J. Maurer. 2001. Incidence of class 1 and 2 integrases in clinical and normal flora bacteria from livestock, companion animals, and exotics. Antimicrob. Agents Chemother. 45:723-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gustafson, R. H., and R. E. Brown. 1997. Antibiotic use in animal agriculture. J. Appl. Microbiol. 83:531-541. [DOI] [PubMed] [Google Scholar]
- 17.Harnett, N. M., and C. L. Gyles. 1984. Resistance to drugs and heavy metals, colicin production, and biochemical characteristics of selected bovine and porcine Escherichia coli strains. Appl. Environ. Microbiol. 48:930-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harnett, N. M., and C. L. Gyles. 1985. Linkage of genes for heat-stable enterotoxins, drug resistance, K99 antigen, and colicin in bovine and porcine strains of enterotoxigenic Escherichia coli. Am. J. Vet. Res. 46:428-433. [PubMed] [Google Scholar]
- 19.Keyes, K., C. Hudson, J. J. Mauer, S. Thayer, D. G. White, and M. D. Lee. 2000. Detection of florfenicol resistance genes in Escherichia coli isolated from sick chickens. Antimicrob. Agents Chemother. 44:421-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khachatourians, G. G. 1998. Agricultural use of antibiotics and the evolution and transfer of antibiotic-resistant bacteria. Can. Med. Assoc. J. 159:1129-1136. [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, E., and T. Aoki. 1996. Sequence analysis of the florfenicol resistance gene encoded in the transferable T-plasmid of a fish pathogen, Pasteruella piscicida. Microbiol. Immunol. 40:665-669. [DOI] [PubMed] [Google Scholar]
- 22.Libal, M. C., and C. E. Gates. 1982. Antimicrobial resistance in Escherichia coli strains isolated from pigs with diarrhea. J. Am. Vet. Med. Assoc. 180:908-909. [PubMed] [Google Scholar]
- 23.Mackinnon, J. D. 1993. The proper use and benefits of veterinary antimicrobial agents in swine practice. Vet. Microbiol. 35:357-367. [DOI] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard (M31-A). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 25.Nijsten, R., N. London, A. van den Bogaard, and E. Stobberingh. 1996. Antibiotic resistance among Escherichia coli isolated from faecal samples of pig farmers and pigs. J. Antimicrob. Chemother. 37:1131-1140. [DOI] [PubMed] [Google Scholar]
- 26.Settepani, J. A. 1984. The hazard of using chloramphenicol in food animals. J. Am. Vet. Med. Assoc. 184:930-931. [PubMed] [Google Scholar]
- 27.Shaw, W. V. 1983. Chloramphenicol acetyltransferase: enzymology and molecular biology. Crit. Rev. Biochem. 14:1-46. [DOI] [PubMed] [Google Scholar]
- 28.Shyrock, T. R. 1999. Relationship between usage of antibiotics in food-producing animals and the appearance of antibiotic resistant bacteria. Int. J. Antimicrob. Agents 12:275-278. [DOI] [PubMed] [Google Scholar]
- 29.Vassort-Bruneau, C., M. Lesage-Descauses, J. Martel, J. Lafont, and E. Chaslus-Dancla. 1996. CAT III chloramphenicol resistance in Pasteurella haemolytica and Pasteurella multocida isolated from calves. J. Antimicrob. Chemother. 38:205-213. [DOI] [PubMed] [Google Scholar]
- 30.White, D. G., C. Hudson, J. J. Maurer, S. Ayers, S. Zhao, M. D. Lee, L. Bolton, T. Foley, and J. Sherwood. 2000. Characterization of chloramphenicol and flofenicol resistance in Escherichia coli associated with bovine diarrhea. J. Clin. Microbiol. 38:4593-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White, D. G., L. J. V. Piddock, J. J. Maurer, S. Zhao, V. Ricci, and S. G. Thayer. 2000. Characterization of fluoroquinolone resistance in veterinary isolates of avian Escherichia coli and cross-resistance to human fluoroquinolones. Antimicrob. Agents Chemother. 44:2897-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams, J. B. 1996. Drug efflux as a mechanism of resistance. Br. J. Biomed. Sci. 53:290-293. [PubMed] [Google Scholar]